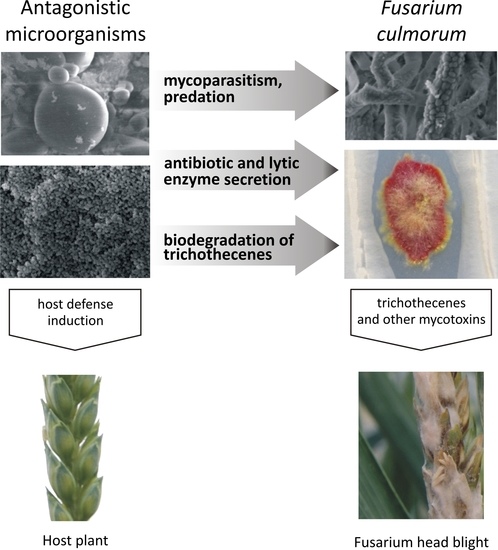
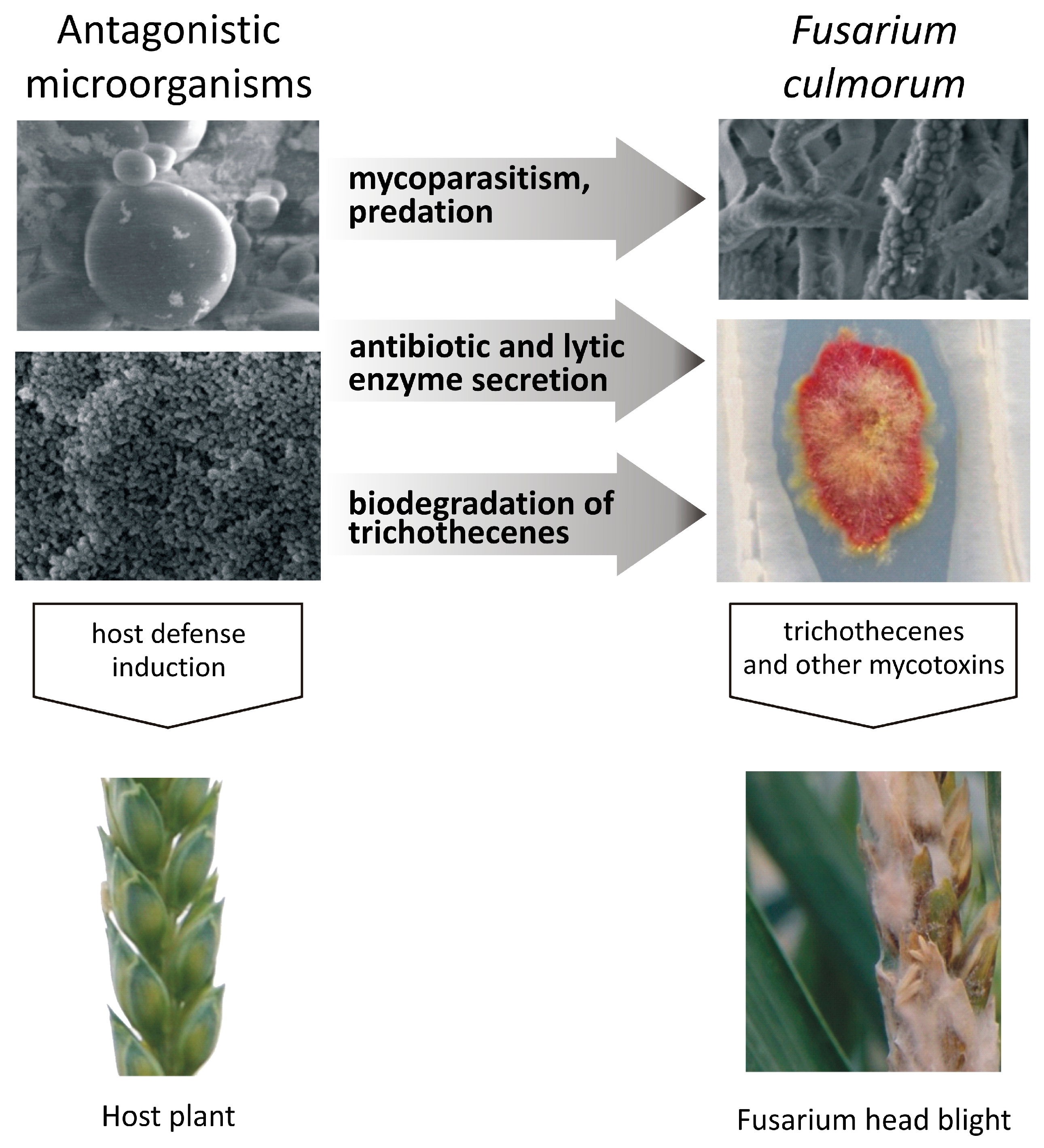
Microbial Inhibition of Fusarium Pathogens and Biological Modification of Trichothecenes in Cereal Grains
Abstract
:1. Introduction
2. Trichothecenes
3. Inhibition of the Growth of Fusarium Pathogens and Trichothecene Production by Microorganisms
4. Trichothecene Degradation by Microorganisms
4.1. Trichothecene-Degrading Bacteria
4.2. Fungi Capable of Degradation and Acetylation of Trichothecenes
5. Conclusions
Conflicts of Interest
Abbreviations
| 15-Ac-DON | 15-Acetyl-deoxynivalenol |
| 3-Ac-DON | 3-Acetyl-deoxynivalenol |
| D3G | Deoxynivalenol-3-β-d-glucopyranoside |
| DOM-1 | De-epoxy-deoxynivalenol |
| DON | Deoxynivalenol |
| FHB | Fusarium head blight |
| FPP | Farnesyl pyrophosphate |
| MAPK | Mitogen-activated protein kinase |
| NEO | Neosolaniol |
| NIV | Nivalenol |
References
- Stoev, S.D. Foodborne mycotoxicoses, risk assessment and underestimated hazard of masked mycotoxins and joint mycotoxin effects or interaction. Environ. Toxicol. Pharmacol. 2015, 39, 794–809. [Google Scholar] [CrossRef] [PubMed]
- Pirgozliev, S.R.; Edwards, S.G.; Hare, M.C.; Jenkinson, P. Effect of dose rate of azoxystrobin and metconazole on the development of Fusarium Head Blight and the accumulation of deoxynivalenol (DON) in wheat grain. Eur. J. Plant Pathol. 2002, 108, 469–478. [Google Scholar] [CrossRef]
