Investigation of Binding Modes and Functional Surface of Scorpion Toxins ANEP to Sodium Channels 1.7
Abstract
:1. Introduction
2. Results
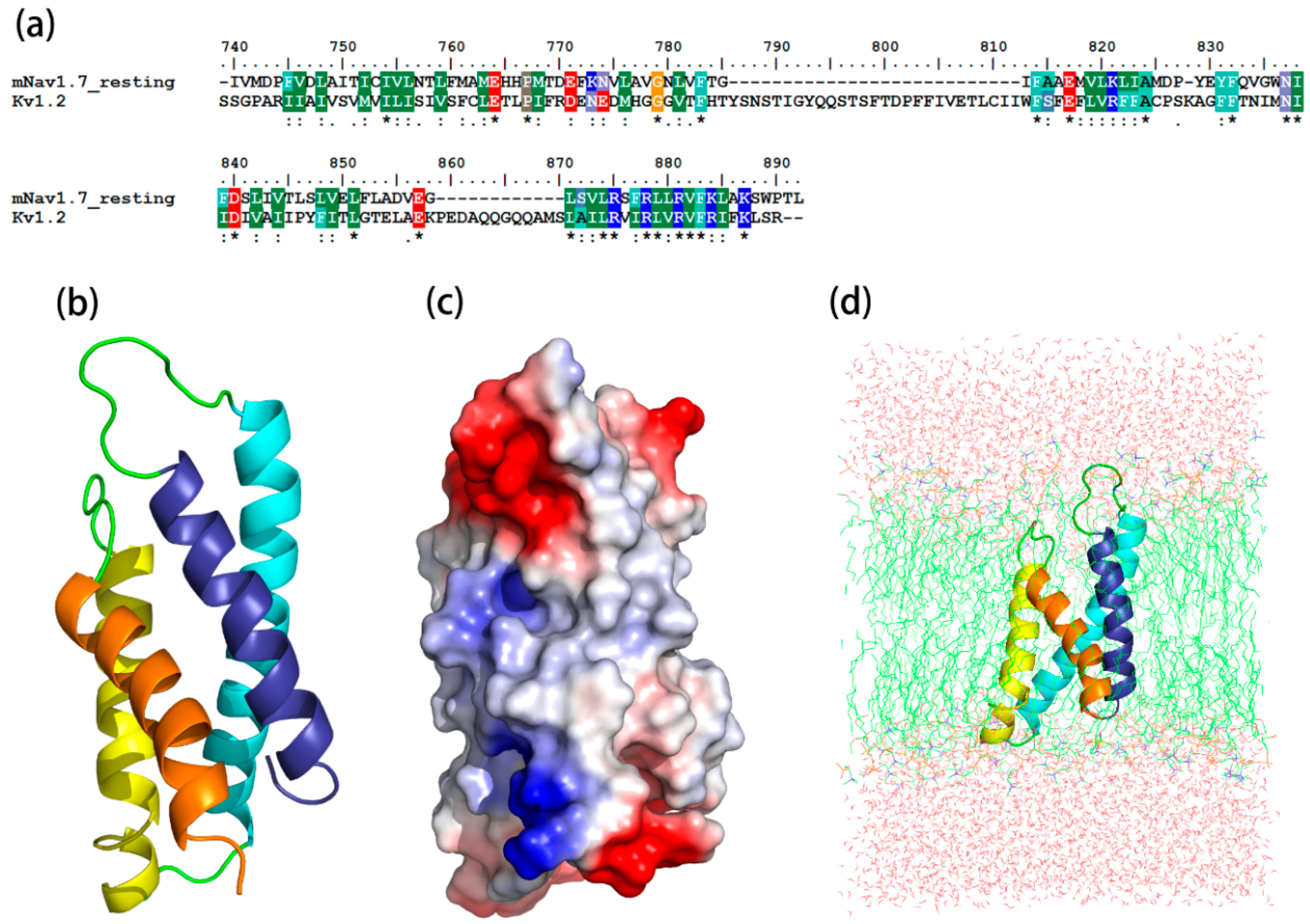
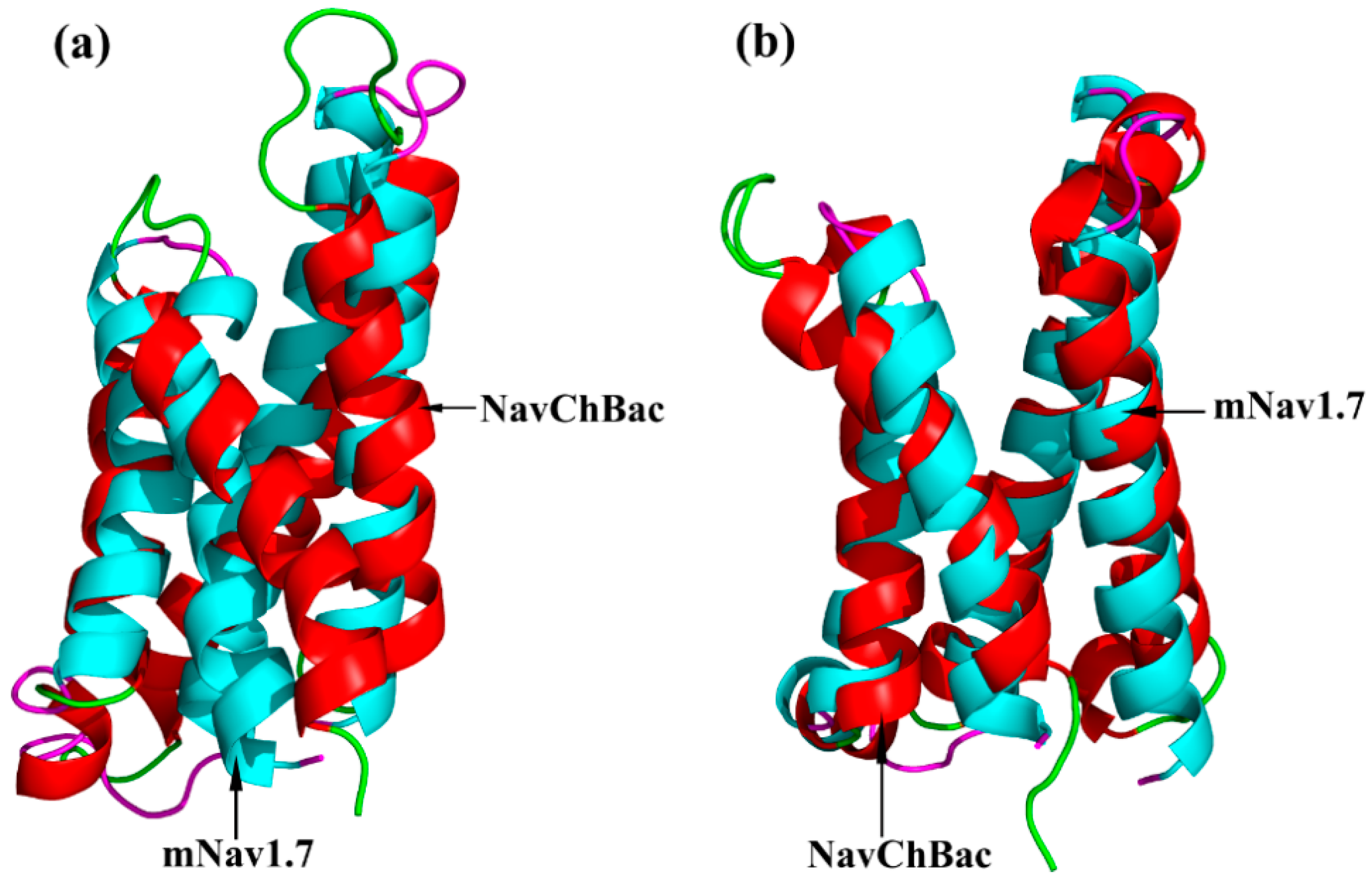
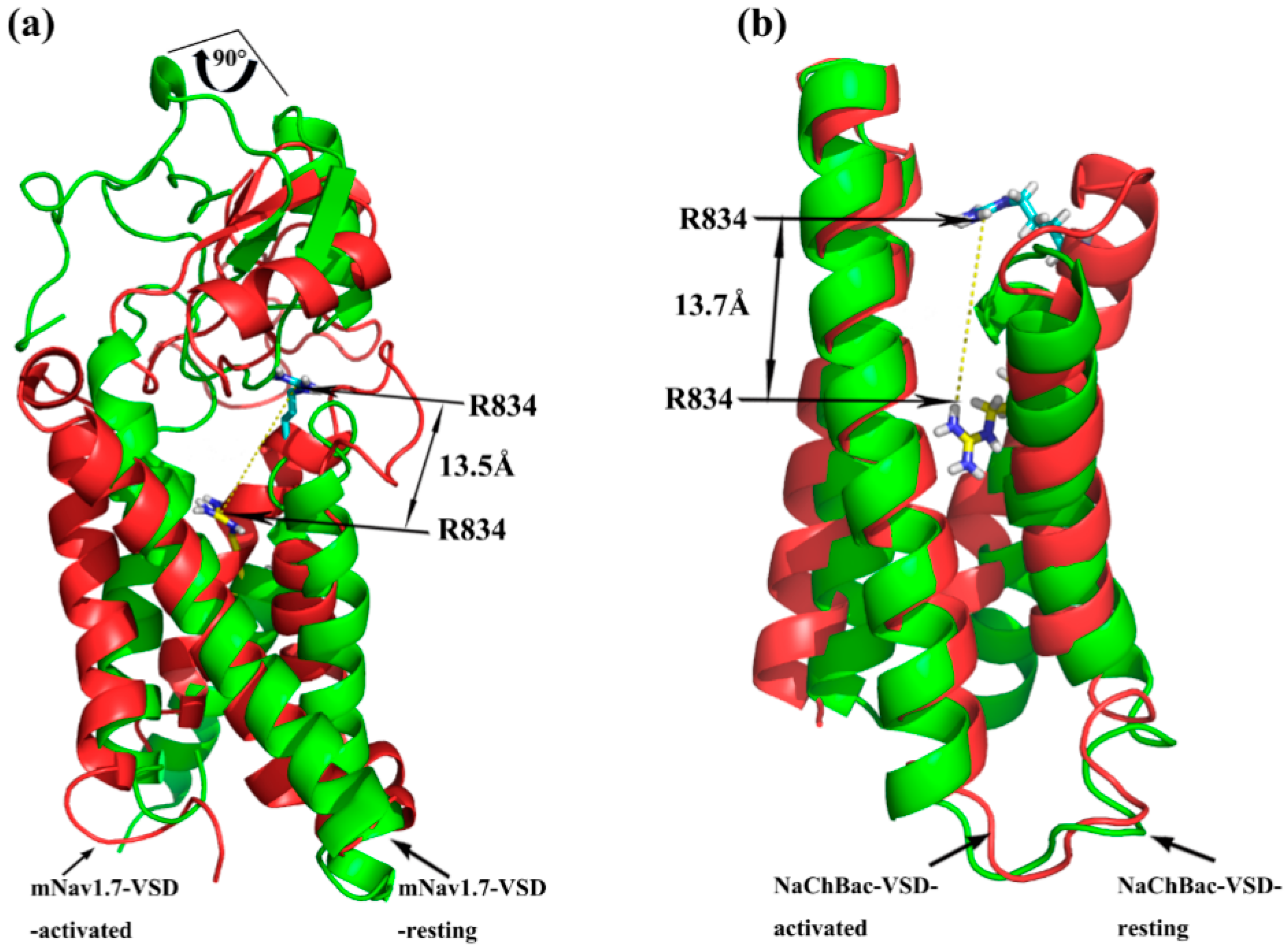
2.1. Molecular Modeling of mNav1.7 (Resting and Activated States)
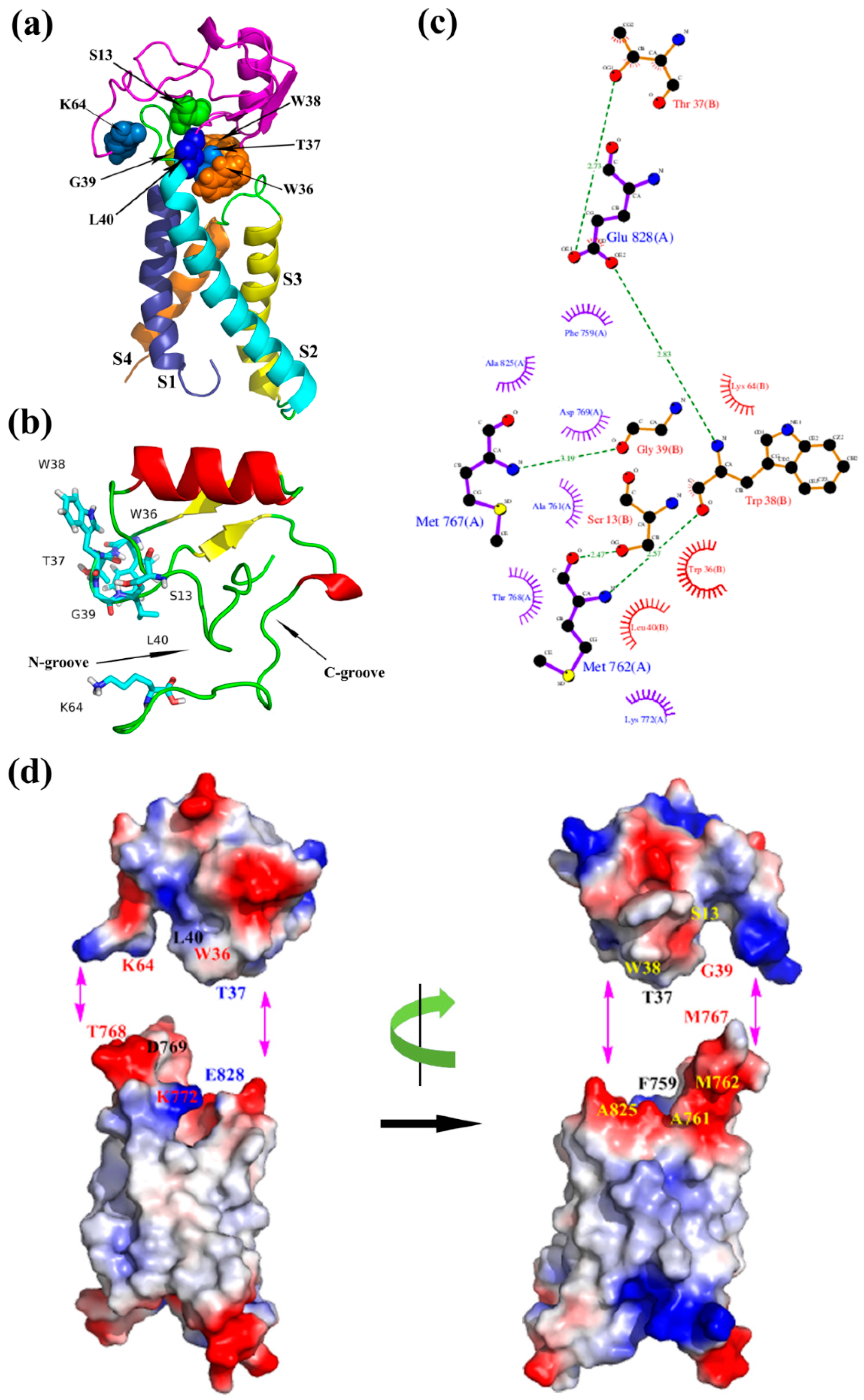
2.2. Molecular Docking Study
2.3. Molecular Dynamics Simulations
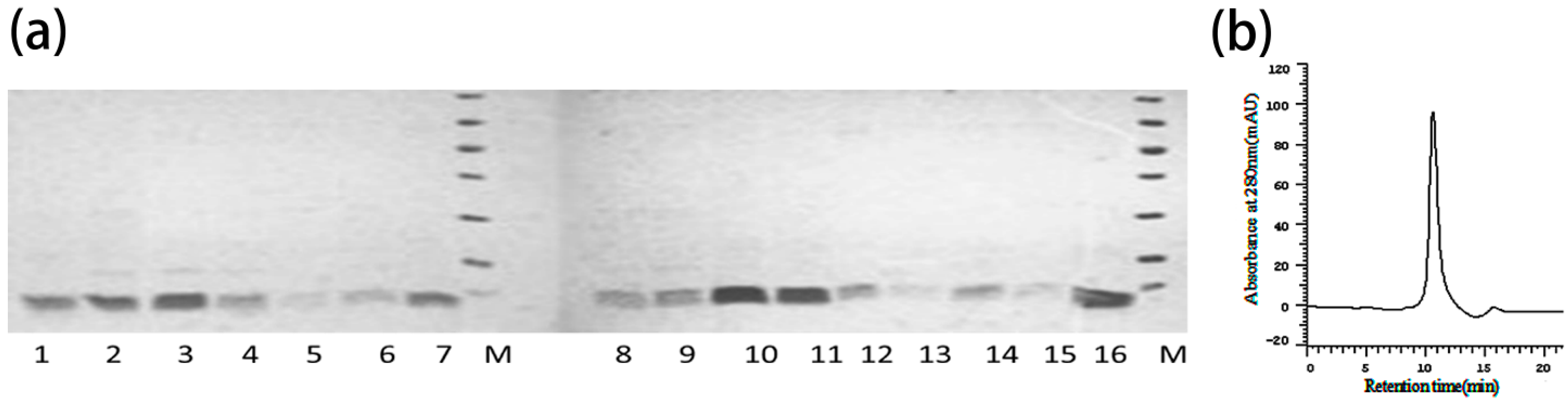
2.4. Construction, Expression and Purification of Mutants
2.5. Analgesic Activities of ANEP Mutants
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Homology Models
5.2. IIS1–S4 Domains Molecular Dynamics Simulations
5.3. Molecular Docking
5.4. The Complex Molecular Dynamics Simulations
5.5. Strains, Materials, and Animals
5.6. Site-Directed Mutagenesis of ANEP
5.7. Expression and Purification of ANEP and Its Mutants
5.8. Analgesic Activity Assays
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Shen, H.; Zhou, Q.; Pan, X.; Li, Z.; Wu, J.; Yan, N. Structure of a eukaryotic voltage-gated sodium channel at near-atomic resolution. Science 2017, 355. [Google Scholar] [CrossRef] [PubMed]
- Mccusker, E.C.; Bagneris, C.; Naylor, C.E.; Cole, A.R.; D’Avanzo, N.; Nichols, C.G.; Wallace, B.A. Structure of a bacterial voltage-gated sodium channel pore reveals mechanisms of opening and closing. Nat. Commun. 