Involvement of FvSet1 in Fumonisin B1 Biosynthesis, Vegetative Growth, Fungal Virulence, and Environmental Stress Responses in Fusarium verticillioides
Abstract
:1. Introduction
2. Results
2.1. In Silico Analysis of FvSet1 in F. verticillioides
2.2. Deletion and Complementation of FvSet1 in F. verticillioides
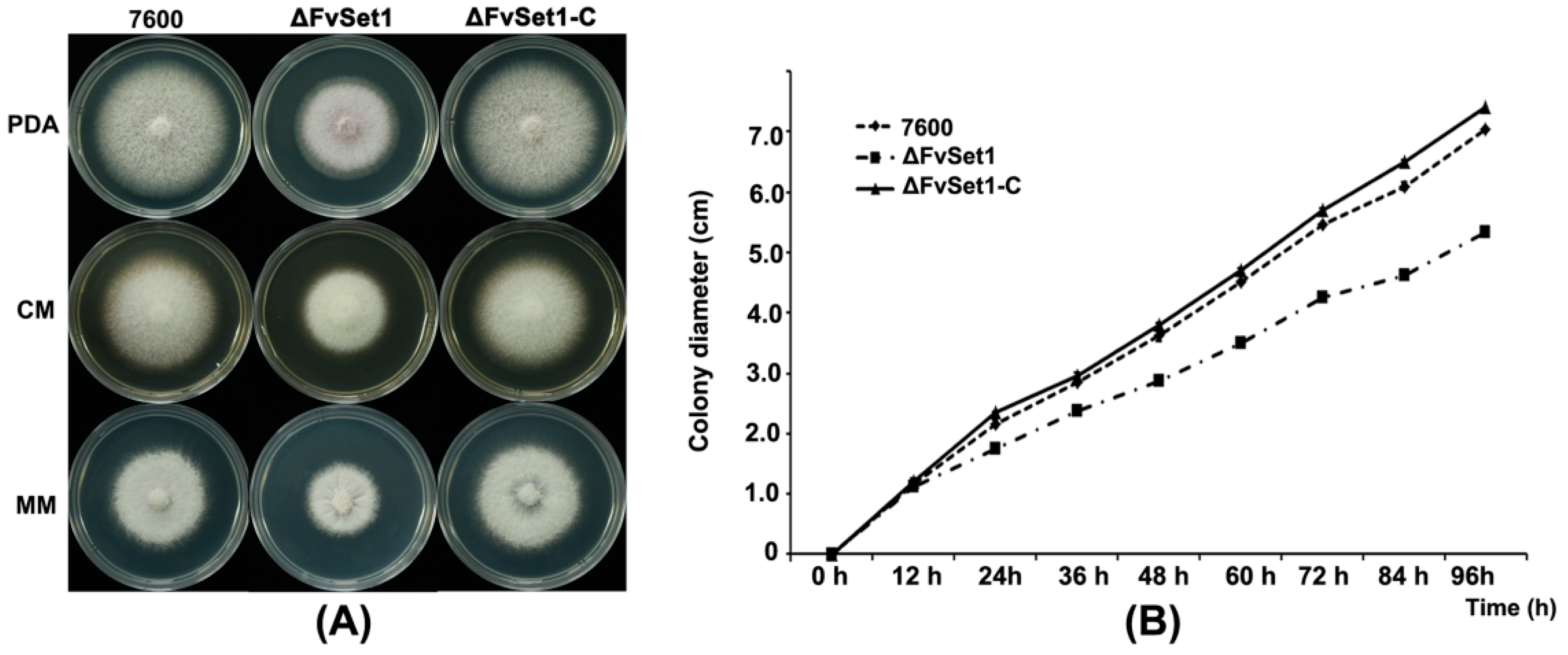
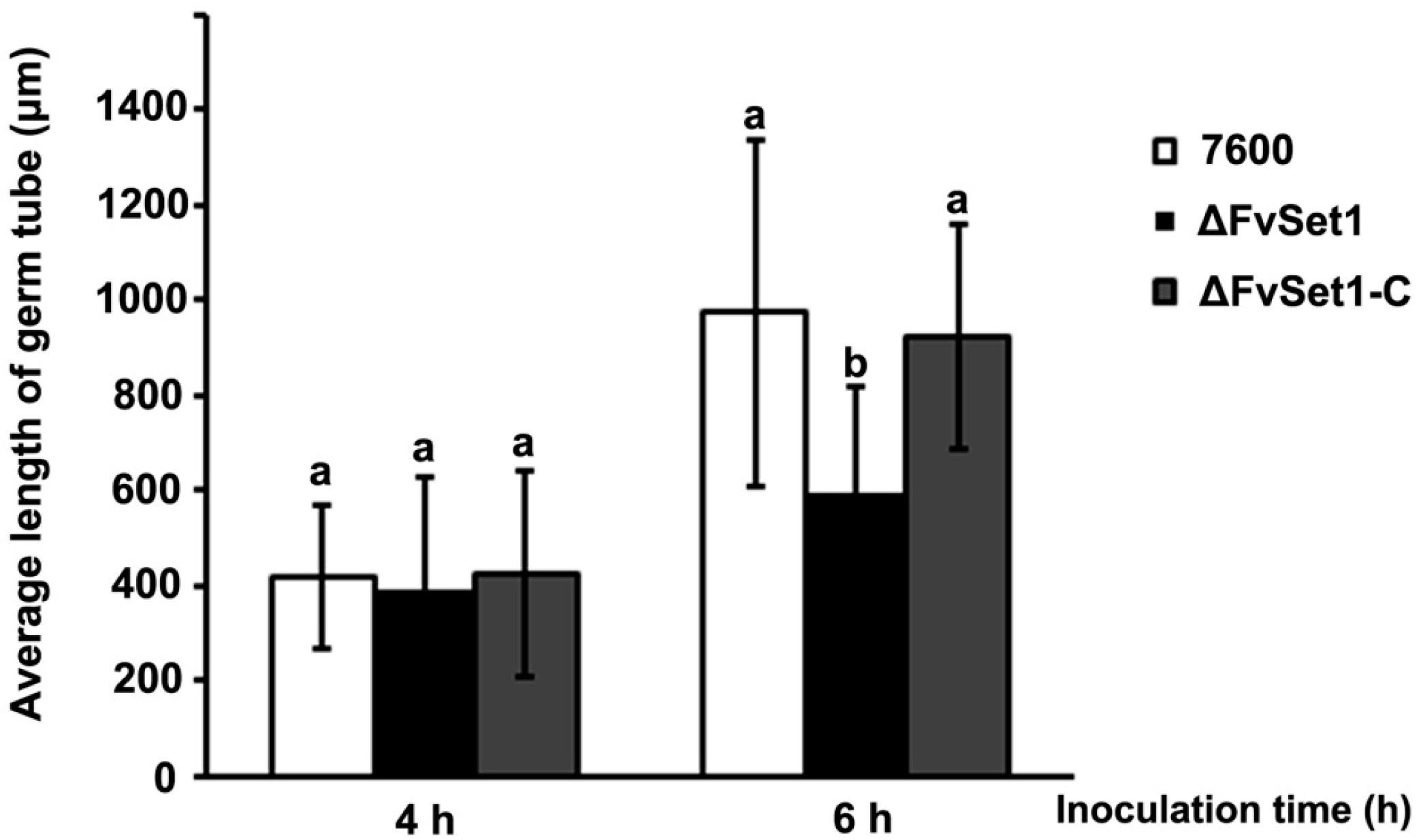
2.3. FvSet1 Is Involved in Fungal Growth and Conidial Germination of F. verticillioides
2.4. FvSet1 Is Indispensable for Full Virulence in F. verticillioides
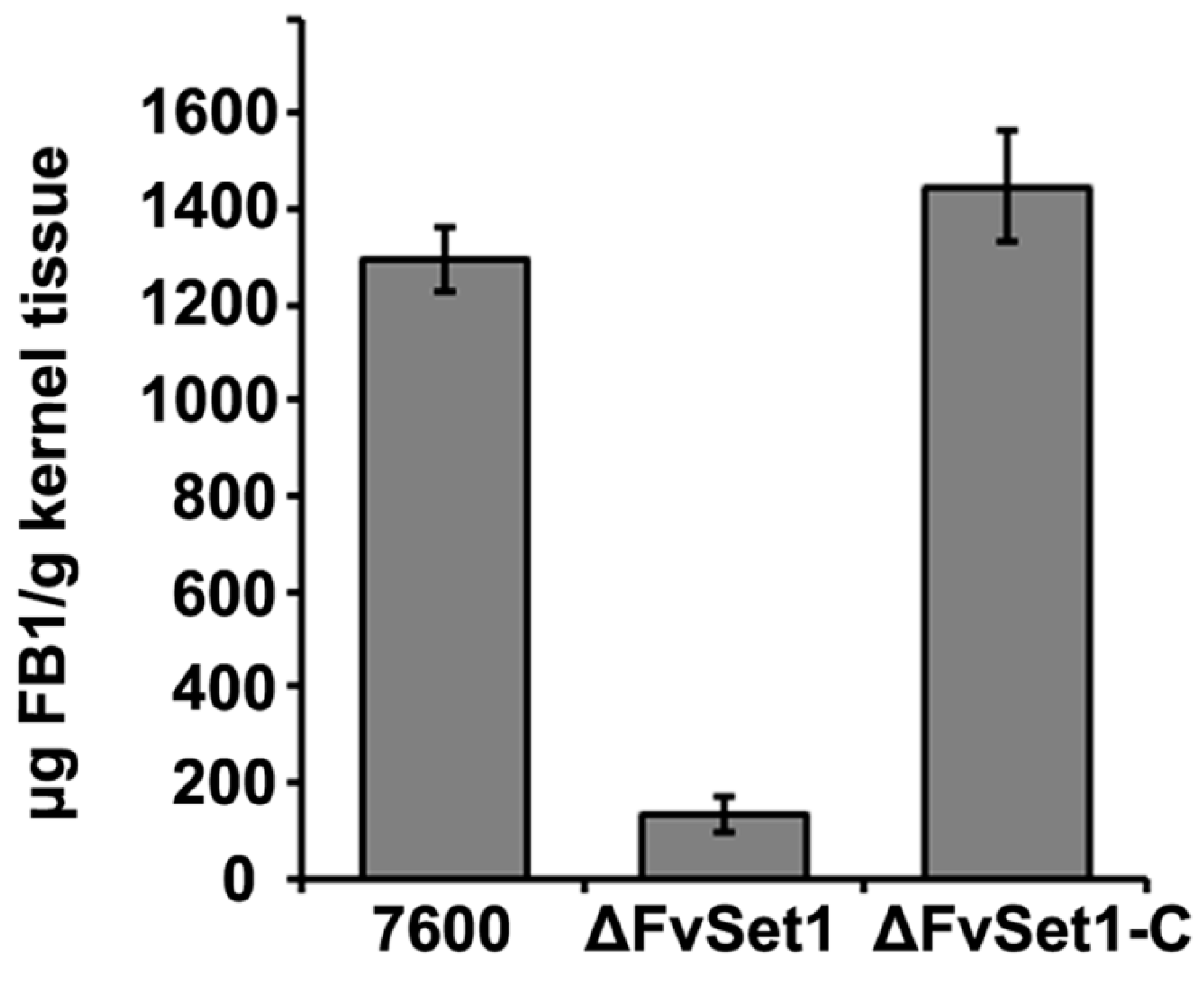
2.5. FvSet1 Is Required for FB1 Biosynthesis in F. verticillioides
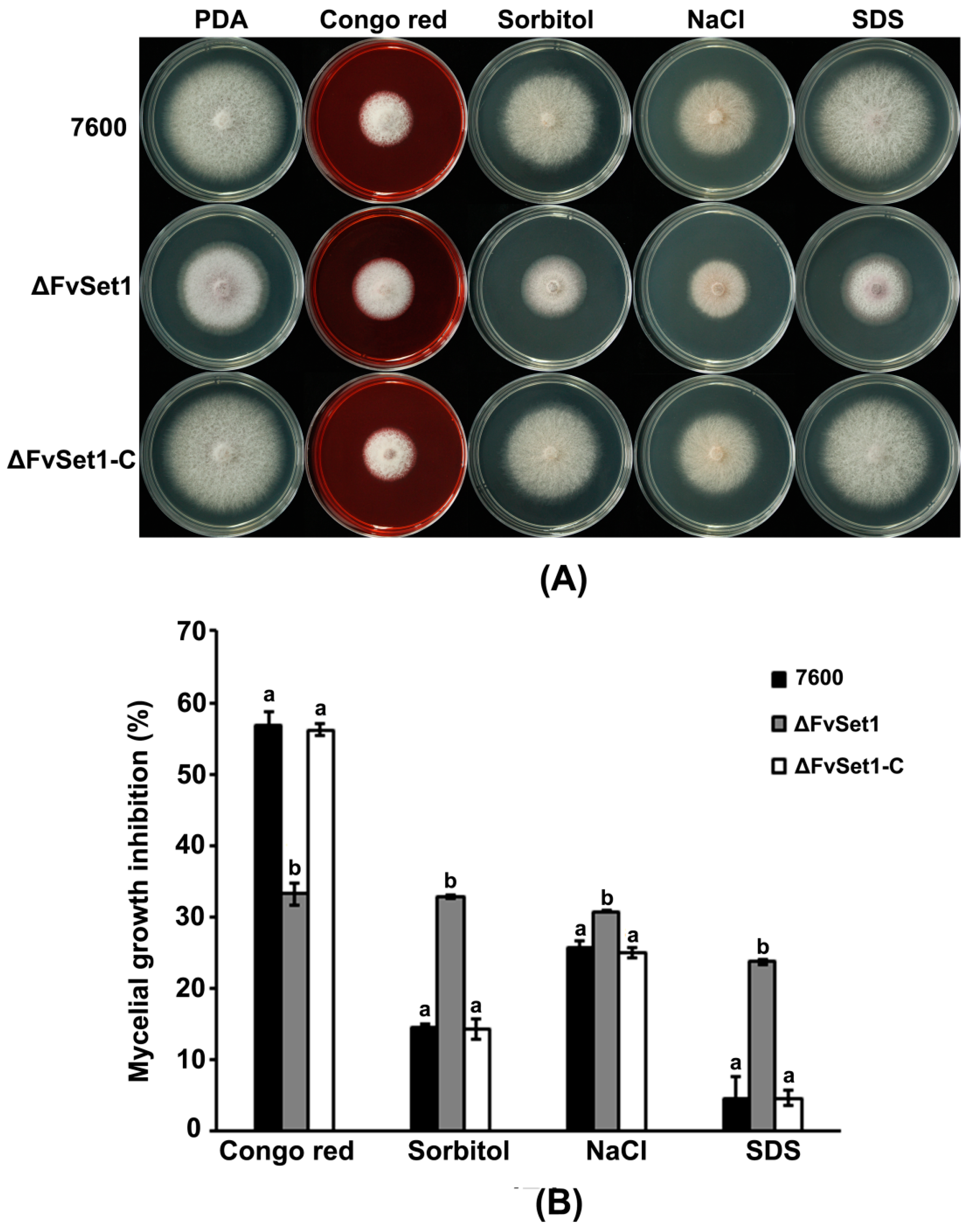
2.6. FvSet1 Is Involved in Responses to Multiple Stresses Response
3. Discussion
4. Materials and Methods
4.1. Fungal Strains, Growth Conditions and Sporulation Tests
4.2. Constructed Gene Deletion and Complemented Strains
4.3. Pathogenicity Assays
4.4. Determination of FB1 Production
4.5. RNA Extraction and Quantitative Real-Time PCR
4.6. Western Blotting Assay
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Munkvold, G.P. Cultural and genetic approaches to managing mycotoxins in maize. Annu. Rev. Phytopathol. 2003, 41, 99–116. [Google Scholar] [CrossRef] [PubMed]