- European Commission. Commission Regulation (EC) No 1107/2009 of 21 October 2009. Concerning the Placing of Plant Protection Products on the Market. Available online: http://eur-lex.europa.eu/legal-content/EN/TXT/?uri=celex%3A32009R1107 (accessed on 2 December 2017).
- European Commission. Commission Regulation (EC) No 889/2008 of 5 September 2008 Laying down Detailed Rules for the Implementation of Council Regulation (EC) No 834/2007 on Organic Production and Labeling of Organic Products with Regard to Organic Production, Labeling and Control. Available online: http://eur-lex.europa.eu/legal-content/en/ALL/?uri=CELEX%3A32008R0889 (accessed on 2 December 2017).
- Commission Regulation EC No 1881/2006 of 19 December 2006 Setting Maximum Levels for Certain Contaminants for Foodstuffs. Available online: http://eur-lex.europa.eu/legal-content/EN/TXT?url=CELEX:02006R1881-20170728 (accessed on 2 December 2017).
- Lindblad, M.; Gidlund, A.; Sulyok, M.; Börjesson, T.; Krska, R.; Olsen, M.; Fredlund, E. Deoxynivalenol and other selected Fusarium toxins in Swedish wheat-occurrence and correlation to specific Fusarium species. Int. J. Food Microbiol. 2013, 167, 284–291. [Google Scholar] [CrossRef] [PubMed]
- Commission Recommendation of 27 March 2013 on the Presence T-2 and HT-2 Toxin in Cereals and Cereal Products. Available online: http://eur-lex.europa.eu/legal-content/EN/ALL/?uri=CELEX:32013H0165 (accessed on 2 December 2017).