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, F.C.; Lewis, R.J. Sodium channels and pain: From toxins to therapies. Br. J. Pharmacol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.J.; Reimann, F.; Nicholas, A.K.; Thornton, G.; Roberts, E.; Springell, K.; Karbani, G.; Jafri, H.; Mannan, J.; Raashid, Y.; et al. An SCN9A channelopathy causes congenital inability to experience pain. Nature 2006, 444, 894–898. [Google Scholar] [CrossRef] [PubMed]
- Dib-Hajj, S.D.; Rush, A.M.; Cummins, T.R.; Hisama, F.M.; Novella, S.; Tyrrell, L.; Marshall, L.; Waxman, S.G. Gain-of-function mutation in Nav1.7 in familial erythromelalgia induces bursting of sensory neurons. Brain 2005, 128, 1847–1854. [Google Scholar] [CrossRef] [PubMed]
- Nassar, M.A.; Stirling, L.C.; Forlani, G.; Baker, M.D.; Matthews, E.A.; Dickenson, A.H.; Wood, J.N. Nociceptor-specific gene deletion reveals a major role for Nav1.7 (PN1) in acute and inflammatory pain. Proc. Natl. Acad. Sci. USA 2004, 101, 12706–12711. [Google Scholar] [CrossRef] [PubMed]
- Cestele, S.; Catterall, W.A. Molecular mechanisms of neurotoxin action on voltage-gated sodium channels. Biochimie 2000, 82, 883–892. [Google Scholar] [CrossRef]
- Zhang, J.Z.; Yarov-Yarovoy, V.; Scheuer, T.; Karbat, I.; Cohen, L.; Gordon, D.; Gurevitz, M.; Catterall, W.A. Mapping the interaction site for a β-scorpion toxin in the pore module of domain III of voltage-gated Na(+) channels. J. Biol. Chem. 2012, 287, 30719–30728. [Google Scholar] [CrossRef] [PubMed]