- Nelson, P.E. Taxonomy and biology of Fusarium moniliforme. Mycopathologia 1992, 117, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Howard, P.C.; Eppley, R.M.; Stack, M.E.; Warbritton, A.; Voss, K.A.; Lorentzen, R.J.; Kovach, R.M.; Bucci, T.J. Fumonisin b1 carcinogenicity in a two-year feeding study using F344 rats and B6C3F1 mice. Environ. Health Perspect. 2001, 109, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Park, D.L.; Troxell, T.C. U.S. Perspective on mycotoxin regulatory issues. Adv. Exp. Med. Biol. 2002, 504, 277–285. [Google Scholar] [PubMed]
- Glenn, A.E.; Zitomer, N.C.; Zimeri, A.M.; Williams, L.D.; Riley, R.T.; Proctor, R.H. Transformation-mediated complementation of a FUM gene cluster deletion in Fusarium verticillioides restores both fumonisin production and pathogenicity on maize seedlings. Mol. Plant-Microbe Interact. 2008, 21, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Alexander, N.J.; Proctor, R.H.; McCormick, S.P. Genes, gene clusters, and biosynthesis of trichothecenes and fumonisins in Fusarium. Toxin Rev. 2009, 28, 198–215. [Google Scholar] [CrossRef]
- Yin, S.; Guo, X.; Li, J.; Fan, L.; Hu, H. Fumonisin B1 induces autophagic cell death via activation of ERN1-MAPK8/9/10 pathway in monkey kidney MARC-145 cells. Arch. Toxicol. 2016, 90, 985–996. [Google Scholar] [CrossRef] [PubMed]
- Strahl, B.D.; Allis, C.D. The language of covalent histone modifications. Nature 2000, 403, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.G.; Fire, A. Partitioning the C. elegans genome by nucleosome modification, occupancy, and positioning. Chromosoma 2010, 119, 73–87. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Y.; Bernatavichute, Y.V.; Cokus, S.; Pellegrini, M.; Jacobsen, S.E. Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 2009, 10, R62. [Google Scholar] [CrossRef] [PubMed]