- Panel, E.C. Scientific opinion on risks for animal and public health related to the presence of nivalenol in food and feed. EFSA J. 2013, 11, 3262. [Google Scholar]
- Okubara, P.A.; Blechl, A.E.; McCormick, S.P.; Alexander, N.J.; Dill-Macky, R.; Hohn, T.M. Engineering deoxynivalenol metabolism in wheat through the expression of a fungal trichothecene acetyltransferase gene. Theor. Appl. Genet. 2002, 106, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Ohsato, S.; Ochiai-Fukuda, T.; Nishiuchi, T.; Takahashi-Ando, N.; Koizumi, S.; Hamamoto, H.; Kudo, T.; Yamaguchi, I.; Kimura, M. Transgenic rice plants expressing trichothecene 3-O-acetyltransferase show resistance to the Fusarium phytotoxin deoxynivalenol. Plant Cell Rep. 2007, 26, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Berthiller, F.; Crews, C.; Dall’Asta, C.; Saeger, S.D.; Haesaert, G.; Karlovsky, P.; Oswald, I.P.; Seefelder, W.; Speijers, G.; Stroka, J. Masked mycotoxins: A review. Mol. Nutr. Food Res. 2013, 57, 165–186. [Google Scholar] [PubMed]
- Rychlik, M.; Humpf, H.U.; Marko, D.; Dänicke, S.; Mally, A.; Berthiller, F.; Klaffke, H.; Lorenz, N. Proposal of a comprehensive definition of modified and other forms of mycotoxins including “masked” mycotoxins. Mycotoxin Res. 2014, 30, 197–205. [Google Scholar] [PubMed]
- Wiwart, M.; Suchowilska, E.; Kandler, W.; Sulyok, M.; Groenwald, P.; Krska, R. Can Polish wheat (Triticum polonicum L.) be an interesting gene source for breeding wheat cultivars with increased resistance to Fusarium head blight? Genet. Resour. Crop Evol. 2013, 60, 2359–2373. [Google Scholar] [CrossRef]
- Kluger, B.; Bueschl, C.; Lemmens, M.; Berthiller, F.; Häubl, G.; Jaunecker, G.; Adam, G.; Krska, R.; Schuhmacher, R. Stable isotopic labelling-assisted untargeted metabolic profiling reveals novel conjugates of the mycotoxin deoxynivalenol in wheat. Anal. Bioanal. Chem. 2013, 405, 5031–5036. [Google Scholar] [PubMed]
- Pan, D.; Mionetto, A.; Tiscornia, S.; Bettucci, L. Endophytic bacteria from wheat grain as biocontrol agents of Fusarium graminearum and deoxynivalenol production in wheat. Mycotoxin Res. 2015, 31, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Turner, T.R.; James, E.K.; Poole, P.S. The plant microbiome. Genome Biol. 2013, 14, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Gdanetz, K.; Frances Trail, F. The Wheat Microbiome under Four Management Strategies, and Potential for Endophytes in Disease Protection. Phytobiomes 2017, 1, 158–168. [Google Scholar] [CrossRef]
- Doty, S.L. Functional Importance of the Plant Microbiome: Implications for Agriculture, Forestry and Bioenergy; Springer: Berlin, Germany, 2017; p. 111. [Google Scholar]
- Wachowska, U.; Głowacka, K. Antagonistic interactions between Aureobasidium pullulans and Fusarium culmorum, a fungal pathogen of winter wheat. BioControl 2014, 59, 635–645. [Google Scholar] [CrossRef]
- Schisler, D.A.; Core, A.B.; Boehm, M.J.; Horst, L.; Krause, C.; Dunlap, C.A.; Rooney, A.P. Population dynamics of the Fusarium head blight biocontrol agent Cryptococcus flavescens OH 182.9 on wheat anthers and heads. Biol. Control 2014, 70, 17–27. [Google Scholar] [CrossRef]
- Kosawang, C.; Karlsson, M.; Jensen, D.F.; Dilokpimol, A.; Collinge, D.B. Transcriptomic profiling to identify genes involved in Fusarium mycotoxin deoxynivalenol and zearalenone tolerance in the mycoparasitic fungus Clonostachys rosea. BMC Genom. 2014, 15, 55. [Google Scholar] [CrossRef] [PubMed]
- Druefors, U.A.; Schnürer, J. Mold-inhibitory activity of different yeast species during airtight storage of wheat grain. FEMS Yeast Res. 2005, 5, 373–378. [Google Scholar]
- Roberti, R.; Veronesi, A.R.; Cesari, A.; Cascone, A.; Berardino, I.D.; Bertini, L.; Caruso, C. Induction of PR proteins and resistance by the biocontrol agent Clonostachys rosea in wheat plants infected with Fusarium culmorum. Plant Sci. 2008, 175, 339–347. [Google Scholar] [CrossRef]