- Bosmans, F.; Martineauclaire, M.F.; Tytgat, J. Differential effects of five ‘classical’ scorpion beta-toxins on rNav1.2a and DmNav1 provide clues on species-selectivity. Toxicol. Appl. Pharmacol. 2007, 218, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Cohen, L.; Karbat, I.; Gilles, N.; Froy, O.; Corzo, G.; Angelovici, R.; Gordon, D.; Gurevitz, M. Dissection of the functional surface of an anti-insect excitatory toxin illuminates a putative “hot spot” common to all scorpion beta-toxins affecting Na+ channels. J. Biol. Chem. 2004, 279, 8206–8211. [Google Scholar] [CrossRef] [PubMed]
- Cohen, L.; Karbat, I.; Gilles, N.; Ilan, N.; Benveniste, M.; Gordon, D.; Gurevitz, M. Common features in the functional surface of scorpion beta-toxins and elements that confer specificity for insect and mammalian voltage-gated sodium channels. J. Biol. Chem. 2005, 280, 5045–5053. [Google Scholar] [CrossRef] [PubMed]
- Karbat, I.; Cohen, L.; Gilles, N.; Gordon, D.; Gurevitz, M. Conversion of a scorpion toxin agonist into an antagonist highlights an acidic residue involved in voltage sensor trapping during activation of neuronal Na+ channels. FASEB J. 2004, 18, 683–689. [Google Scholar] [PubMed]
- Cui, Y.; Guo, G.-L.; Ma, L.; Hu, N.; Song, Y.-B.; Liu, Y.-F.; Wu, C.-F.; Zhang, J.-H. Structure and function relationship of toxin from Chinese scorpion Buthus martensii Karsch (BmKAGAP): Gaining insight into related sites of analgesic activity. Peptides 2010, 31, 995–1000. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hao, Z.; Shao, J.; Song, Y.; Li, C.; Li, C.; Zhao, Y.; Liu, Y.; Wei, T.; Wu, C.; Zhang, J. The role of Ser54 in the antinociceptive activity of BmK9, a neurotoxin from the scorpion Buthus martensii Karsch. Toxicon 