The Biology of Pichia membranifaciens Killer Toxins
Abstract
:1. Introduction
2. Isolation of Pichia membranifaciens Killer Strains
3. Production of PMKT and PMKT2
4. Homogeneity Purification and Biochemical Characterization of PMKT and PMKT2
5. Killing Mechanism of Action
5.1. Primary Receptors in Sensitive Yeast Cells
5.2. Secondary Receptors for P. membranifaciens Killer Toxins
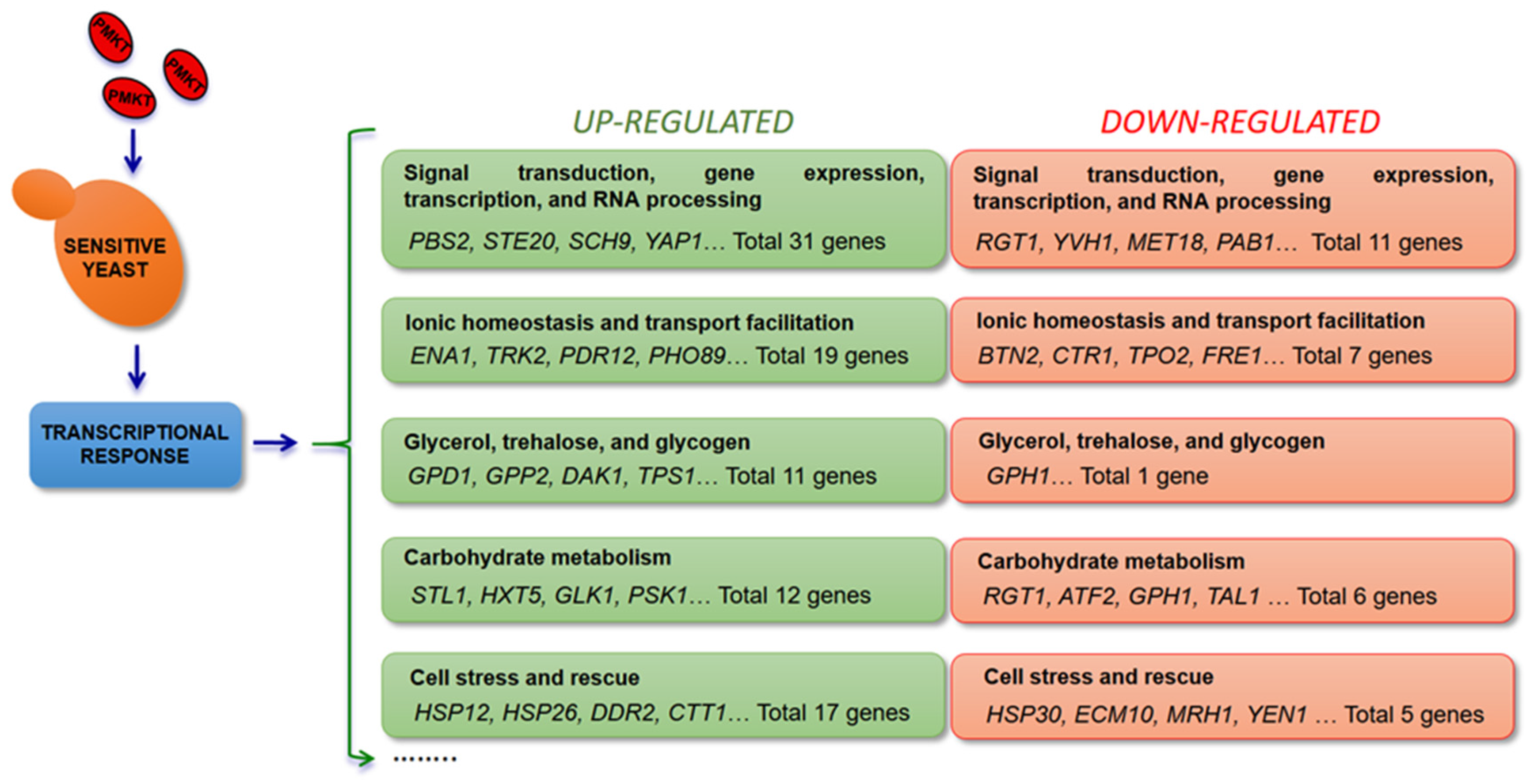
5.3. Cellular Responses to Killer Toxins
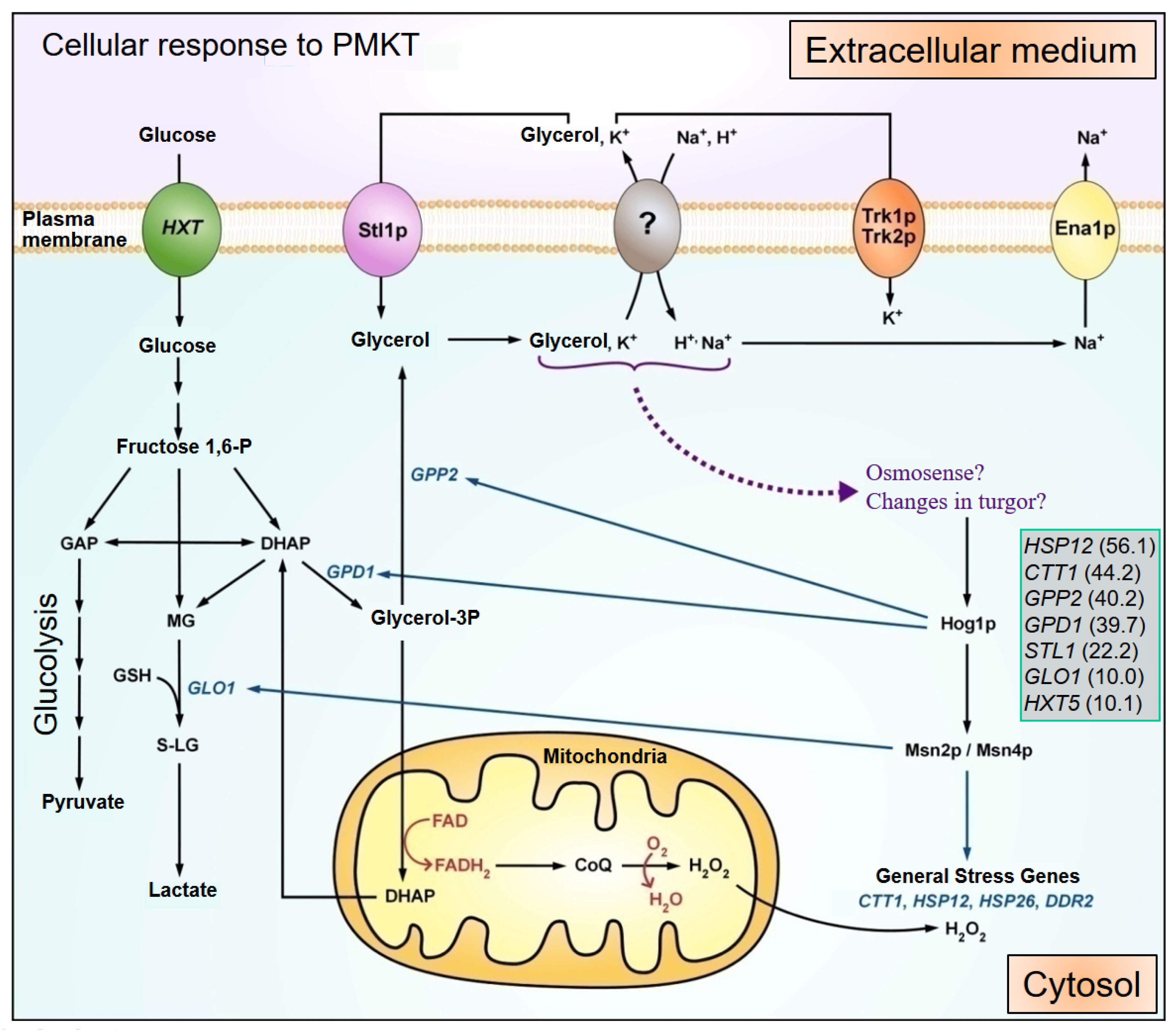
5.3.1. PMKT
5.3.2. PMKT2
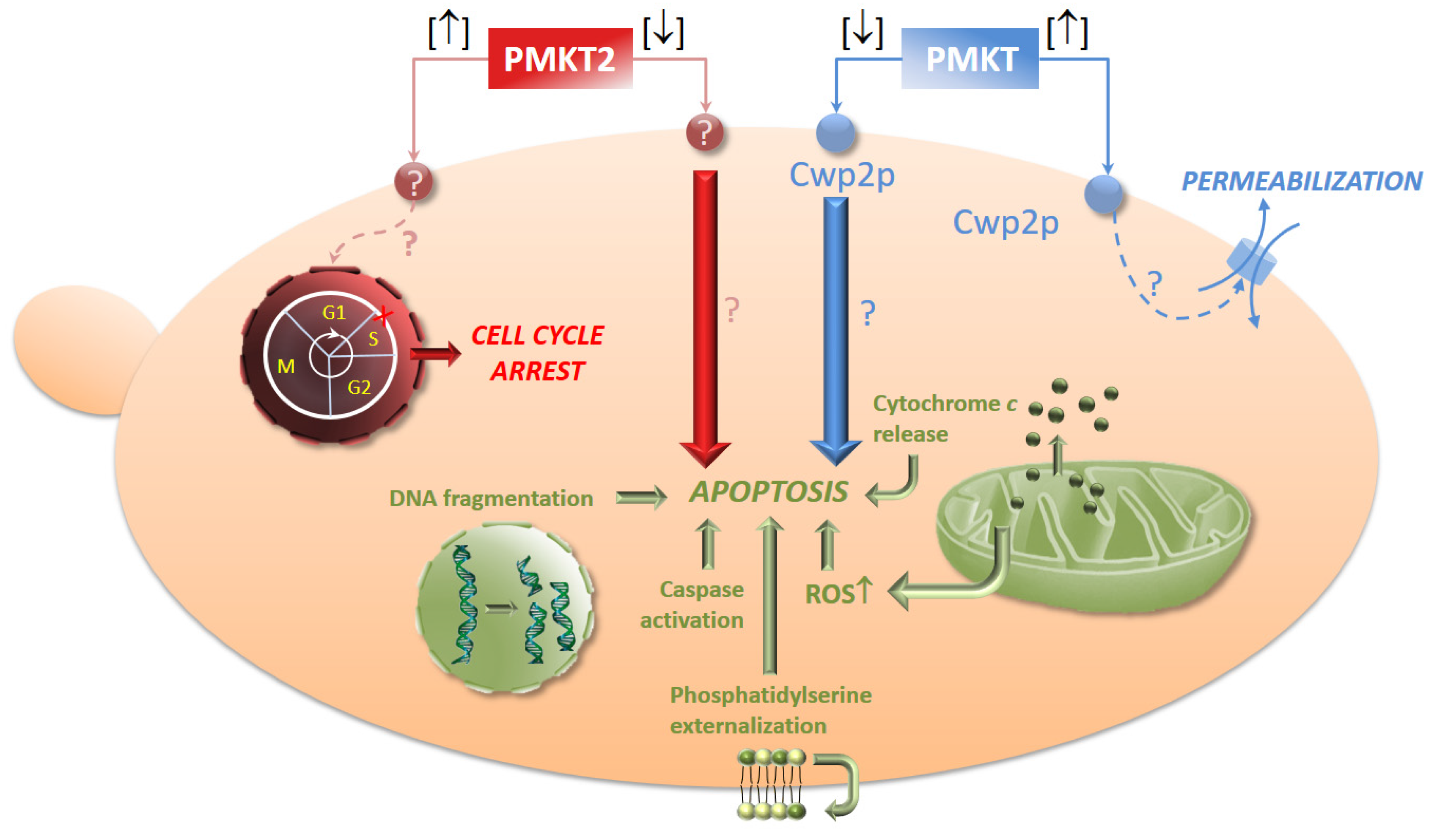
5.4. The Dual Mechanism of Action of PMKT and PMKT2
6. Applications, Conclusions and Future Perspectives of P. membranifaciens Killer Toxins
Acknowledgments
Conflicts of Interest
References
- Bevan, E.; Makower, M. The physiological basis of the killer character in yeast. Proc. Int. Congr. Genet. 1963, 1, 202–203. [Google Scholar]
- Marquina, D.; Santos, A.; Peinado, J. Biology of killer yeasts. Int. Microbiol. 2002, 5, 65–71. [Google Scholar] [CrossRef] [PubMed]
- El-Benna, A.; El-Sahn, M.; Shehata, M. Yeasts producing killer toxins: An overview. Alex J. Food Sci. Technol. 2011, 8, 41–53. [Google Scholar]
- Liu, Y.; Rousseaux, S.; Tourdot-Marechal, R.; Sadoudi, M.; Gougeon, R.; Schmitt-Kopplin, P.; Alexandre, H. Wine microbiome, a dynamic world of microbial interactions. Crit. Rev. Food Sci. Nutr. 2017, 57, 856–873. [Google Scholar] [PubMed]
- Golubev, W. Antagonistic interactions among yeasts. In Biodiversity and Ecophysiology of Yeasts; Péter, G., Rosa, C., Eds.; Springer: Berlin, Germany, 2006; pp. 197–219. [Google Scholar]
- Schmitt, M.J.; Breinig, F. The viral killer system in yeast: From molecular biology to application. FEMS Microbiol. Lett. 2002, 26, 257–276. [Google Scholar] [CrossRef]
- Meinhardt, F.; Klassen, R. Yeast killer toxins: Fundamentals and applications. In Physiology and Genetics; Anke, T., Weber, D., Eds.; Springer: Berlin, Germany, 2009; Volume 15, pp. 107–130. [Google Scholar]
- Labbani, F.-Z.K.; Turchetti, B.; Bennamoun, L.; Dakhmouche, S.; Roberti, R.; Corazzi, L.; Meraihi, Z.; Buzzini, P. A novel killer protein from Pichia kluyveri isolated from an algerian soil: Purification and characterization of its in vitro activity against food and beverage spoilage yeasts. Antonie Leeuwenhoek 2015, 107, 961–970. [Google Scholar] [CrossRef] [PubMed]
- Villalba, M.L.; Sáez, J.S.; del Monaco, S.; Lopes, C.A.; Sangorrín, M.P. TdKT, a new killer toxin produced by Torulaspora delbrueckii effective against wine spoilage yeasts. Int. J. Food Microbiol. 2016, 217, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.; Navascués, E.; Bravo, E.; Marquina, D. Ustilago maydis killer toxin as a new tool for the biocontrol of the wine spoilage yeast Brettanomyces bruxellensis. Int. J. Food Microbiol. 2011, 145, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.L.; Leu, J.Y.; Chang, T.H. A population study of killer viruses reveals different evolutionary histories of two closely related Saccharomyces sensu stricto yeasts. Mol. Ecol. 2015, 24, 4312–4322. [Google Scholar] [CrossRef] [PubMed]
- Breinig, F.; Sendzik, T.; Eisfeld, K.; Schmitt, M.J. Dissecting toxin immunity in virus-infected killer yeast uncovers an intrinsic strategy of self-protection. Proc. Natl. Acad. Sci. USA 2006, 103, 3810–3815. [Google Scholar] [CrossRef] [PubMed]
- Kast, A.; Voges, R.; Schroth, M.; Schaffrath, R.; Klassen, R.; Meinhardt, F. Autoselection of cytoplasmic yeast virus like elements encoding toxin/antitoxin systems involves a nuclear barrier for immunity gene expression. PLoS Genet. 2015, 11, e1005005. [Google Scholar] [CrossRef] [PubMed]
- Wickner, R.B.; Edskes, H.K. Yeast killer elements hold their hosts hostage. PLoS Genet. 2015, 11, e1005139. [Google Scholar] [CrossRef] [PubMed]
- Fuentefria, A.M.; Faganello, J.; Pazzini, F.; Schrank, A.; Valente, P.; Vainstein, M. Typing and patterns of cellular morphological alterations in Cryptococcus neoformans and Cryptococcus gattii isolates exposed to a panel of killer yeasts. Med. Mycol. 2007, 45, 503–512. [Google Scholar] [CrossRef] [PubMed]
- Keszthelyi, A.; Ohkusu, M.; Takeo, K.; Pfeiffer, I.; Litter, J.; Kucsera, J. Characterisation of the anticryptococcal effect of the Fc-1 toxin produced by Filobasidium capsuligenum. Mycoses 2006, 49, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Pérez, M.F.; Contreras, L.; Garnica, N.M.; Fernández-Zenoff, M.V.; Farías, M.E.; Sepulveda, M.; Ramallo, J.; Dib, J.R. Native killer yeasts as biocontrol agents of postharvest fungal diseases in lemons. PLoS ONE 2016, 11, e0165590. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, M.; Velázquez, R.; Maqueda, M.; Zamora, E.; López-Piñeiro, A.; Hernández, L.M. Influence of the dominance of must fermentation by Torulaspora delbrueckii on the malolactic fermentation and organoleptic quality of red table wine. Int. J. Food Microbiol. 2016, 238, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.; Marquina, D. Killer toxin of Pichia membranifaciens and its possible use as a biocontrol agent against grey mould disease of grapevine. Microbiology 2004, 150, 2527–2534. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.; San Mauro, M.; Bravo, E.; Marquina, D. PMKT2, a new killer toxin from Pichia membranifaciens, and its promising biotechnological properties for control of the spoilage yeast Brettanomyces bruxellensis. Microbiology 2009, 155, 624–634. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, M.J.; Breinig, F. Yeast viral killer toxins: Lethality and self-protection. Nat. Rev. Microbiol. 2006, 4, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.; Fell, J.W.; Boekhout, T. The Yeasts: A Taxonomic Study, 5th ed.; Elsevier: Amsterdam, The Netherland, 2011. [Google Scholar]
- Hansen, E.C. Grundlinien zur Systematik der Saccharomyceten. Zentralbl. Bakteriol. Parasitenk. Abt. II 1904, 12, 529–538. [Google Scholar]
- Noronha-da-Costa, P.; Rodrigues, C.; Spencer-Martins, I.; Loureiro, V. Fatty acid patterns of film-forming yeasts and new evidence for the heterogeneity of Pichia membranaefaciens. Lett. Appl. Microbiol. 1996, 23, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Villa-Carvajal, M.; Querol, A.; Belloch, C. Identification of species in the genus Pichia by restriction of the internal transcribed spacers (ITS1 and ITS2) and the 5.8 S ribosomal DNA gene. Antonie Leeuwenhoek 2006, 90, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Deak, T.; Beuchat, L. Handbook of Food Spoilage Yeasts, Crc Series in Contemporary Food Science, 2nd ed.; CRC Press: Boca Raton, FL, USA, 1996. [Google Scholar]
- Boekhout, T.; Robert, V. Yeasts in Food, 1st ed.; Elsevier: Amsterdam, The Netherland, 2003. [Google Scholar]