- Petti, C.; Khan, M.; Doohan, F. Lipid transfer proteins and protease inhibitors as key factors in the priming of barley responses to Fusarium head blight disease by a biocontrol strain of Pseudomonas fluorescens. Funct. Integr. Genom. 2010, 10, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Cserháti, M.; Kriszt, B.; Krifaton, C.; Szoboszlay, S.; Hahn, J.; Toth, S.; Nagy, I.; Kukolya, J. Mycotoxin-degradation profile of Rhodococcus strains. Int. J. Food Microbiol. 2013, 166, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Pierron, A.; Mimoun, S.; Murate, L.S.; Loiseau, N.; Lippi, Y.; Bracarense, A.P.F.L.; Schatzmayr, G.; He, J.W.; Zhou, T.; Moll, W.D.; et al. Microbial biotransformation of DON: Molecular basis for reduced toxicity. Sci. Rep. 2016, 6, 29105. [Google Scholar] [CrossRef] [PubMed]
- Grove, J.F. The Trichothecenes and Their Biosynthesis. In Progress in the Chemistry of Organic Natural Products; Herz, W., Falk, H., Kirby, G.W., Eds.; Springer: Vienna, Austria, 2007; pp. 63–130. ISBN 978-3-211-20688-1. [Google Scholar]
- McCormick, S.P.; Stanley, A.M.; Stover, N.A.; Alexander, N.J. Trichothecenes: From Simple to Complex Mycotoxins. Toxins 2011, 3, 802–814. [Google Scholar] [CrossRef] [PubMed]
- Arunachalam, C.; Doohan, F.M. Trichothecene toxicity in eukaryotes: Cellular and molecular mechanisms in plants and animals. Toxicol. Lett. 2013, 217, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Stępień, Ł. The use of Fusarium secondary metabolite biosynthetic genes in chemotypic and phylogenetic studies. Crit. Rev. Microbiol. 2014, 40, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Pestka, J. Toxicological mechanisms and potential health effects of deoxynivalenol and nivalenol. World Mycotoxin J. 2010, 3, 323–347. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Z.; Beier, R.C.; Shen, J.; Smet, D.; Saeger, S.; Zhang, S. T-2 toxin, a trichothecene mycotoxins: Review of toxicity, metabolism, and analytical methods. J. Agric. Food Chem. 2011, 8, 3441–3453. [Google Scholar] [CrossRef] [PubMed]
- Goossens, J.; Devreese, M.; Pasmans, F.; Osselaere, A.; De Baere, S.; Verbrugghe, E.; Haesebrouck, F.; De Backer, P.; Croubels, S. Chronic exposure to the mycotoxin T-2 promotes oral absorption of chlortetracycline in pigs. J. Vet. Pharmacol. Ther. 2013, 36, 621–624. [Google Scholar] [CrossRef] [PubMed]
- Garreau de Loubresse, N.; Prokhorova, I.; Holtkamp, W.; Rodnina, M.V.; Yusupova, G.; Yusupov, M. Structural basis for the inhibition of the eukaryotic ribosome. Nature 2014, 513, 517–522. [Google Scholar] [CrossRef] [PubMed]
- McCormick, S.P.; Price, N.P.J.; Kurtzman, C.P. Glucosylation and other biotransformations of T-2 toxin by yeasts of the Trichomonascus clade. Appl. Environ. Microbiol. 2012, 78, 8694–8702. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.; Lee, S.H.; Shin, J.Y.; Kim, H.K.; Yun, S.H.; Kim, H.Y.; Lee, S.; Ryu, J.G. Comparison of Trichothecene Biosynthetic Gene Expression between Fusarium graminearum and Fusarium asiaticum. Plant Pathol. J. 2014, 30, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Cardoza, R.E.; Malmierca, M.G.; Hermosa, M.R.; Alexander, N.J.; McCormick, S.P.; Proctor, R.H.; Tijerino, A.M.; Rumbero, A.; Monte, E.; Gutierrez, S. Identification of Loci and Functional Characterization of Trichothecene Biosynthesis Genes in Filamentous Fungi of the Genus Trichoderma. Appl. Environ. Microbiol. 2011, 77, 4867–4877. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M.; Tokai, T.; Takahashi-Ando, N.; Ohsato, S.; Fujimura, M. Molecular and genetic studies of Fusarium trichothecene biosynthesis: Pathways, genes and evolution. Biosci. Biotechnol. Biochem. 2007, 71, 2105–2123. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.W.; McCormick, S.P.; Alexander, N.J.; Proctor, R.H.; Desjardins, A.E. A genetic and biochemical approach to study trichothecene diversity in Fusarium sporotrichioides and Fusarium graminearum. Fungal Genet. Biol. 2001, 32, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.W.; McCormick, S.P.; Alexander, N.J.; Proctor, R.H.; Desjardins, A.E. Inactivation of a cytochrome P-450 is a determinant of trichothecene diversity in Fusarium species. Fungal Genet. Biol. 2002, 36, 224–233. [Google Scholar] [CrossRef]