2011, 58, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Song, Y.B.; Ma, L.; Liu, Y.F.; Li, G.D.; Wu, C.F.; Zhang, J.H. Site-directed mutagenesis of the toxin from the Chinese scorpion Buthus martensii Karsch (BmKAS): Insight into sites related to analgesic activity. Arch. Pharm. Res. 2010, 33, 1633–1639. [Google Scholar] [CrossRef] [PubMed]
- Tong, X.; Yao, J.; He, F.; Chen, X.; Zheng, X.; Xie, C.; Wu, G.; Zhang, N.; Ding, J.; Wu, H. NMR solution structure of BmK-betaIT, an excitatory scorpion beta-toxin without a ‘hot spot’ at the relevant position. Biochem. Biophys. Res. Commun. 2006, 349, 890–899. [Google Scholar] [CrossRef] [PubMed]
- Han, T.; Ming, H.; Deng, L.; Zhu, H.; Liu, Z.; Zhang, J.; Song, Y. A novel expression vector for the improved solubility of recombinant scorpion venom in Escherichia coli. Biochem. Biophys. Res. Commun. 2016, 482, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Yarov-Yarovoy, V.; DeCaen, P.G.; Westenbroek, R.E.; Pan, C.Y.; Scheuer, T.; Baker, D.; Catterall, W.A. Structural basis for gating charge movement in the voltage sensor of a sodium channel. Proc. Natl. Acad. Sci. USA 2012, 109, E93–E102. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Z.; Yarov-Yarovoy, V.; Scheuer, T.; Karbat, I.; Cohen, L.; Gordon, D.; Gurevitz, M.; Catterall, W.A. Structure-function map of the receptor site for β-scorpion toxins in domain II of voltage-gated sodium channels. J. Biol. Chem. 2011, 286, 33641–33651. [Google Scholar] [CrossRef] [PubMed]