- Bakır, M.; Cerikcioğlu, N.; Tırtır, A.; Berrak, S.; Özek, E.; Canpolat, C. Pichia anomala fungaemia in immunocompromised children. Mycoses 2004, 47, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Otag, F.; Kuyucu, N.; Erturan, Z.; Sen, S.; Emekdas, G.; Sugita, T. An outbreak of Pichia ohmeri infection in the paediatric intensive care unit: Case reports and review of the literature. Mycoses 2005, 48, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Kast, A.; Klassen, R.; Meinhardt, F. RNA fragmentation induced by a yeast killer toxin. Mol. Microbiol. 2014, 91, 606–617. [Google Scholar] [CrossRef] [PubMed]
- Satwika, D.; Klassen, R.; Meinhardt, F. Anticodon nuclease encoding virus-like elements in yeast. Appl. Microbiol. Biotechnol. 2012, 96, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Wemhoff, S.; Klassen, R.; Meinhardt, F. DNA damage induced by the anticodon nuclease from a Pichia acaciae killer strain is linked to ribonucleotide reductase depletion. Cell. Microbiol. 2016, 18, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Coelho, A.R.; Tachi, M.; Pagnocca, F.C.; Nobrega, G.M.A.; Hoffmann, F.L.; Harada, K.-I.; Hirooka, E.Y. Purification of Candida guilliermondii and Pichia ohmeri killer toxin as an active agent against Penicillium expansum. Food Addit. Contam 2009, 26, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Worsham, P.; Bolen, P. Killer toxin production in Pichia acaciae is associated with linear DNA plasmids. Curr. Genet. 1990, 18, 77–80. [Google Scholar] [CrossRef] [PubMed]
- Bevan, E.; Herring, A.; Mitchell, D.J. Preliminary characterization of two species of dsRNA in yeast and their relationship to the “killer“ character. Nature 1973, 245, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.-B.; Han, Y.-F.; Jong, S.-C.; Chang, S.-C. Isolation, purification, and characterization of a killer protein from Schwanniomyces occidentalis. Appl. Environ. Microbiol. 2000, 66, 5348–5352. [Google Scholar] [CrossRef] [PubMed]
- Middelbeek, E.; Hermans, J.; Stumm, C. Production, purification and properties of a Pichia kluyveri killer toxin. Antonie Leeuwenhoek 1979, 45, 437–450. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, P.; Radler, F. Comparison of the killer toxin of several yeasts and the purification of a toxin of type K2. Arch. Microbiol. 1984, 137, 357–361. [Google Scholar] [CrossRef] [PubMed]
- İzgü, F.; Altınbay, D.; Türeli, A.E. In vitro susceptibilities of Candida spp. To Panomycocin, a novel exo-β-1,3-glucanase isolated from Pichia anomala NCYC 434. Microbiol. Immunol. 2007, 51, 797–803. [Google Scholar] [CrossRef] [PubMed]
- Klassen, R.; Paluszynski, J.P.; Wemhoff, S.; Pfeiffer, A.; Fricke, J.; Meinhardt, F. The primary target of the killer toxin from Pichia acaciae is tRNAgln. Mol. Microbiol. 2008, 69, 681–697. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.; Marquina, D. Ion channel activity by Pichia membranifaciens killer toxin. Yeast 2004, 21, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.; Alonso, A.; Belda, I.; Marquina, D. Cell cycle arrest and apoptosis, two alternative mechanisms for PmKT2 killer activity. Fungal Genet. Biol. 2013, 50, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Marquina, D.; Peres, C.; Caldas, F.; Marques, J.; Peinado, J.; Spencer-Martins, I. Characterization of the yeast population in olive brines. Lett. Appl. Microbiol. 1992, 14, 279–283. [Google Scholar] [CrossRef]
- Llorente, P.; Marquina, D.; Santos, A.; Peinado, J.; Spencer-Martins, I. Effect of salt on the killer phenotype of yeasts from olive brines. Appl. Environ. Microbiol. 1997, 63, 1165–1167. [Google Scholar] [PubMed]
- Da Silva, S.; Calado, S.; Lucas, C.; Aguiar, C. Unusual properties of the halotolerant yeast Candida nodaensis killer toxin, CnKT. Microbiol. Res. 2008, 163, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Klassen, R.; Meinhardt, F. Linear plasmids pWR1A and pWR1B of the yeast Wingea robertsiae are associated with a killer phenotype. Plasmid 2002, 48, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, M.J.; Neuhausen, F. Killer toxin-secreting double-stranded RNA mycoviruses in the yeasts Hanseniaspora uvarum and Zygosaccharomyces bailii. J. Virol. 1994, 68, 1765–1772. [Google Scholar] [PubMed]
- Gunge, N.; Tamaru, A.; Ozawa, F.; Sakaguchi, K. Isolation and characterization of linear deoxyribonucleic acid plasmids from Kluyveromyces lactis and the plasmid-associated killer character. J. Bacteriol. 1981, 145, 382–390. [Google Scholar] [PubMed]
- Comitini, F.; Di Pietro, N.; Zacchi, L.; Mannazzu, I.; Ciani, M. Kluyveromyces phaffii killer toxin active against wine spoilage yeasts: Purification and characterization. Microbiology 2004, 150, 2535–2541. [Google Scholar] [CrossRef] [PubMed]
- Young, T.; Yagiu, M. A comparison of the killer character in different yeasts and its classification. Antonie Leeuwenhoek 1978, 44, 59–77. [Google Scholar] [CrossRef] [PubMed]
- Dignard, D.; Whiteway, M.; Germain, D.; Tessier, D.; Thomas, D.Y. Expression in yeast of a cDNA copy of the K2 killer toxin gene. Mol. Gen. Genet. 1991, 227, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Lukša, J.; Podoliankaitė, M.; Vepštaitė, I.; Strazdaitė-Žielienė, Ž.; Urbonavičius, J.; Servienė, E. Yeast β-1,6-glucan is a primary target for the Saccharomyces cerevisiae K2 toxin. Eukaryot. Cell 2015, 14, 406–414. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, M.J.; Tipper, D.J. K28, a unique double-stranded RNA killer virus of Saccharomyces cerevisiae. Mol. Cell. Biol. 1990, 10, 4807–4815. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Cousiño, N.; Maqueda, M.; Ambrona, J.; Zamora, E.; Esteban, R.; Ramírez, M. A new wine Saccharomyces cerevisiae killer toxin (Klus), encoded by a double-stranded RNA virus, with broad antifungal activity is evolutionarily related to a chromosomal host gene. Appl. Environ. Microbiol. 2011, 77, 1822–1832. [Google Scholar] [CrossRef] [PubMed]
- Goto, K.; Iwatuki, Y.; Kitano, K.; Obata, T.; Hara, S. Cloning and nucleotide sequence of the KHR killer gene of Saccharomyces cerevisiae. Agric. Biol. Chem. 1990, 54, 979–984. [Google Scholar] [CrossRef] [PubMed]
- Park, C.M.; Banerjee, N.; Koltin, Y.; Bruenn, J.A. The Ustilago maydis virally encoded KP1 killer toxin. Mol. Microbiol. 1996, 20, 957–963. [Google Scholar] [CrossRef] [PubMed]
- Park, C.M.; Bruenn, J.A.; Ganesa, C.; Flurkey, W.F.; Bozarth, R.F.; Koltin, Y. Structure and heterologous expression of the Ustilago maydis viral toxin KP4. Mol. Microbiol. 1994, 11, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.; Ginsberg, I.; Banerjee, N.; Held, W.; Koltin, Y.; Bruenn, J. Ustilago maydis KP6 killer toxin: Structure, expression in Saccharomyces cerevisiae, and relationship to other cellular toxins. Mol. Cell. Biol. 1990, 10, 1373–1381. [Google Scholar] [CrossRef] [PubMed]
- Ashida, S.; Shimazaki, T.; Kitano, K.; Hara, S. New killer toxin of Hansenula mrakii. Agric. Biol. Chem. 1983, 47, 2953–2955. [Google Scholar] [CrossRef]
- Kasahara, S.; Inoue, S.B.; Mio, T.; Yamada, T.; Nakajima, T.; Ichishima, E.; Furuichi, Y.; Yamada, H. Involvement of cell wall β-glucan in the action of HM-1 killer toxin. FEBS Lett. 1994, 348, 27–32. [Google Scholar] [CrossRef]