- Lee, T.; Han, Y.K.; Kim, K.H.; Yun, S.H.; Lee, Y.W. Tri13 and Tri17 determine deoxynivalenol- and nivalenol-producing chemotypes of Gibberella zeae. Appl. Environ. Microbiol. 2002, 68, 2148–2154. [Google Scholar] [CrossRef] [PubMed]
- Moretti, A.; Susca, A.; Mule, G.; Logrieco, A.F.; Proctor, R.H. Molecular biodiversity of mycotoxigenic fungi that threaten food safety. Int. J. Food Microbiol. 2013, 167, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Lia, H.P.; Wu, A.B.; Zhao, C.S.; Scholten, O.; Löffler, H.; Lia, Y.C. Development of a generic PCR detection of deoxynivalenol- and nivalenol-chemotypes of Fusarium graminearum. FEMS Microbiol. Lett. 2005, 243, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Varga, E.; Wiesenberger, G.; Hametner, C.; Ward, T.J.; Dong, Y.; Schöfbeck, D.; McCormick, S.; Broz, K.; Stückler, R.; Schuhmacher, R.; et al. New tricks an old enemy: Isolates of Fusarium graminearum produce a type A trichothecene mycotoxin. Environ. Microbiol. 2015, 17, 2588–2600. [Google Scholar] [CrossRef] [PubMed]
- Lutz, M.P.; Feichtinger, G.; Defago, G.; Duffy, B. Mycotoxigenic Fusarium and deoxynivalenol production repress chitinase gene expression in the biocontrol agent Trichoderma atroviride P1. Appl. Environ. Microbiol. 2003, 69, 3077–3084. [Google Scholar] [CrossRef] [PubMed]
- Dunlap, C.A.; Schisler, D.A.; Price, N.P.; Vaughn, S.F. Cyclic lipopeptide profile of three Bacillus subtilis strains; antagonists of Fusarium head blight. J. Microbiol. 2011, 49, 603–609. [Google Scholar] [CrossRef] [PubMed]
- Palazzini, J.M.; Dunlap, C.A.; Bowman, M.J.; Sofía, N.; Chulze, S.N. Bacillus velezensis RC 218 as a biocontrol agent to reduce Fusarium head blight and deoxynivalenol accumulation: Genome sequencing and secondary metabolite cluster profiles. Microbiol. Res. 2016, 192, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Vegi, A.; Wolf-Hall, C. Screening of lactic acid bacteria for anti-Fusarium activity and optimization of incubation conditions. J. Food Prot. 2017, 80, 1648–1656. [Google Scholar] [CrossRef] [PubMed]
- Baffoni, L.; Gaggia, F.; Dalanaj, N.; Prodi, A.; Nipoti, P.; Pisi, A.; Biavati, B.; Di Gioia, D. Microbial inoculants for the biocontrol of Fusarium spp. in durum wheat. BMC Microbiol. 2015, 15, 242. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.K.; Savard, M.E.; Reid, L.M.; Cyr, T.; McCormick, W.A.; Seguin, C. Identification of lipopeptide antibiotics of a Bacillus subtilis isolate and their control of Fusarium graminearum diseases in maize and wheat. BioControl 2009, 54, 567–574. [Google Scholar] [CrossRef]
- Shi, C.; Yan, P.; Li, J.; Wu, H.; Li, Q.; Guan, S. Biocontrol of Fusarium graminearum growth and deoxynivalenol production in wheat kernels with bacterial antagonists. Int. J. Environ. Res. Public Health 2014, 11, 1094–1105. [Google Scholar] [CrossRef]
- Nagaraja, H.; Chennappa, G.; Rakesh, S.; Naik, M.K.; Amaresh, Y.S.; Sreenivasa, M.Y. Antifungal activity of Azotobacter nigricans against trichothecene-producing Fusarium species associated with cereals. Food Sci. Biotechnol. 2016, 25, 1197–1204. [Google Scholar] [CrossRef]
- Mnasri, N.; Chennaoui, C.; Gargouri, S.; Mhamdi, R.; Hessini, K.; Elkahoui, S.; Djébali, N. Efficacy of some rhizospheric and endophytic bacteria in vitro and as seed coating for the control of Fusarium culmorum infecting durum wheat in Tunisia. Eur. J. Plant Pathol. 2017, 147, 501–515. [Google Scholar] [CrossRef]
- Alimi, M.; Soleimani, M.J.; Darzi, M.T. Characterization and application of microbial antagonists for control of Fusarium head blight of wheat caused by Fusarium graminearum using single and mixture strain of antagonistic bacteria on resistance and susceptible cultivars. Afr. J. Microbiol. Res. 2012, 6, 326–334. [Google Scholar]