- Catterall, W.A.; Cestèle, S.; Yarov-Yarovoy, V.; Yu, F.H.; Konoki, K.; Scheuer, T. Voltage-gated ion channels and gating modifier toxins. Toxicon 2007, 49, 124–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, R.; Chung, S.H. Conserved functional surface of antimammalian scorpion β-toxins. J. Phys. Chem. B 2012, 116, 4796–4800. [Google Scholar] [CrossRef] [PubMed]
- Karbat, I.; Turkov, M.; Cohen, L.; Kahn, R.; Gordon, D.; Gurevitz, M.; Frolow, F. X-ray structure and mutagenesis of the scorpion depressant toxin LqhIT2 reveals key determinants crucial for activity and anti-insect selectivity. J. Mol. Biol. 2007, 366, 586–601. [Google Scholar] [CrossRef] [PubMed]
- Payandeh, J.; Scheuer, T.; Zheng, N.; Catterall, W.A. The crystal structure of a voltage-gated sodium channel. Nature 2011, 475, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Catterall, W.A. Voltage-dependent gating of sodium channels: Correlating structure and function. Trends Neurosci. 1986, 9, 7–10. [Google Scholar] [CrossRef]
- Yarov-Yarovoy, V.; Baker, D.; Catterall, W.A. Voltage sensor conformations in the open and closed states in ROSETTA structural models of K(+) channels. Proc. Natl. Acad. Sci. USA 2006, 103, 7292–7297. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Guan, R.J.; Xiang, Y.; Zhang, Y.; Wang, D.C. Structure of an excitatory insect-specific toxin with an analgesic effect on mammals from the scorpion Buthus martensii Karsch. Acta Crystallogr. 2005, 61, 14–21. [Google Scholar]