- Komiyama, T.; Furuichi, Y.; Tsukada, Y. Structure and activity of HYI killer toxin from Hansenula saturnus. Biol. Pharm. Bull. 1995, 18, 1057–1059. [Google Scholar] [CrossRef] [PubMed]
- Komiyama, T.; Shirai, T.; Urakami, H.; Furuichi, Y.; Tsukada, Y. Action properties of HYI killer toxin from Williopsis saturnus var. Saturnus, and antibiotics, Aculeacin a and Papulacandin b. Biol. Pharm. Bull. 1998, 21, 1013–1019. [Google Scholar] [CrossRef] [PubMed]
- Radler, F.; Herzberger, S.; Schönig, I.; Schwarz, P. Investigation of a killer strain of Zygosaccharomyces bailii. Microbiology 1993, 139, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, M.; Velázquez, R.; Maqueda, M.; López-Piñeiro, A.; Ribas, J.C. A new wine Torulaspora delbrueckii killer strain with broad antifungal activity and its toxin-encoding double-stranded RNA virus. Front. Microbiol. 2015, 6, 983. [Google Scholar] [CrossRef] [PubMed]
- Izgü, F.; Altinbay, D. Isolation and characterization of the k5-type yeast killer protein and its homology with an exo-β-1,3-glucanase. Biosci. Biotechnol. Biochem. 2004, 68, 685–693. [Google Scholar] [CrossRef] [PubMed]
- Guyard, C.; Seguy, N.; Lange, M.; Ricard, I.; Polonelli, L.; Cailliez, J. First steps in the purification and characterization of a Pichia anomala killer toxin. J. Eukaryot. Microbiol. 1998, 46, 144S. [Google Scholar]
- De Ingeniis, J.; Raffaelli, N.; Ciani, M.; Mannazzu, I. Pichia anomala DBVPG 3003 secretes a ubiquitin-like protein that has antimicrobial activity. Appl. Environ. Microbiol. 2009, 75, 1129–1134. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chi, Z.; Yue, L.; Li, J. Purification and characterization of killer toxin from a marine yeast Pichia anomala YF07B against the pathogenic yeast in crab. Curr. Microbiol. 2007, 55, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Farkas, Z.; Márki-Zay, J.; Kucsera, J.; Vágvölgyi, C.; Golubev, W.; Pfeiffer, I. Characterization of two different toxins of Wickerhamomyces anomalus (Pichia anomala) VKM Y-159. Acta Biol. Hung. 2012, 63, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, C.; Nikkuni, S. The primary and subunit structure of a novel type killer toxin produced by a halotolerant yeast, Pichia farinosa. J. Biol. Chem. 1994, 269, 3041–3046. [Google Scholar] [PubMed]
- Klassen, R.; Meinhardt, F. Structural and functional analysis of the killer element pPin1—3 from Pichia inositovora. Mol. Genet. Genom. 2003, 270, 190–199. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, B.K.; Raina, S.; Singh, S. Killer toxin from a novel killer yeast Pichia kudriavzevii RY55 with idiosyncratic antibacterial activity. J. Basic Microbiol. 2013, 53, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.; del Mar Álvarez, M.; San Mauro, M.; Abrusci, C.; Marquina, D. The transcriptional response of Saccharomyces cerevisiae to Pichia membranifaciens killer toxin. J. Biol. Chem. 2005, 280, 41881–41892. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.; Marquina, D. The transcriptional response of Saccharomyces cerevisiae to proapoptotic concentrations of Pichia membranifaciens killer toxin. Fungal. Genet. Biol. 2011, 48, 979–989. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.; Marquina, D.; Leal, J.; Peinado, J. (1→6)-β-d-glucan as cell wall receptor for Pichia membranifaciens killer toxin. Appl. Environ. Microbiol. 2000, 66, 1809–1813. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.; San Mauro, M.; Abrusci, C.; Marquina, D. Cwp2p, the plasma membrane receptor for Pichia membranifaciens killer toxin. Mol. Microbiol. 2007, 64, 831–843. [Google Scholar] [CrossRef] [PubMed]
- Arroyo-López, F.N.; Medina, E.; Ruiz-Bellido, M.Á.; Romero-Gil, V.; Montes-Borrego, M.; Landa, B.B. Enhancement of the knowledge on fungal communities in directly brined Aloreña de Málaga green olive fermentations by metabarcoding analysis. PLoS ONE 2016, 11, e0163135. [Google Scholar] [CrossRef]
- Leventdurur, S.; Sert-Aydın, S.; Boyaci-Gunduz, C.P.; Agirman, B.; Ben Ghorbal, A.; Francesca, N.; Martorana, A.; Erten, H. Yeast biota of naturally fermented black olives in different brines made from cv. Gemlik grown in various districts of the Cukurova region of Turkey. Yeast 2016, 33, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Arroyo-López, F.; Romero-Gil, V.; Bautista-Gallego, J.; Rodríguez-Gómez, F.; Jiménez-Díaz, R.; García-García, P.; Querol, A.; Garrido-Fernández, A. Yeasts in table olive processing: Desirable or spoilage microorganisms? Int. J. Food Microbiol. 2012, 160, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Vaughn, R.H.; Jakubczyk, T.; MacMillan, J.D.; Higgins, T.E.; Davé, B.A.; Crampton, V.M. Some pink yeasts associated with softening of olives. Appl. Microbiol. 1969, 18, 771–775. [Google Scholar] [PubMed]
- Vaughn, R.H.; Stevenson, K.E.; Davé, B.A.; Park, H.C. Fermenting yeasts associated with softening and gas-pocket formation in olives. Appl. Microbiol. 1972, 23, 316–320. [Google Scholar] [PubMed]
- Arroyo-López, F.; Querol, A.; Bautista-Gallego, J.; Garrido-Fernández, A. Role of yeasts in table olive production. Int. J. Food Microbiol. 2008, 128, 189–196. [Google Scholar] [PubMed]
- Hernández, A.; Martín, A.; Córdoba, M.G.; Benito, M.J.; Aranda, E.; Pérez-Nevado, F. Determination of killer activity in yeasts isolated from the elaboration of seasoned green table olives. Int. J. Food Microbiol. 2008, 121, 178–188. [Google Scholar] [CrossRef] [PubMed]
- Barandica, J.; Santos, A.; Marquina, D.; López, F.; Acosta, F.; Peinado, J. A mathematical model for toxin accumulation by killer yeasts based on the yeast population growth. J. Appl. Microbiol. 1999, 86, 805–811. [Google Scholar] [CrossRef] [PubMed]
- Palfree, R.G.; Bussey, H. Yeast killer toxin: Purification and characterisation of the protein toxin from Saccharomyces cerevisiae. Eur. J. Biochem. 1979, 93, 487–493. [Google Scholar] [CrossRef] [PubMed]
- Woods, D.; Bevan, E. Studies on the nature of the killer factor produced by Saccharomyces cerevisiae. Microbiology 1968, 51, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Hutchins, K.; Bussey, H. Cell wall receptor for yeast killer toxin: Involvement of (1→6)-β-d-glucan. J. Bacteriol. 1983, 154, 161–169. [Google Scholar] [PubMed]
- Brown, J.L.; Kossaczka, Z.; Jiang, B.; Bussey, H. A mutational analysis of killer toxin resistance in Saccharomyces cerevisiae identifies new genes involved in cell wall (1→6)-beta-glucan synthesis. Genetics 1993, 133, 837–849. [Google Scholar] [PubMed]
- Ohta, Y.; Tsukada, Y.; Sugimori, T. Production, purification and characterization of HYI, an antiyeast substance, produced by Hansenula saturnus. Agric. Biol. Chem. 1984, 48, 903–908. [Google Scholar] [CrossRef]