- Méndez-Gómez, M.; Castro-Mercado, E.; Alexandre, G.; García-Pineda, E. Oxidative and antioxidative responses in the wheat-Azospirillum brasilense interaction. Protoplasma 2016, 253, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Hilber-Bodmer, M.; Schmid, M.; Ahrens, C.H.; Freimoser, F.M. Competition assays and physiological experiments of soil and phyllosphere yeasts identify Candida subhashii as a novel antagonist of filamentous fungi. BMC Microbiol. 2017, 17, 4. [Google Scholar] [CrossRef] [PubMed]
- Vujanovic, V.; Goh, Y.K. Sphaerodes mycoparasitica biotrophic mycoparasite of 3-acetyldeoxynivalenol- and 15-acetyldeoxynivalenol-producing toxigenic Fusarium graminearum chemotypes. FEMS Microbiol. Lett. 2011, 316, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, A.F. Genetic Variation and Biological Control of Fusarium graminearum Isolated from Wheat in Assiut-Egypt. Plant Pathol. J. 2016, 32, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Mamarabadi, M.; Dan Funck, J.; Mette, L. An N-acetyl-β-d-glucosaminidase gene, cr-nag1, from the biocontrol agent Clonostachys rosea is up-regulated in antagonistic interactions with Fusarium culmorum. Mycol. Res. 2009, 1, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Ferre, F.S.; Santamarina, M.P. Efficacy of Trichoderma harzianum in suppression of Fusarium culmorum. Ann. Microbiol. 2010, 60, 335–340. [Google Scholar] [CrossRef]
- Zou, C.S.; Mo, M.H.; Gu, Y.Q.; Zhou, J.P.; Zhang, K.Q. Possible contributions of volatile-producing bacteria to soil fungistases. Soil Biol. Biochem. 2007, 39, 2371–2379. [Google Scholar] [CrossRef]
- Chi, Z.; Wang, X.X.; Ma, Z.C.; Buzdar, M.A.; Chiet, Z.M. The unique role of siderophore in marine-derived Aureobasidium pullulans HN6.2. Biometals 2012, 25, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Ai, C.; Xin, L.; Zhou, G. The siderophore-producing bacterium, Bacillus subtilis CAS15, has a biocontrol effect on Fusarium wilt and promotes the growth of pepper. Eur. J. Soil Biol. 2011, 47, 138–145. [Google Scholar] [CrossRef]
- Alexander, N.J.; McCormick, S.P.; Hohn, T.M. The identification of the Saccharomyces cerevisiae gene AYT1(ORF-YLL063c) encoding an acetyltransferase. Yeast 2002, 19, 1425–1430. [Google Scholar] [CrossRef] [PubMed]
- Beeton, S.; Bull, A.T. Biotransformation and detoxification of T-2 toxin by soil and freshwater bacteria. Appl. Environ. Microbiol. 1989, 55, 190–197. [Google Scholar] [PubMed]
- Ikunaga, Y.; Sato, I.; Grond, S.; Numaziri, N.; Yoshida, S.; Yamaya, H.; Hiradate, S.; Hasegawa, M.; Toshima, H.; Koitabashi, M.; et al. Nocardioides sp. strain WSN05-2, isolated from a wheat field, degrades deoxynivalenol, producing the novel intermediate 3-epi-deoxynivalenol. Appl. Microbiol. Biotechnol. 2011, 89, 419–427. [Google Scholar] [CrossRef] [PubMed]
- McCormick, S.P. Microbial Detoxification of Mycotoxins. J. Chem. Ecol. 2013, 39, 907–918. [Google Scholar] [CrossRef] [PubMed]
- Meng-Reiterer, J.; Buesch, C.; Rechhaler, J.; Berthiller, F.; Lemmens, M.; Schuhmacher, R. Metabolism of HT-2 toxin and T-2-toxin in oats. Toxins 2016, 8, 364. [Google Scholar] [CrossRef] [PubMed]
- Mc Cormick, S.P.; Kato, T.; Maragos, C.M.; Busman, M.; Lattanzio, V.M.T.; Galaverna, G.; Dall-Asta, C.; Crich, D.; Price, N.P.J.; Kurtzman, C.P. Anomericity of T-2 toxin-glucoside: Masked mycotoxin in cereal crops. J. Agric. Food Chem. 2015, 63, 731–738. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, H. Research on mycotoxin glucosides (masked mycotoxins). JSM Mycotoxins 2016, 66, 21–25. [Google Scholar] [CrossRef]
- He, C.; Fan, Y.; Liu, G.; Zhang, H. Isolation and identification of a strain of Aspergillus tubingensis with deoxynivalenol biotransformation capability. Int. J. Mol. Sci. 2008, 9, 2366–2375. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Zhou, T.; Gong, J.; Young, C.; Su, X.; Li, X.Z.; Zhu, H.; Tsao, R.; Yang, R. Isolation of deoxynivalenol-transforming bacteria from the chicken intestines using the approach of PCR-DGGE guided microbial selection. BMC Microbiol. 2010, 10, 182. [Google Scholar] [CrossRef] [PubMed]