- Pinheiro, C.B.; Marangoni, S.; Toyama, M.H.; Polikarpov, I. Structural analysis of Tityus serrulatus Ts1 neurotoxin at atomic resolution: Insights into interactions with Na+ channels. Acta Crystallogr. 2003, 59, 405–415. [Google Scholar]
- Webb, B.; Sali, A. Comparative Protein Structure Modeling Using MODELLER; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2016. [Google Scholar]
- Song, Y.B.; Ma, L.; Yang, W.Y.; Wang, J.; Cheng, M.S.; Wu, C.F.; Zhang, J.H. Study of the binding residues between ANEPII and insect sodium channel receptor. C. R. Biol. 2010, 333, 637–641. [Google Scholar] [CrossRef] [PubMed]
- Pathak, M.M.; Yarov-Yarovoy, V.; Agarwal, G.; Roux, B.; Barth, P.; Kohout, S.; Tombola, F.; Isacoff, E.Y. Closing in on the resting state of the Shaker K(+) channel. Neuron 2007, 56, 124–140. [Google Scholar] [CrossRef] [PubMed]
- Hess, B.; Kutzner, C.; van der Spoel, D.; Lindahl, E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 2008, 4, 435–447. [Google Scholar] [CrossRef] [PubMed]
- Oostenbrink, C.; Villa, A.; Mark, A.E.; van Gunsteren, W.F. A biomolecular force field based on the free enthalpy of hydration and solvation: The GROMOS force-field parameter sets 53A5 and 53A6. J. Comput. Chem. 2004, 25, 1656–1676. [Google Scholar] [CrossRef] [PubMed]
- Kandt, C.; Ash, W.L.; Tieleman, D.P. Setting up and running molecular dynamics simulations of membrane proteins. Methods 2007, 41, 475–488. [Google Scholar] [CrossRef] [PubMed]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Miyamoto, S.; Kollman, P.A. SETTLE: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992, 13, 952–962. [Google Scholar] [CrossRef]