- Lowes, K.; Shearman, C.; Payne, J.; MacKenzie, D.; Archer, D.; Merry, R.; Gasson, M. Prevention of yeast spoilage in feed and food by the yeast mycocin HMK. Appl. Environ. Microbiol. 2000, 66, 1066–1076. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Hiratani, T.; Hirata, H.; Imai, M.; Yamaguchi, H. Killer toxin from Hansenula mrakii selectively inhibits cell wall synthesis in a sensitive yeast. FEBS Lett. 1986, 197, 50–54. [Google Scholar] [CrossRef]
- Yamamoto, T.; Imai, M.; Tachibana, K.; Mayumi, M. Application of monoclonal antibodies to the isolation and characterization of a killer toxin secreted by Hansenula mrakii. FEBS Lett. 1986, 195, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Golubev, W. Killer activity of Tilletiopsis albescens gokhale: Taxonomic and phylogenetic implication. Syst. Appl. Microbiol. Title 1998, 21, 429–432. [Google Scholar] [CrossRef]
- De, S.; Bubnys, A.; Alonzo, F.; Hyun, J.; Lary, J.W.; Cole, J.L.; Torres, V.J.; Olson, R. The relationship between glycan binding and direct membrane interactions in Vibrio cholerae Cytolysin, a channel-forming toxin. J. Biol. Chem. 2015, 290, 28402–28415. [Google Scholar] [CrossRef] [PubMed]
- Al-Aidroos, K.; Bussey, H. Chromosomal mutants of Saccharomyces cerevisiae affecting the cell wall binding site for killer factor. Can. J. Microbiol. 1978, 24, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, M.; Radler, F. Mannoprotein of the yeast cell wall as primary receptor for the killer toxin of Saccharomyces cerevisiae strain 28. Microbiology 1987, 133, 3347–3354. [Google Scholar] [CrossRef] [PubMed]
- Takita, M.A.; Castilho-Valavicius, B. Absence of cell wall chitin in Saccharomyces cerevisiae leads to resistance to Kluyveromyces lactis killer toxin. Yeast 1993, 9, 589–598. [Google Scholar] [CrossRef] [PubMed]
- Breinig, F.; Tipper, D.J.; Schmitt, M.J. Kre1p, the plasma membrane receptor for the yeast K1 viral toxin. Cell 2002, 108, 395–405. [Google Scholar] [CrossRef]
- Novotná, D.; Flegelová, H.; Janderová, B. Different action of killer toxins K1 and K2 on the plasma membrane and the cell wall of Saccharomyces cerevisiae. FEMS Yeast Res. 2004, 4, 803–813. [Google Scholar] [CrossRef] [PubMed]
- Lesage, G.; Bussey, H. Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2006, 70, 317–343. [Google Scholar] [CrossRef] [PubMed]
- Kollár, R.; Petráková, E.; Ashwell, G.; Robbins, P.W.; Cabib, E. Architecture of the yeast cell wall the linkage between chitin and β-(1,3)-glucan. J. Biol. Chem. 1995, 270, 1170–1178. [Google Scholar] [PubMed]
- Radler, F.; Schmitt, M.; Meyer, B. Killer toxin of Hanseniaspora uvarum. Arch. Microbiol. 1990, 154, 175–178. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, M.; Radler, F. Molecular structure of the cell wall receptor for killer toxin KT28 in Saccharomyces cerevisiae. J. Bacteriol. 1988, 170, 2192–2196. [Google Scholar] [CrossRef] [PubMed]
- Weiler, F.; Schmitt, M.J. Zygocin, a secreted antifungal toxin of the yeast Zygosaccharomyces bailii, and its effect on sensitive fungal cells. FEMS Yeast Res. 2003, 3, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Boone, C.; Sommer, S.; Hensel, A.; Bussey, H. Yeast KRE genes provide evidence for a pathway of cell wall-glucan assembly. J. Cell. Biol. 1990, 110, 1833–1843. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, M.J.; Radler, F. Blockage of cell wall receptors for yeast killer toxin KT28 with antimannoprotein antibodies. Antimicrob. Agents Chemother. 1990, 34, 1615–1618. [Google Scholar] [PubMed]
- Bussey, H. K1 killer toxin, a pore-forming protein from yeast. Mol. Microbiol. 1991, 5, 2339–2343. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Bussey, H. Mutational analysis of the functional domains of yeast K1 killer toxin. Mol. Cell. Biol. 1991, 11, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Jones, G.H.; Ballou, C.E. Studies on the structure of yeast mannan. I. Purification and some properties of an α-mannosidase from an arthrobacter species. J. Biol. Chem. 1969, 244, 1043–1051. [Google Scholar] [PubMed]
- Lipke, P.N.; Ovalle, R. Cell wall architecture in yeast: New structure and new challenges. J. Bacteriol. 1998, 180, 3735–3740. [Google Scholar] [PubMed]
- Bussey, H.; Sherman, D.; Somers, J. Action of yeast killer factor: A resistant mutant with sensitive spheroplasts. J. Bacteriol. 1973, 113, 1193–1197. [Google Scholar] [PubMed]
- Breinig, F.; Schleinkofer, K.; Schmitt, M.J. Yeast Kre1p is gpi-anchored and involved in both cell wall assembly and architecture. Microbiology 2004, 150, 3209–3218. [Google Scholar] [CrossRef] [PubMed]
- Becker, B.; Blum, A.; Gießelmann, E.; Dausend, J.; Rammo, D.; Müller, N.C.; Tschacksch, E.; Steimer, M.; Spindler, J.; Becherer, U. H/KDEL receptors mediate host cell intoxication by a viral A/B toxin in yeast. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Zink, S.; Mehlgarten, C.; Kitamoto, H.K.; Nagase, J.; Jablonowski, D.; Dickson, R.C.; Stark, M.J.; Schaffrath, R. Mannosyl-diinositolphospho-ceramide, the major yeast plasma membrane sphingolipid, governs toxicity of Kluyveromyces lactis zymocin. Eukaryot. Cell 2005, 4, 879–889. [Google Scholar] [CrossRef] [PubMed]
- Sawant, A.; Abdelal, A.; Ahearn, D. Purification and characterization of the anti-candida toxin of Pichia anomala WC 65. Antimicrob. Agents Chemother. 1989, 33, 48–52. [Google Scholar] [CrossRef] [PubMed]
- Sawant, A.; Ahearn, D. Involvement of a cell wall receptor in the mode of action of an anti-candida toxin of Pichia anomala. Antimicrob. Agents Chemother. 1990, 34, 1331–1335. [Google Scholar] [CrossRef] [PubMed]
- Diep, D.B.; Nelson, K.L.; Raja, S.M.; Pleshak, E.N.; Buckley, J.T. Glycosylphosphatidylinositol anchors of membrane glycoproteins are binding determinants for the channel-forming toxin aerolysin. J. Biol. Chem. 1998, 273, 2355–2360. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Schmidt, W.G.; Hou, Y.; Williams, A.F.; Jacobson, K. Spontaneous incorporation of the glycosyl-phosphatidylinositol-linked protein Thy-1 into cell membranes. Proc. Natl. Acad. Sci. USA 1992, 89, 5231–5235. [Google Scholar] [CrossRef] [PubMed]
- Hua, M.-X.; Chi, Z.; Liu, G.-L.; Buzdar, M.A.; Chi, Z.-M. Production of a novel and cold-active killer toxin by Mrakia frigida 2E00797 isolated from sea sediment in Antarctica. Extremophiles 2010, 14, 515–521. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-X.; Chi, Z.; Peng, Y.; Wang, X.-H.; Ru, S.-G.; Chi, Z.-M. Purification, characterization and gene cloning of the killer toxin produced by the marine-derived yeast Williopsis saturnus WC91–2. Microbiol. Res. 2012, 167, 558–563. [Google Scholar] [CrossRef] [PubMed]
- De la Pena, P.; Barros, F.; Gascon, S.; Lazo, P.; Ramos, S. Effect of yeast killer toxin on sensitive cells of Saccharomyces cerevisiae. J. Biol. Chem. 1981, 256, 10420–10425. [Google Scholar] [PubMed]
- Martinac, B.; Zhu, H.; Kubalski, A.; Zhou, X.; Culbertson, M.; Bussey, H.; Kung, C. Yeast K1 killer toxin forms ion channels in sensitive yeast spheroplasts and in artificial liposomes. Proc. Natl. Acad. Sci. USA 1990, 87, 6228–6232. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.; Sesti, F.; Ilan, N.; Shih, T.M.; Sturley, S.L.; Goldstein, S.A. A molecular target for viral killer toxin: Tok1 potassium channels. Cell 1999, 99, 283–291. [Google Scholar] [CrossRef]