- Islam, R.; Zhou, T.; Young, J.C.; Goodwin, P.H.; Pauls, K.P. Aerobic and anaerobic de-epoxidation of mycotoxin deoxynivalenol by bacteria originating from agricultural soil. World J. Microbiol. Biotechnol. 2012, 28, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Sato, I.; Ito, M.; Ishizaka, M.; Ikunaga, Y.; Sato, Y.; Yoshida, S.; Koitabashi, M.; Tsushima, S. Thirteen novel deoxynivalenol-degrading bacteria are classified within two genera with distinct degradation mechanisms. FEMS Microbiol. Lett. 2012, 327, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Ito, M.; Sato, I.; Koitabashi, M.; Yoshida, S.; Imai, M.; Tsushima, S. A novel actinomycete derived from wheat heads degrades deoxynivalenol in the grain of wheat and barley affected by Fusarium head blight. Appl. Microbiol. Biotechnol. 2012, 96, 1059–1070. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, E.; Binder, E.M.; Heidler, D.; Krska, R. Structural characterization of metabolites after the microbial degradation of A- and B-trichothecenes by the bacterial strain BBSH 797. Mycotoxin Res. 2002, 16, 66–69. [Google Scholar] [CrossRef] [PubMed]
- Vanhoutte, I.; Audenaert, K.; De Gelder, L. Biodegradation of Mycotoxins: Tales from Known and Unexplored Worlds. Front. Microbiol. 2016, 7, 561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueno, Y.; Nakayama, K.; Ishii, K.; Tashiro, F.; Minoda, Y.; Omori, T.; Komagata, K. Metabolism of T-2 toxin in Curtobacterium sp. strain 114-2. Appl. Environ. Microbiol. 1983, 46, 120–127. [Google Scholar] [PubMed]
- Tran, S.T.; Smith, T.K. Conjugation of deoxynivalenol by Alternaria alternata (54028 NRRL), Rhizopus microsporus var. rhizopodiformis (54029 NRRL) and Aspergillus oryzae (5509 NRRL). Mycotoxin Res. 2014, 28, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Tan, Y.; Liu, N.; Yan, Z.; Liao, Y.; Chen, J.; de Saeger, S. Detoxification of Deoxynivalenol via Glycosylation Represents Novel Insights on Antagonistic Activities of Trichoderma when Confronted with Fusarium graminearum. Toxins 2016, 8, 335. [Google Scholar] [CrossRef] [PubMed]
- Ahad, R.; Zhou, T.; Lepp, D.; Pauls, K.P. Microbial detoxification of eleven food and feed contaminating trichothecene mycotoxins. BMC Biotechnol. 2017, 17, 30. [Google Scholar] [CrossRef] [PubMed]
- Vanhoutte, I.; De Mets, L.; De Boevre, M.; Uka, V.; Di Mavungu, J.D.; De Saeger, S.; De Gelder, L.; Audenaert, K. Microbial detoxification of deoxynivalenol (DON), assessed via a Lemna minor L. bioassay, through biotransformation to 3-epi-DON and 3-epi-DOM-1. Toxins 2017, 9, 63. [Google Scholar] [CrossRef] [PubMed]
- Khatibi, P.A.; Newmister, S.A.; Rayment, I.; McCormick, S.P.; Alexander, N.J.; Schmale, D.G. Bioprospecting for trichothecene-3-O-acetyltransferases in the fungal genus Fusarium yields functional enzymes that vary in their ability to modify the mycotoxin deoxynivalenol. Appl. Environ. Microbiol. 2011, 77, 1162–1170. [Google Scholar] [CrossRef] [PubMed]
- Eriksen, G.S.; Pettersson, H.; Lundh, T. Comparative cytotoxicity of deoxynivalenol, nivalenol, their acetylated derivatives and de-epoxy metabolites. Food Chem. Toxicol. 2004, 42, 619–624. [Google Scholar] [CrossRef] [PubMed]

| Microorganisms | Microbial Mechanisms of Action | Pathogens | Reduction in DON Content | Reference |
|---|---|---|---|---|
| Bacteria | ||||
| Bacillus megaterium, Bacillus subtilis | Not studied | Fusarium graminearum | 50–89% | [15] |
| Bacillus subtilis | Fungicidal or fungistatic metabolites: surfactin, iturin and fengycin lipopeptides | F. graminearum | not studied | [46] |
| Bacillus subtilis | Antifungal lipopeptides, fengycin-like isoforms | F. graminearum | 51% | [50] |
| Bacillus amyloliquefaciens | Production of antibiotics or competition for nutrients | F. graminearum | 16–90% | [51] |
| Azotobacter nigricans | Antifungal activity | F. sporotrichioides, F. poae, F. crookwellense, F. equiseti, F. graminearum, F. sambucinum, F. culmorum | not studied | [52] |
| Bacillus, Pseudomonas, Microbacterium | Promotion of plant growth | F. culmorum | not studied | [53] |
| Bacillus cereus | Volatile antagonistic compounds | F. graminearum | not studied | [54] |