- Peters, A.H.; Kubicek, S.; Mechtler, K.; O’Sullivan, R.J.; Derijck, A.A.; Perez-Burgos, L.; Kohlmaier, A.; Opravil, S.; Tachibana, M.; Shinkai, Y.; et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 2003, 12, 1577–1589. [Google Scholar] [CrossRef]
- Yao, T.T.; Cohen, R.E. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 2002, 419, 403–407. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, N.; Yin, Y.; Chen, Y.; Jiang, J.; Ma, Z. Histone H3K4 methylation regulates hyphal growth, secondary metabolism and multiple stress responses in Fusarium graminearum. Environ. Microbiol. 2015, 17, 4615–4630. [Google Scholar] [CrossRef] [PubMed]
- Pham, K.T.M.; Mo Inoue, Y.; Vu, B.V.; Nguyen, H.H.; Nakayashiki, T.; Ikeda, K.; Nakayashiki, H. MoSet1 (Histone H3K4 methyltransferase in Magnaporthe oryzae) regulates global gene expression during infection-related morphogenesis. PLoS Genet. 2015, 11, e1005385. [Google Scholar]
- Roguev, A.; Schaft, D.; Shevchenko, A.; Pijnappe, W.W.; Wilm, M.; Aasland, R.; Stewart, A.F. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 2001, 20, 7137–7148. [Google Scholar] [CrossRef] [PubMed]
- Krogan, N.J.; Dover, J.; Khorrami, S.; Greenblatt, J.F.; Schneider, J.; Johnston, M.; Shilatifard, A. COMPASS, a histone H3 (lysine 4) methyltransferase required for telomeric silencing of gene expression. J. Biol. Chem. 2002, 277, 10753–10755. [Google Scholar] [CrossRef] [PubMed]