- Kurzweilová, H.; Sigler, K. Comparison of three different methods for determining yeast killer toxin K1 activity and standardisation of units. Experientia 1995, 51, 26–28. [Google Scholar] [PubMed]
- Kagan, B.L. Mode of action of yeast killer toxins: Channel formation in lipid bilayer membranes. Nature 1983, 302, 709–711. [Google Scholar] [CrossRef] [PubMed]
- Kapteyn, J.; Ter Riet, B.; Vink, E.; Blad, S.; De Nobel, H.; Van Den Ende, H.; Klis, F. Low external pH induces HOG1-dependent changes in the organization of the Saccharomyces cerevisiae cell wall. Mol. Microbiol. 2001, 39, 469–480. [Google Scholar] [CrossRef] [PubMed]
- García-Marqués, S.; Randez-Gil, F.; Prieto, J.A. Nuclear versus cytosolic activity of the yeast Hog1 MAP kinase in response to osmotic and tunicamycin-induced ER stress. FEBS Lett. 2015, 589, 2163–2168. [Google Scholar] [CrossRef] [PubMed]
- Nadal, E.; Posas, F. Osmostress-induced gene expression—A model to understand how stress-activated protein kinases (SAPKs) regulate transcription. FEBS J. 2015, 282, 3275–3285. [Google Scholar] [CrossRef] [PubMed]
- Rep, M.; Krantz, M.; Thevelein, J.M.; Hohmann, S. The transcriptional response of Saccharomyces cerevisiae to osmotic shock. Hot1p and Msn2p/Msn4p are required for the induction of subsets of high osmolarity glycerol pathway-dependent genes. J. Biol. Chem. 2000, 275, 8290–8300. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.A.; Nicholls, S.; Morgan, B.A.; Brown, A.J.; Quinn, J. A conserved stress-activated protein kinase regulates a core stress response in the human pathogen Candida albicans. Mol. Biol. Cell. 2004, 15, 4179–4190. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Monge, R.; Navarro-García, F.; Román, E.; Negredo, A.I.; Eisman, B.; Nombela, C.; Pla, J. The Hog1 mitogen-activated protein kinase is essential in the oxidative stress response and chlamydospore formation in Candida albicans. Eukaryot. Cell 2003, 2, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Monge, R.; Román, E.; Arana, D.M.; Prieto, D.; Urrialde, V.; Nombela, C.; Pla, J. The Sko1 protein represses the yeast-to-hypha transition and regulates the oxidative stress response in Candida albicans. Fungal Genet. Biol. 2010, 47, 587–601. [Google Scholar] [CrossRef] [PubMed]
- Enjalbert, B.; Smith, D.A.; Cornell, M.J.; Alam, I.; Nicholls, S.; Brown, A.J.; Quinn, J. Role of the Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol. Biol. Cell. 2006, 17, 1018–1032. [Google Scholar] [CrossRef] [PubMed]
- Pagé, N.; Gérard-Vincent, M.; Ménard, P.; Beaulieu, M.; Azuma, M.; Dijkgraaf, G.J.; Li, H.; Marcoux, J.; Nguyen, T.; Dowse, T. A Saccharomyces cerevisiae genome-wide mutant screen for altered sensitivity to K1 killer toxin. Genetics 2003, 163, 875–894. [Google Scholar] [PubMed]
- Schmitt, M.J.; Reiter, J. Viral induced yeast apoptosis. Biochim. Biophys. Acta Mol. Cell. Res. 2008, 1783, 1413–1417. [Google Scholar] [CrossRef] [PubMed]
- Somers, J.; Bevan, E. The inheritance of the killer character in yeast. Genet. Res. 1969, 13, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Cailliez, J.; Séguy, N.; Aliouat, E.; Polonelli, L.; Camus, D.; Del-Cas, E. The yeast killer phenomenon: A hypothetical way to control Pneumocystis carinii pneumonia. Med. Hypotheses 1994, 43, 167–171. [Google Scholar] [CrossRef]
- Polonelli, L.; Conti, S.; Gerloni, M.; Magliani, W.; Castagnola, M.; Morace, G.; Chezzi, C. Antibiobodies‘: Antibiotic-like anti-idiotypic antibodies. J. Med. Vet. Mycol. 1991, 29, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Séguy, N.; Cailliez, J.-C.; Polonelli, L.; Dei-Cas, E.; Camus, D. Inhibitory effect of a Pichia anomala killer toxin on Pneumocystis carinii infectivity to the scid mouse. Parasitol. Res. 1996, 82, 114–116. [Google Scholar] [CrossRef] [PubMed]
- Javadekar, V.; SivaRaman, H.; Gokhale, D. Industrial yeast strain improvement: Construction of a highly flocculent yeast with a killer character by protoplast fusion. J. Ind. Microbiol. 1995, 15, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Palpacelli, V.; Ciani, M.; Rosini, G. Activity of different ‘killer‘yeasts on strains of yeast species undesirable in the food industry. FEMS Microbiol. Lett. 1991, 84, 75–78. [Google Scholar] [CrossRef]
- Petering, J.; Symons, M.; Langridge, P.; Henschke, P. Determination of killer yeast activity in fermenting grape juice by using a marked Saccharomyces wine yeast strain. Appl. Environ. Microbiol. 1991, 57, 3232–3236. [Google Scholar] [PubMed]
- Seki, T.; Choi, E.-H.; Ryu, D. Construction of killer wine yeast strain. Appl. Environ. Microbiol. 1985, 49, 1211–1215. [Google Scholar] [PubMed]
- Kitamoto, H.; Hasebe, A.; Ohmomo, S.; Suto, E.; Muraki, M.; Iimura, Y. Prevention of aerobic spoilage of maize silage by a genetically modified killer yeast, Kluyveromyces lactis, defective in the ability to grow on lactic acid. Appl. Environ. Microbiol. 1999, 65, 4697–4700. [Google Scholar] [PubMed]
- Kitamoto, H.K.; Ohmomo, S.; Nakahara, T. Selection of killer yeasts (Kluyveromyces lactis) to prevent aerobic deterioration in silage making. J. Dairy res. 1993, 76, 803–811. [Google Scholar] [CrossRef]
- Walker, G.M.; Mcleod, A.H.; Hodgson, V.J. Interactions between killer yeasts and pathogenic fungi. FEMS Microbiol. Lett. 1995, 127, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Uchida, K.; Hiratani, T.; Miyazaki, T.; Yagiu, J.; Yamaguchi, H. In vitro activity of the killer toxin from yeast Hansenula mrakii against yeasts and molds. J. Antibiot. 1988, 41, 398–403. [Google Scholar] [PubMed]
- Magliani, W.; Conti, S.; Gerloni, M.; Bertolotti, D.; Polonelli, L. Yeast killer systems. Clin. Microbiol. Rev. 1997, 10, 369–400. [Google Scholar] [PubMed]
- Magliani, W.; Conti, S.; Salati, A.; Vaccari, S.; Ravanetti, L.; Maffei, D.L.; Polonelli, L. Therapeutic potential of yeast killer toxin-like antibodies and mimotopes. FEMS Yeast Res. 2004, 5, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Theisen, S.; Molkenau, E.; Schmitt, M.J. Wicaltin, a new protein toxin secreted by the yeast Williopsis californica and its broad-spectrum antimycotic potential. J. Microbiol. Biotechnol. Res. 2000, 10, 547–550. [Google Scholar]
- Buzzini, P.; Corazzi, L.; Turchetti, B.; Buratta, M.; Martini, A. Characterization of the in vitro antimycotic activity of a novel killer protein from Williopsis saturnus DBVPG 4561 against emerging pathogenic yeasts. FEMS Microbiol. Lett. 2004, 238, 359–365. [Google Scholar] [PubMed]