| Pseudomonas fluorescens | Elicit host defense responses | F. culmorum | not studied | [24] |
| Azospirillium brasilense | Elicit host defense responses | root pathogens | not studied | [55] |
| Yeasts | ||||
| Cryptococcus flavescens | Competitive inhibition via aggressive colonization of plant | F. graminearum | 15–18% | [20] |
| Metschnikowia pulcherrima, Hanseniaspora sp., Cyberlindnera sargentensis, Aureobasidium pullulans, Candida subhashii, Pichia kluyveri | Strong competition for micronutrients and/or macronutrients | F. poae, F. langsethiae, F. graminearum, F. culmorum, F. crookwellense, F. oxysporum | not studied | [56] |
| Sphaerodes mycoparasitica | Hyperparasitism | F. graminearum | not studied | [57] |
| Trichoderma harzianum | Antagonistic activity | F. graminearum | not studied | [58] |
| Trichoderma atroviride | Hyperparasitism | F. graminearum | not studied | [45] |
| Clonostachys rosea | Cell wall degrading enzymes: chitinases, glucanases and proteases | F. culmorum | not studied | [59] |
| Trichoderma harzianum | Hyperparasitism elicit host defense responses | F. culmorum | not studied | [60] |
| Clonostachys rosea | F. culmorum | not studied | [23] |
| Microorganisms | Origin of Isolates | Mycotoxins | Efficiency of Biodegradation | Biodegradation Products | Reference |
|---|---|---|---|---|---|
| Bacteria | |||||
| Lactobacillus plantarum and other lactic acid bacteria | Culture collection | DON, T-2 toxin | 28–35% | Physical adsorption | [61] |
| Nocardioides sp. | Soil | DON | 90% | 3-epi-DON | [66] |
| Marmoricola sp. | Wheat spikes | DON | 100% | Not studied | [75] |
| Serratia, Clostridium, Citrobacter, Enterococcus, Stenotrophomonas, Streptomyces | Soil | DON | 100% | DOM-1 | [73] |
| Rhodococcus erythropolis, Rh. coprophilus, Rh. rhodochrous, Rh. globerulus | Soil/Soil contaminated with oil | T-2 toxin | 90% | Not studied | [25] |
| Pseudomonas sp., Blastobacter sp., Arthrobacter sp., Rhizobiaceae | Leaves, soil, water | T-2 toxin | 100% | NEO, T-2 tetraol, T-2 triol | [65] |
| Curtobacterium sp. | No data | T-2 toxin | 100% | T-2 triol | [78] |
| Devosia sp., Nocardioides sp. | Soil, wheat leaves | DON | 100% | 3-epi DON and other unidentified compounds | [74] |
| Bacillus, Anaerofilum, Collinsella, Clostridiales | Chicken intestines | DON | 32–100% | Not studied | [72] |
| Eubacterium sp. | Rumen | DON | DOM-1 | [76] | |
| Yeasts | |||||
| Blastobotrys muscicola, B. robertii, B. peoriensis | Culture collection | T-2 toxin | T-2 toxin 3-β-d-glucoside | [35] | |
| B. capitulata, B. adeninivorans, B. mokoenaii, B. malaysiensis, B. raffinofermentas, Trichomonascus petasosporum, B. indianensis | 48–100% | NEO | |||
| T. petasosporun, T. ciferri, B. indianensis, B. adeninivorans, B. raffinofermentas, B. mokoenaii, B. malaysiensis, B. nivea, B. terestis, B. arbuscula, B. attinorum, B. parvus, B. serpentis, B. illinoisensis | 3-acetyl T-2 toxin | ||||
| Filamentous fungi | |||||
| Alternaria alternata | Culture collection | DON | 91–97% | DON glutathione conjugates | [79] |
| Rhizopus microsporus var. rhizopodiformis | 93–96% | ||||
| Aspergillus oryzae | 84–97% | ||||
| Aspergillus tubingenesis | Soil | DON | 94% | Hydrolysis | [71] |
| Trichoderma harzianum, T. koningii, T. longibranchiatum, T. atroviride, T. asperellum, T. virens | Culture collection | DON | 70–90% | D3G | [80] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wachowska, U.; Packa, D.; Wiwart, M. Microbial Inhibition of Fusarium Pathogens and Biological Modification of Trichothecenes in Cereal Grains. Toxins 2017, 9, 408. https://doi.org/10.3390/toxins9120408
Wachowska U, Packa D, Wiwart M. Microbial Inhibition of Fusarium Pathogens and Biological Modification of Trichothecenes in Cereal Grains. Toxins. 2017; 9(12):408. https://doi.org/10.3390/toxins9120408
Chicago/Turabian StyleWachowska, Urszula, Danuta Packa, and Marian Wiwart. 2017. "Microbial Inhibition of Fusarium Pathogens and Biological Modification of Trichothecenes in Cereal Grains" Toxins 9, no. 12: 408. https://doi.org/10.3390/toxins9120408