| Group 1 | Dosage (mg/kg) | Number of Writhes (Mean ± SEM) | Inhibition Efficiency (%) 1 | Relative Activity (%) |
|---|---|---|---|---|
| NS | - | 47.67 ± 1.77 | - | - |
| rANEP | 1.5 | 29.00 ± 3.41 * | 39.17% | 100 |
| rANEP-N8A | 1.5 | 21.60 ± 3.7 *,2 | 54.68% | 132 |
| rANEP-V12A | 1.5 | 20.33 ± 2.98 *,2 | 57.35% | 143 |
| rANEP-S13A | 1.5 | 29.70 ± 2.5 * | 37.69% | 96 |
| rANEP-L15A | 1.5 | 22.00 ± 1.25 *,2 | 53.85% | 132 |
| rANEP-W16A | 1.5 | 23.88 ± 1.55 * | 49.90% | 121 |
| rANEP-E24A | 1.5 | 20.50 ± 3.26 *,2 | 57.00% | 141 |
| rANEP-Y28A | 1.5 | 24.75 ± 2.6 * | 48.06% | 114 |
| rANEP-W36A | 1.5 | 13.56 ± 2.36 *,2 | 71.55% | 214 |
| rANEP-T37A | 1.5 | 15.89 ± 3.11 *,2 | 66.67% | 183 |
| rANEP-W38A | 1.5 | 15.56 ± 2.47 *,2 | 67.36% | 186 |
| rANEP-G39A | 1.5 | 21.93 ± 2.1 *,2 | 55.12% | 132 |
| rANEP-L40A | 1.5 | 30.33 ± 2.8 * | 35.71% | 93 |
| rANEP-C60A | 1.5 | 35.21 ± 2.1 *,2 | 26.14% | 67 |
| rANEP-K64⊿ | 1.5 | 14.98 ± 3.16 *,2 | 68.58% | 175 |
| Name | Nucleotide Sequence (5′-3′) | Orientation |
|---|---|---|
| ANEP-F | CATGCCATGGGACATCATCATCATCATCACGATGGATATATAAGAGGAAGTAACGGATG | Sense |
| ANEP-R | CGGGATCCTTACTTTTTGCCACCGCATGTATTACTTTCA | Antisense |
| N8A | CATGCCATGGGACACCACCACCACCACCACGATGGATATATAAGAGGAAGTGCGGGATG | Sense |
| V12A | AACGGATGCAAGGCGTCATGC | Sense |
| ATGACGCTTGCATCCGTTACT | Antisense | |
| S13A | CAAGGTTGCGTGCTTATGGGGA | Sense |
| CATAAGCACGCAACCTTGCATCC | Antisense | |
| L15A | TCATGCGCGTGGGGAAATG | Sense |
| TCCCCACGCGCATGAAACCTT | Antisense | |