- Masih, E.; Slezack-Deschaumes, S.; Marmaras, I.; Barka, E.A.; Vernet, G.; Charpentier, C.; Adholeya, A.; Paul, B. Characterisation of the yeast Pichia membranifaciens and its possible use in the biological control of Botrytis cinerea, causing the grey mould disease of grapevine. FEMS Microbiol. Lett. 2001, 202, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Masih, E.I.; Paul, B. Secretion of β-1,3-glucanases by the yeast Pichia membranifaciens and its possible role in the biocontrol of Botrytis cinerea causing grey mold disease of the grapevine. Curr. Microbiol. 2002, 44, 391–395. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.; Sanchez, A.; Marquina, D. Yeasts as biological agents to control Botrytis cinerea. Microbiol. Res. 2004, 159, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Lutz, M.C.; Lopes, C.A.; Rodriguez, M.E.; Sosa, M.C.; Sangorrín, M.P. Efficacy and putative mode of action of native and commercial antagonistic yeasts against postharvest pathogens of pear. Int J. Food Microbiol. 2013, 164, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Parafati, L.; Cirvilleri, G.; Restuccia, C.; Wisniewski, M. Potential role of exoglucanase genes (WaEXG1 and WaEXG2) in the biocontrol activity of Wickerhamomyces anomalus. Microb. Ecol. 2016, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hua, S.S.T.; Beck, J.J.; Sarreal, S.B.L.; Gee, W. The major volatile compound 2-phenylethanol from the biocontrol yeast, Pichia anomala, inhibits growth and expression of aflatoxin biosynthetic genes of Aspergillus flavus. Mycotoxin Res. 2014, 30, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.; Enkegaard, A.; Osborne, L.S.; Ramakers, P.M.; Messelink, G.J.; Pijnakker, J.; Murphy, G. The banker plant method in biological control. Crit. Rev. Plant Sci. 2011, 30, 259–278. [Google Scholar] [CrossRef]
- Huang, X.; Zhang, N.; Yong, X.; Yang, X.; Shen, Q. Biocontrol of Rhizoctonia solani damping-off disease in cucumber with Bacillus pumilus SQR-N43. Microbiol. Res. 2012, 167, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Druvefors, U.Ä.; Schnürer, J. Mold-inhibitory activity of different yeast species during airtight storage of wheat grain. FEMS Yeast Res. 2005, 5, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Lemos Junior, W.J.F.; Bovo, B.; Nadai, C.; Crosato, G.; Carlot, M.; Favaron, F.; Giacomini, A.; Corich, V. Biocontrol ability and action mechanism of Starmerella bacillaris (synonym Candida zemplinina) isolated from wine musts against gray mold disease agent Botrytis cinerea on grape and their effects on alcoholic fermentation. Front. Microbiol. 2016, 7, 1249. [Google Scholar] [CrossRef] [PubMed]
- Nally, M.; Pesce, V.; Maturano, Y.; Assaf, L.R.; Toro, M.; de Figueroa, L.C.; Vazquez, F. Antifungal modes of action of Saccharomyces and other biocontrol yeasts against fungi isolated from sour and grey rots. Int. J. Food Microbiol. 2015, 204, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Yuan, Y.; Hu, Z.; Zheng, Y. Combination of Pichia membranifaciens and ammonium molybdate for controlling blue mould caused by Penicillium expansum in peach fruit. Int. J. Food Microbiol. 2010, 141, 173–176. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Zheng, Y.; Tang, S.; Wang, K. Improved control of anthracnose rot in loquat fruit by a combination treatment of Pichia membranifaciens with CaCl2. Int. J. Food Microbiol. 2008, 126, 216–220. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Deng, L.; Zhou, Y.; Wang, W.; Yao, S.; Zeng, K. Optimization of a protective medium for freeze-dried Pichia membranifaciens and application of this biocontrol agent on citrus fruit. J. Appl. Microbiol. 2016, 121, 234–243. [Google Scholar] [CrossRef] [PubMed]
- Carroll, S.Y.; Stirling, P.C.; Stimpson, H.E.; Gießelmann, E.; Schmitt, M.J.; Drubin, D.G. A yeast killer toxin screen provides insights into A/B toxin entry, trafficking, and killing mechanisms. Dev. Cell 2009, 17, 552–560. [Google Scholar] [PubMed]
- Buzzini, P.; Martini, A. Utilisation of differential killer toxin sensitivity patterns for fingerprinting and clustering yeast strains belonging to different genera. Syst. Appl. Microbiol. 2000, 23, 450–457. [Google Scholar] [CrossRef]
- Corte, L.; Lattanzi, M.; Buzzini, P.; Bolano, A.; Fatichenti, F.; Cardinali, G. Use of RAPD and killer toxin sensitivity in Saccharomyces cerevisiae strain typing. J. Appl. Microbiol. 2005, 99, 609–617. [Google Scholar] [CrossRef] [PubMed]
- Boekhout, T.; Van Belkum, A.; Leenders, A.C.; Verbrugh, H.A.; Mukamurangwa, P.; Swinne, D.; Scheffers, W.A. Molecular typing of Cryptococcus neoformans: Taxonomic and epidemiological aspects. Int. J. Syst. Evol. Microbiol. 1997, 47, 432–442. [Google Scholar] [CrossRef] [PubMed]
- Lopes, C.A.; Lavalle, T.L.; Querol, A.; Caballero, A.C. Combined use of killer biotype and mtDNA-RFLP patterns in a patagonian wine Saccharomyces cerevisiae diversity study. Antonie Leeuwenhoek 2006, 89, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Vaughan-Martini, A.; Cardinali, G.; Martini, A. Differential killer sensitivity as a tool for fingerprinting wine-yeast strains of Saccharomyces cerevisiae. J. Ind. Microbiol. Biotechnol. 1996, 17, 124–127. [Google Scholar]
- Provost, F.; Polonelli, L.; Conti, S.; Fisicaro, P.; Gerloni, M.; Boiron, P. Use of yeast killer system to identify species of the Nocardia asteroides complex. J. Clin. Microbiol. 1995, 33, 8–10. [Google Scholar] [PubMed]
- Polonelli, L.; Conti, S. Biotyping of Candida albicans and other fungi by yeast killer toxins sensitivity. Candida Albicans Methods Mol. Biol. 2009, 97–115. [Google Scholar]
- Séguy, N.; Polonelli, L.; Dei-Cas, E.; Cailliez, J.-C. Effect of a killer toxin of Pichia anomala to pneumocystis. Perspectives in the control of pneumocystosis. FEMS Immunol. Med. Microbiol. 1998, 22, 145–149. [Google Scholar] [CrossRef]
- Fuentefria, A.M.; Franskoviaki, I.M.; Mercado, L.W.; Ramos, J.P.; Vainstein, M.H.; Valente, P. Inhibition of clinical and environmental Cryptococcus neoformans isolates by two brazilian killer yeasts. J. Basic Microbiol. 2006, 46, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Keszthelyi, A. Characterisation of anticryptococcal Fc-1 toxin of Filobasidium capsuligenum. Acta Biol. Szeged. 2006, 50, 155. [Google Scholar]
- Cenci, E.; Mencacci, A.; Spreca, A.; Montagnoli, C.; Bacci, A.; Perruccio, K.; Velardi, A.; Magliani, W.; Conti, S.; Polonelli, L. Protection of killer antiidiotypic antibodies against early invasive aspergillosis in a murine model of allogeneic T-cell-depleted bone marrow transplantation. Infect. Immun. 2002, 70, 2375–2382. [Google Scholar] [CrossRef] [PubMed]
- Guyard, C.; Séguy, N.; Cailliez, J.-C.; Drobecq, H.; Polonelli, L.; Dei-Cas, E.; Mercenier, A.; Menozzi, F.D. Characterization of a Williopsis saturnus var. mrakii high molecular weight secreted killer toxin with broad-spectrum antimicrobial activity. J. Antimicrob. Chemother. 2002, 49, 961–971. [Google Scholar] [CrossRef] [PubMed]
- Buzdar, M.A.; Chi, Z.; Wang, Q.; Hua, M.-X.; Chi, Z.-M. Production, purification, and characterization of a novel killer toxin from Kluyveromyces siamensis against a pathogenic yeast in crab. Appl. Microbiol. Biotechnol. 2011, 91, 1571–1579. [Google Scholar] [CrossRef] [PubMed]