- Schneider, J.; Wood, A.; Lee, J.S.; Schuster, R.; Dueker, J.; Maguire, C.; Swanson, S.K.; Florens, L.; Washburn, M.P.; Shilatifard, A. Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol. Cell 2005, 19, 849–856. [Google Scholar] [CrossRef] [PubMed]
- Nislow, C.; Ray, E.; Pillus, L. SET1, a yeast member of the Trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol. Biol. Cell 1997, 8, 2421–2436. [Google Scholar] [CrossRef] [PubMed]
- Santos-Rosa, H.; Schneider, R.; Bannister, A.J.; Sherriff, J.; Bernstein, B.E.; Emre, N.C.; Schreiber, S.L.; Mellor, J.; Kouzarides, T. Active genes are tri-methylated at K4 of histone H3. Nature 2002, 419, 407–411. [Google Scholar] [CrossRef] [PubMed]
- Ng, H.H.; Robert, F.; Young, R.A.; Struhl, K. Targeted recruitment of set1 histone methylase by elongating pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 2003, 11, 709–719. [Google Scholar] [CrossRef]
- Cheng, H.L.; He, X.Y.; Moore, C. The essential WD repeat protein Swd2 has dual functions in RNA polymerase II transcription termination and lysine 4 methylation of histone H3. Mol. Cell. Biol. 2004, 24, 2932–2943. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.M.; Keller, N.P. Secondary metabolism in fungi: Does chromosomal location matter? Curr. Opin. Microbiol. 2010, 13, 431–436. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.M.; Bok, J.W.; Lee, S.; Dagenais, T.R.; Andes, D.R.; Kontoyiannis, D.P.; Keller, N.P. Loss of CclA, required for histone 3 lysine 4 methylation, decreases growth but increases secondary metabolite production in Aspergillus fumigatus. PeerJ 2013, 1, e4. [Google Scholar] [CrossRef] [PubMed]
- Noma, K.; Grewal, S.I.S. Histone H3 lysine 4 methylation is mediated by Set1 and promotes maintenance of active chromatin states in fission yeast. Proc. Natl. Acad. Sci. USA 2002, 99, 16438–16445. [Google Scholar] [CrossRef] [PubMed]
- Klittich, C.J.R.; Leslie, J.F. Chlorate-Resistant, Nitrate-Utilizing (Crn) Mutants in Fusarium-Moniliforme (Gibberella-Fujikuroi). Phytopathology 1989, 135, 721–727. [Google Scholar] [CrossRef]
- Shim, W.B.; Woloshuk, C.P. Regulation of fumonisin B(1) biosynthesis and conidiation in Fusarium verticillioides by a cyclin-like (C-type) gene, FCC1. Appl. Environ. Microbiol. 2001, 67, 1607–1612. [Google Scholar] [CrossRef] [PubMed]
- Plattner, R.D.; Desjardins, A.E.; Leslie, J.F.; Nelson, P.E. Identification and characterization of strains of Gibberella fujikuroi mating population A with rare fumonisin production phenotypes. Mycologia 1996, 88, 416–424. [Google Scholar] [CrossRef]
- Plattner, R.D.; Weisleder, D.; Poling, S.M. Analytical determination of fumonisins and other metabolites produced by Fusarium moniliforme and related species on corn. Fumonisins Food 1996, 392, 57–64. [Google Scholar]