| W16A | TCATGCTTAGCGGGAAATGAC | Sense |
| GACATTTCCCGCTAAGCATGAA | Antisense | |
| E24A | CAATAAAGCGTGCAGAGC | Sense |
| TGCACGCTTTATTGCAACCGTCATT | Antisense | |
| Y28A | AGAGCGGCGGGTGCCTCTT | Sense |
| GAGGCACCCGCCGCTCTG | Antisense | |
| W36A | TATGGTTATTGCGCGACCTGGGGA | Sense |
| AAGTCCCCAGGTCGCGCAATAACCATAAGA | Antisense | |
| T37A | TGCTGGGCGTGGGGACT | Sense |
| TCCCCACGCCCAGCAATAA | Antisense | |
| W38A | ACCGCGGGACTTGCATGC | Sense |
| TCCCGCGGTCCAGCAATAAC | Antisense | |
| G39A | ACCTGGGCGCTTGCATGCTG | sense |
| CAGCATGCAAGCGCCCAGGT | Antisense | |
| L40A | TGGGGAGCGGCATGCTG | Sense |
| CAGCATGCCGCTCCCCA | Antisense | |
| C60A | CGGGATCCTTACTTTTTGCCACCCGCTGTATTACT | Antisense |
| K64⊿ | CGGGATCCTTATTTGCCACCGCATGTATTACTT | Antisense |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, Y.; Liu, Z.; Zhang, Q.; Li, C.; Jin, W.; Liu, L.; Zhang, J.; Zhang, J. Investigation of Binding Modes and Functional Surface of Scorpion Toxins ANEP to Sodium Channels 1.7. Toxins 2017, 9, 387. https://doi.org/10.3390/toxins9120387
Song Y, Liu Z, Zhang Q, Li C, Jin W, Liu L, Zhang J, Zhang J. Investigation of Binding Modes and Functional Surface of Scorpion Toxins ANEP to Sodium Channels 1.7. Toxins. 2017; 9(12):387. https://doi.org/10.3390/toxins9120387
Chicago/Turabian StyleSong, Yongbo, Zeyu Liu, Qi Zhang, Chunming Li, Wei Jin, Lili Liu, Jianye Zhang, and Jinghai Zhang. 2017. "Investigation of Binding Modes and Functional Surface of Scorpion Toxins ANEP to Sodium Channels 1.7" Toxins 9, no. 12: 387. https://doi.org/10.3390/toxins9120387