- Pérez, M.A.; Gallego, F.J.; Hidalgo, P. Evaluation of molecular techniques for the genetic characterization of Saccharomyces cerevisiae strains. FEMS Microbiol. Lett. 2001, 205, 375–378. [Google Scholar] [PubMed]
- Velázquez, R.; Zamora, E.; Álvarez, M.; Álvarez, M.L.; Ramírez, M. Using mixed inocula of Saccharomyces cerevisiae killer strains to improve the quality of traditional sparkling-wine. Food Microbiol. 2016, 59, 150–160. [Google Scholar] [CrossRef] [PubMed]
- Ciani, M.; Fatichenti, F. Killer toxin of Kluyveromyces phaffii DBVPG 6076 as a biopreservative agent to control apiculate wine yeasts. Appl. Environ. Microbiol. 2001, 67, 3058–3063. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.-Q.; Tsao, M. Inhibition of spoilage yeasts in cheese by killer yeast Williopsis saturnus var. saturnus . Int. J. Food Microbiol. 2009, 131, 280–282. [Google Scholar] [CrossRef] [PubMed]
- Oro, L.; Ciani, M.; Bizzaro, D.; Comitini, F. Evaluation of damage induced by KwKT and PiKT zymocins against Brettanomyces/Dekkera spoilage yeast, as compared to sulphur dioxide. J. Appl. Microbiol. 2016, 121, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Mehlomakulu, N.; Prior, K.; Setati, M.; Divol, B. Candida pyralidae killer toxin disrupts the cell wall of Brettanomyces bruxellensis in red grape juice. J. Appl. Microbiol. 2016, 121, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Alonso, A.; Belda, I.; Santos, A.; Navascués, E.; Marquina, D. Advances in the control of the spoilage caused by Zygosaccharomyces species on sweet wines and concentrated grape musts. Food Control 2015, 51, 129–134. [Google Scholar] [CrossRef]
- Todd, B.E.; Fleet, G.H.; Henschke, P.A. Promotion of autolysis through the interaction of killer and sensitive yeasts: Potential application in sparkling wine production. Am. J. Enol. Vitic. 2000, 51, 65–72. [Google Scholar]
- Satora, P.; Tarko, T.; Sroka, P.; Blaszczyk, U. The influence of Wickerhamomyces anomalus killer yeast on the fermentation and chemical composition of apple wines. FEMS Yeast Res. 2014, 14, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Yap, N.; de Barros Lopes, M.; Langridge, P.; Henschke, P. The incidence of killer activity of non-Saccharomyces yeasts towards indigenous yeast species of grape must: Potential application in wine fermentation. J. Appl. Microbiol. 2000, 89, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Oro, L.; Ciani, M.; Comitini, F. Antimicrobial activity of Metschnikowia pulcherrima on wine yeasts. J. Appl. Microbiol. 2014, 116, 1209–1217. [Google Scholar] [CrossRef] [PubMed]
- Steensels, J.; Snoek, T.; Meersman, E.; Nicolino, M.P.; Voordeckers, K.; Verstrepen, K.J. Improving industrial yeast strains: Exploiting natural and artificial diversity. FEMS Microbiol. Rev. 2014, 38, 947–995. [Google Scholar] [CrossRef] [PubMed]
- Polonelli, L.; Magliani, W.; Ciociola, T.; Giovati, L.; Conti, S. From Pichia anomala killer toxin through killer antibodies to killer peptides for a comprehensive anti-infective strategy. Antonie. Leeuwenhoek 2011, 99, 35–41. [Google Scholar] [CrossRef] [PubMed]


| Killer Yeast | Strain (Killer Toxin) | Molecular Mass (kDa) | Glycoprotein Nature | Genetic Basis | Primary Receptors | Mode of Action | References |
|---|---|---|---|---|---|---|---|
| Candida nodaensis | PYCC 3198 (CnKT) | - | No | Chromosome | - | - | [45] |
| Debaryomyces robertsiae | CBS6693 (-) | >100.0 | dsDNA (pWR1A pWR1B) | Chitin | - | [46] | |
| Hanseniaspora uvarum | 470 (-) | 18.0 | No | dsRNA | (1→6)-β-d-glucans | - | [47] |
| Kluyveromyces lactis | IFO1267 (Zymocin) | α (97.0); β (31.0); γ (28.0) | Yes (α), No (β, γ) | dsDNA (pGKL1) | Chitin <β2> | rRNA fragmentation | [48] |
| Kluyveromyces phaffii | DBVPG 6076 (KpKt) | 33.0 | Yes | Chromosome | (1→6)-β-d-glucans; (1→3)-β-d-glucans | Glucanase activity | [49] |
| Saccharomyces cerevisiae | KL88 (K1) | α (9,5); β (9.0) | No | M1-dsRNA | (1→6)-β-d-glucans | Ionic channels formation in plasma membrane | [50] |
| - (K2) | α (21.0); β (5.0) | Yes | M2-dsRNA | (1→6)-β-d-glucans | Increase in plasma membrane permeability | [51,52] | |
| CBS8112 (K28) | α (10,5); β (11.0) | Yes | M28-dsRNA | Mannoproteins | Toxin entry by endocytosis and cell cycle arresting in S phase | [53] | |
| - (Klus) | α (?), β (?) | Yes | Mlus-dsRNA | - | - | [54] | |
| 111 (KHR) | 20.0 | - | Chromosome | - | - | [55] | |
| - (KHS) | 75.0 | - | - | - | Increase of membrane permeability to ions | [3] | |
| Schwanniomyces occidentalis | ATCC 44252 (-) | α (7.4); β (4.9) | No | Chromosome | Mannoproteins | Plasma membrane damage | [36] |
| Ustilago maydis | P1 (KP1) | (19.0) α, β | - | dsRNA (P1) | - | Increase of membrane permeability to ions | [56] |
| P2 (KP2) | 11.1 | - | - | - | Increase of membrane permeability to ions | [3] | |
| P4 (KP4) | 13.6 | - | dsRNA (P4) | - | Inhibition of Ca+ channels | [57] | |
| P6 (KP6) | α (8.6); β (9.1) | - | dsRNA (P6) | - | K+ depletion | [58] | |
| Williopsis saturnus var. mrakii | IFO 0895 (HM-1) | 10.7 | No | Chromosome | (1→6)-β-d-glucans; (1→3)-β-d-glucans | Inhibition of b-1,3-glucan synthase | [59,60] |
| Williopsis saturnus var. saturnus | IFO 0117 (HYI) | 9.5 | No | Chromosome | - | (1→3)-b-D-glucans sintase inhibition | [61,62] |
| Zygosaccharomyces bailii | 412 (KT412) | 10.0 | No | dsRNA | Mannoproteins | Plasma membrane damage | [63] |
| Torulaspora delbrueckii | Kbarr (Kbarr-1) | α (?), β (?), γ (?) | Yes | dsRNA | - | - | [64] |
| NPCC 1033 (TdKT) | >30.0 | - | - | (1→3)-β-d-glucans | Cell wall disruption and apoptotic death processes | [9] |
| Killer Yeast | Strain/s (Killer Toxin) | Molecular Mass (kDa) | Glycoprotein Nature | Genetic Basis | Primary Receptors | Mode of Action | Reference |
|---|---|---|---|---|---|---|---|
| P. acaciae | NRRLY-18665 (PaT) | α (110), β (39), γ (38) | Yes | dsDNA (pPac1-2) | Chitin | Cell cycle arrest in G1, chitinase activity | [30] |
| P. anomala | NCYC434 (Panomycocin) | 49.0 | Yes | ? | (1→3)-β-d-glucans | (1→3)-β-d-glucan hydrolysis | [65] |
| ATCC 96603/K36/UP25F (PaKT) | 85.0 | - | Chromosome | β-Glucan | - | [66] | |
| DBVPG 3003 (Pikt) | 8.0 | - | Chromosome | (1→6)-β-d-glucans | - | [67] | |
| YF07b (-) | 47.0 | - | Chromosome | - | (1→3)-β-d-glucanase activity | [68] | |
| VKM-Y (WAKTa/b) | - | - | - | - | - | [69] | |
| P. farinosa | KK1 (SMKT) | α (6.6), β (7.9) | Yes | Chromosome | - | Membrane permeabilization | [70] |
| P. inositovora | NRRL Y-18709 (-) | 3 subunits > 100.0 | - | dsDNA (pPin1-3) | Chitin | rRNA fragmentation | [71] |
| P. kluyveri | 1002 (-) | 19.0 | Yes | Chromosome | - | Membrane permeabilization | [37] |
| DBVPG 5826 (Pkkp) | 54.0 | - | - | Cell wall receptor | - | [8] | |
| P. kudriavzevii | RY55 (-) | 39.8 | - | - | - | - | [72] |
| P. membranifaciens | CYC 1106 (PMKT) | 18.0 | No | Chromosome | (1→6)-β-d-glucans | Membrane permeabilization/apoptosis | [19,41,73,74,75,76] |