- Gu, Q.; Zhang, C.; Yu, F.; Yin, Y.; Shim, W.B.; Ma, Z. Protein kinase FgSch9 serves as a mediator of the target of rapamycin and high osmolarity glycerol pathways and regulates multiple stress responses and secondary metabolism in Fusarium graminearum. Environ. Microbiol. 2015, 17, 2661–2676. [Google Scholar] [CrossRef] [PubMed]
- Gu, Q.; Chen, Y.; Liu, Y.; Zhang, C.; Ma, Z. The transmembrane protein FgSho1 regulates fungal development and pathogenicity via the MAPK module Ste50-Ste11-Ste7 in Fusarium graminearum. New Phytol. 2015, 206, 315–328. [Google Scholar] [CrossRef] [PubMed]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, Q.; Tahir, H.A.S.; Zhang, H.; Huang, H.; Ji, T.; Sun, X.; Wu, L.; Wu, H.; Gao, X. Involvement of FvSet1 in Fumonisin B1 Biosynthesis, Vegetative Growth, Fungal Virulence, and Environmental Stress Responses in Fusarium verticillioides. Toxins 2017, 9, 43. https://doi.org/10.3390/toxins9020043
Gu Q, Tahir HAS, Zhang H, Huang H, Ji T, Sun X, Wu L, Wu H, Gao X. Involvement of FvSet1 in Fumonisin B1 Biosynthesis, Vegetative Growth, Fungal Virulence, and Environmental Stress Responses in Fusarium verticillioides. Toxins. 2017; 9(2):43. https://doi.org/10.3390/toxins9020043
Chicago/Turabian StyleGu, Qin, Hafiz Abdul Samad Tahir, Hao Zhang, Hai Huang, Tiantian Ji, Xiao Sun, Liming Wu, Huijun Wu, and Xuewen Gao. 2017. "Involvement of FvSet1 in Fumonisin B1 Biosynthesis, Vegetative Growth, Fungal Virulence, and Environmental Stress Responses in Fusarium verticillioides" Toxins 9, no. 2: 43. https://doi.org/10.3390/toxins9020043