| CYC 1086 (PMKT2) | 30.0 | - | - | Mannoproteins | Cell cycle arrest/apoptosis | [20,38] | |
| Pichia ohmeri | 158 (-) | <3.0 | - | Chromosome | - | Loss of cellular integrity | [33] |
| Applications | Organism/s | Toxin | General Description | References |
|---|---|---|---|---|
| Biological models | Saccharomyces cerevisiae | K28 | Model for the study of proteins, lipids, and mechanisms required on endocytosis and retrograde trafficking in A/B toxins (as Ricin, Shiga, and Cholera toxins) | [166] |
| Biotyping | Killer strains panel | - | Fingerprinting and clustering yeast strains (genera Debaryomyces, Kluyveromyces, Saccharomyces, and Zygosaccharomyces) by use of killer toxin sensitive patterns | [167] |
| Saccharomyces cerevisiae | - | Typing S. cerevisiae strain by combined use of RAPD and killer toxin sensitivity patterns | [168] | |
| Cryptococcus laurentii (CBS 139) | - | Biotyping varieties of Cryptococcus neomorfans (var. neoformans and var. gattii) by differential killer toxin sensitivity patterns | [169] | |
| Killer strains panel | - | Combined use of mtDNA-RFLP patterns and killer toxin biotype to study wine S. cerevisiae strain diversity in different viticulture regions | [170] | |
| Killer strains panel | - | Fingerprinting of Saccharomyces wine yeast by differential killer sensitivity | [171] | |
| Pichia mrakii | K9 | Use Pichia genera killer toxins to differentiate members belonging to the Nocardia asteroides complex (N. asteroides, N. farcinica, and N. nova) | [172] | |
| Pichia lynferdii | K76 | |||
| Killer strains panel | - | Use of toxins produced by a selected panel of killer yeast to discriminate strains belonging to genus Candida by their killer sensitive patterns | [173] | |
| Antimycotics for the treatment of human infections | Pichia anomala | PaKT | Using killer toxin-like anti-idiotypic antibodies of P. anomala killer toxin (PaKT-antilds) to treat treating human Pneumocystis carinii pneumonia. | [174] |
| Zygosaccharomyces bailii | Zygocin | Antifungal effect of Zygocin, a killer toxin produced by Z. bailii, on a broad-spectrum of sensitive fungal cells (Candida, Pichia, Hanseniaspora, Fusarium genera) | [104] | |
| Kodamaea ohmeri (HB55 and HB88) | - | Inhibition of C. neoformans (vars. neoformans, grubii and gattii), both clinical and environmental isolates, by two Brazilian yeasts selected for being able to inhibit human pathogenic fungi | [175] | |
| Filobasidium capsuligenum | Fc-1 | Use of a novel toxin with activity against the opportunistic fungal pathogen C. neoformans, as a therapeutic agent for the treatment of cryptococcosis | [176] | |
| Pichia anomala | K10 MAbs | Apply of antiidiotypic monoclonal antibodies (KT MAbs) from P. anomala against Aspergillus fumigatus. | [177] | |
| Williopsis mrakii var. mrakii (IFO 0895) | HM-1 | Activity of the killer toxin HM-1 from W. markii var. mrakii (formerly known as Hansenula mrakii) against Candida genus yeasts | [147] | |
| W. saturnus var. mrakii (MUCL 41968) | WmKT | Application of killer toxin (WmKT) secreted by W. saturnus var. mrakii, which is active against a wide range of pathogens. | [178] | |
| Antimycotics for treatment of animal infections | Kluyveromyces siamensis (HN12-1) | - | Use of a killer toxin of K. siamensis against Metschnikowia bicuspidata WCY, agent for the milky disease in crab | [179] |
| Pichia anomala (YF07b) | - | Apply of a killer toxin produced by the marine yeast P. anomala, against pathogenic yeast cells in crab, artemia, and shrimp | [68] | |
| Prevention of contaminants in fermentation industries | Saccharomyces cerevisiae | - | Effect of killer strains of S. cerevisiae on the growth of sensitive strains during wine fermentation, to improve the selected wine strain implantation. Sparkling wines quality improvement | [180,181] |
| Kluyveromyces phaffii (DBVPG 6076) | - | Use of K. phaffii killer toxin to control apiculate spoilage wine yeast (Hanseniaspora uvarum) | [182] | |
| Torulaspora delbrueckii (NPCC 1033) | TdKT | Application of a novel killer toxin (TdKT) from T. delbrueckii as a biocontrol tool in winemaking against different wine spoilage yeasts | [9] | |
| Ustilago maydis (CYC 1410) | - | Use of U. maydis toxins for the biocontrol of Brettanomyces bruxellensis for the reduction of aroma defects caused by this spoilage yeast | [10] | |
| Candida nodaensis (PYCC 3198) | CnKT | Application of CnKT in the preservation of salt-fermented foods, because of its high stability to salinity | [45] | |
| D. hansenii, K. marxianus, P. anomala, P. guilliermondii, S. cerevisiae | - | Control of olive table fermentation by the selection of killer yeast and their toxins which are able to suppress indigenous spoilage yeast growth | [86] | |
| Williopsis saturnus var. saturnus | - | Use of W. saturnus var. saturnus to inhibit spoilage yeast, as S. cerevisiae and K. marxianus, as a biopreservative agent on cheese making (in laboratory conditions) | [183] | |
| Wickerhamomyces anomalus (D2) | PiKT | Application of KwKT killer toxin produced by K. wickerhamii and PiKT produced by W. anomalus killer against B. bruxellensis during wine fermentation. | [184] | |
| Kluyveromyces wickerhamii (D15) | KwKT | |||
| Candida pyralidae (IWBT Y1140) | CpKT1 | Control of B. bruxellensis by C. pyralidae killer toxin in wine fermentation. | [185] | |
| Pichia membranifaciens | PMKT | Use of PMKT to control Zygosaccharomyces spp. contamination in winemaking. | [186] | |
| Wine quality improvement | S. cerevisiae (AWRI 796) | K2 | Interaction of S. cerevisiae killer strain (AWRI 796) and sensitive strain (Tyr 303) to accelerate the autolysis in the sparkling wine production to improve the final wine quality | [187] |
| Wickerhamomyces anomalus (CBS 1982, CBS5759) | - | Application of W. anomalus and S. cerevisiae in a mixed culture to positively influence chemical composition (higher amounts of polyphenol compounds and lower amounts of malic acid) and sensory features of produced apple wines | [188] | |
| Biological control | Kluyveromyces lactis (PCK27) | - | Use of killer strain K. lactis PCK27, to inhibit P. anomala, aerobic spoilage yeast in silage making | [144] |
| Williopsis mrakii | Mycocin HMK | Utilization of killer toxin HMK of W. mrakii, expressed in Aspergillus niger, to control both silage spoilage and yoghurt spoilage caused by yeasts | [90] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Belda, I.; Ruiz, J.; Alonso, A.; Marquina, D.; Santos, A. The Biology of Pichia membranifaciens Killer Toxins. Toxins 2017, 9, 112. https://doi.org/10.3390/toxins9040112
Belda I, Ruiz J, Alonso A, Marquina D, Santos A. The Biology of Pichia membranifaciens Killer Toxins. Toxins. 2017; 9(4):112. https://doi.org/10.3390/toxins9040112
Chicago/Turabian StyleBelda, Ignacio, Javier Ruiz, Alejandro Alonso, Domingo Marquina, and Antonio Santos. 2017. "The Biology of Pichia membranifaciens Killer Toxins" Toxins 9, no. 4: 112. https://doi.org/10.3390/toxins9040112






