Differences in Ribosome Binding and Sarcin/Ricin Loop Depurination by Shiga and Ricin Holotoxins
Abstract
:1. Introduction
2. Results
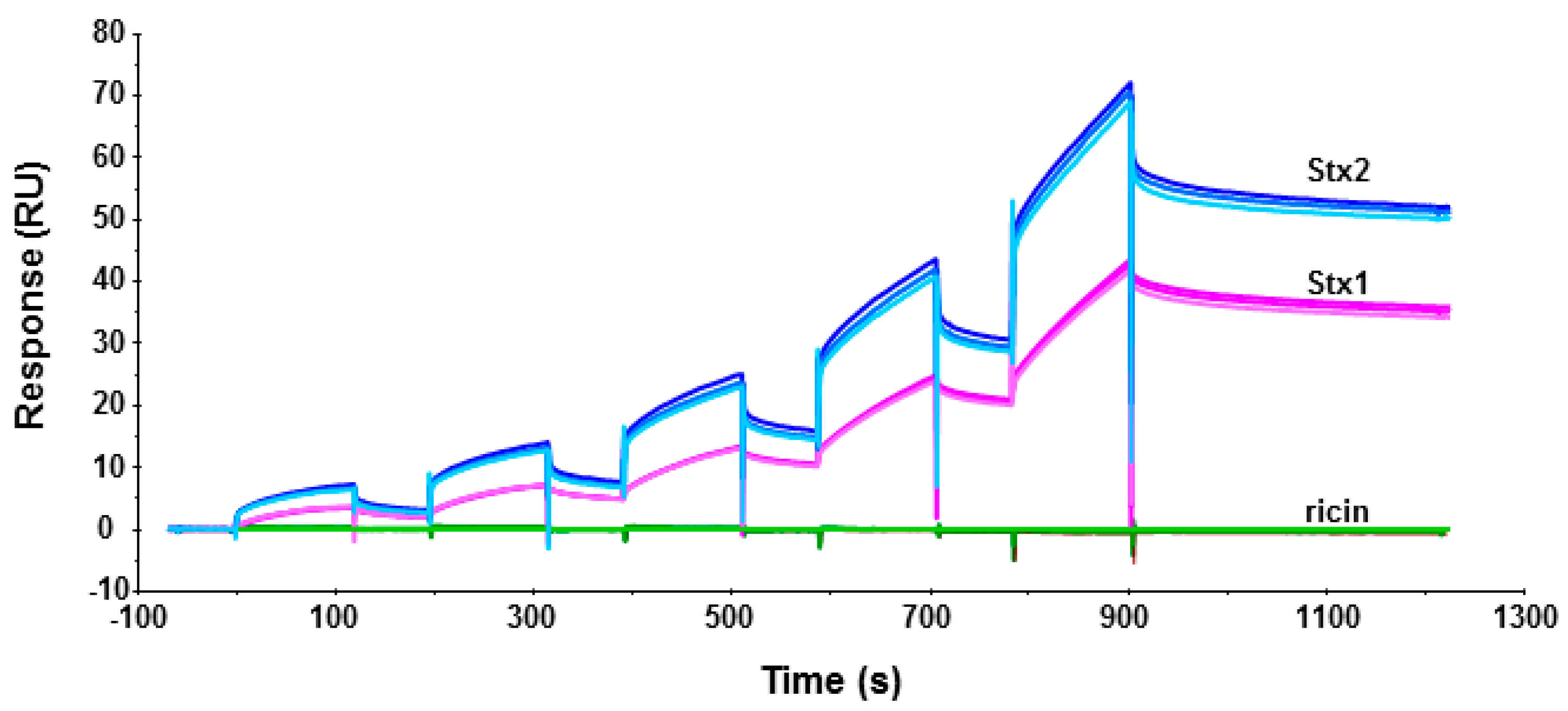
2.1. Stx1 and Stx2 Holotoxins Can Bind the Ribosome
2.2. Stx Holotoxins Are Inactive Towards the Ribosome, but Are Active After Separation of the A1 from the A2 Chain and the B Subunit
3. Discussion
4. Materials and Methods
4.1. Materials
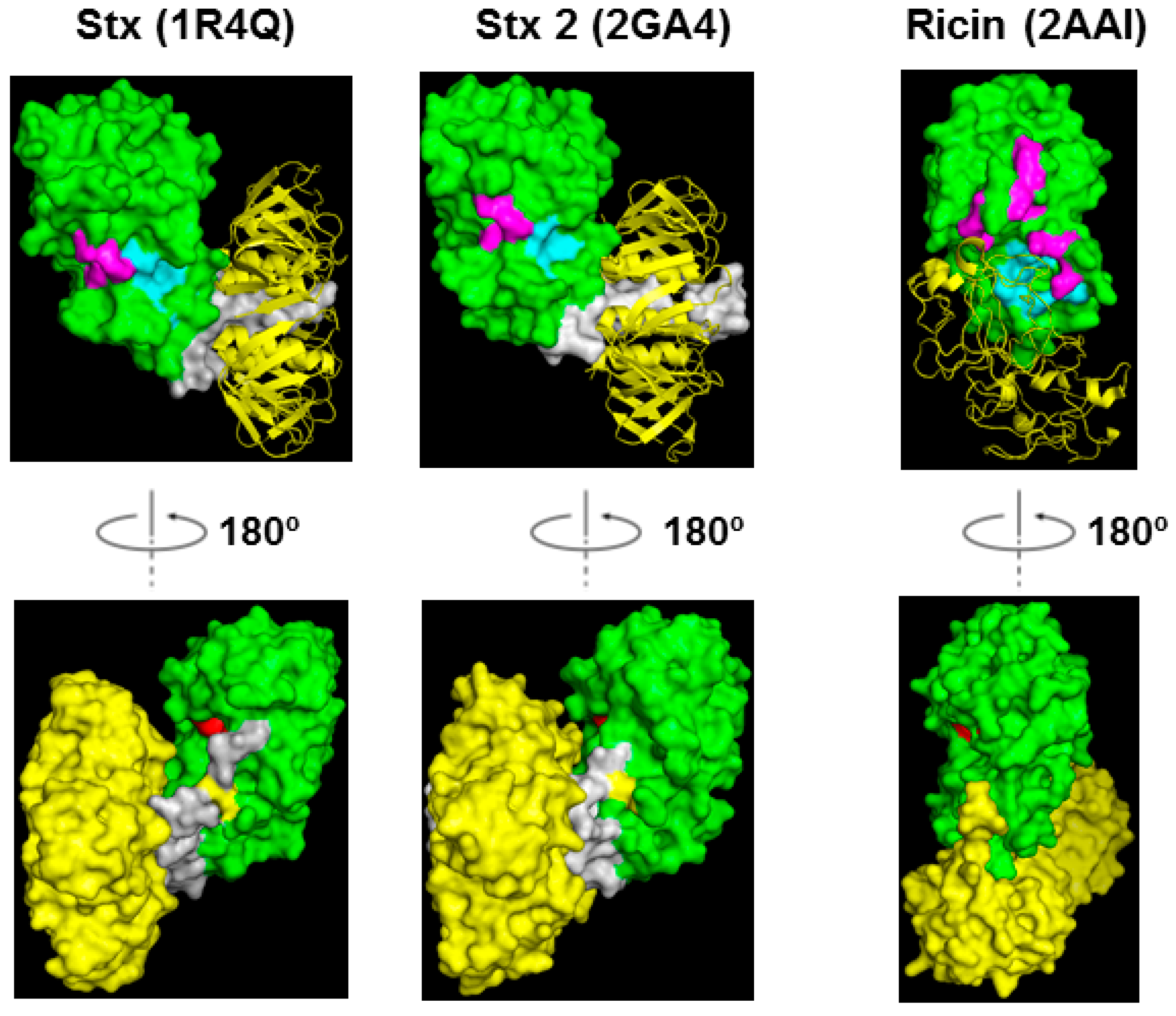
4.2. Toxin Structures
4.3. SDS-PAGE Analysis
4.4. Holotoxin-Ribosome Interaction
4.5. Toxin Activation
4.6. Ribosome Depurination
4.7. SRL Depurination
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Tesh, V.L.; O’Brien, A.D. The pathogenic mechanisms of Shiga toxin and the Shiga-like toxins. Mol. Microbiol. 1991, 5, 1817–1822. [Google Scholar] [CrossRef] [PubMed]
- Olsnes, S. The history of ricin, abrin and related toxins. Toxicon 2004, 44, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Bergan, J.; Dyve Lingelem, A.B.; Simm, R.; Skotland, T.; Sandvig, K. Shiga toxins. Toxicon 2012, 60, 1085–1107. [Google Scholar] [CrossRef] [PubMed]
- Lapadula, W.J.; Sanchez Puerta, M.V.; Juri Ayub, M. Revising the taxonomic distribution, origin and evolution of ribosome inactivating protein genes. PLoS ONE 2013, 8, e72825. [Google Scholar] [CrossRef] [PubMed]
- Sandvig, K.; van Deurs, B. Endocytosis, intracellular transport, and cytotoxic action of Shiga toxin and ricin. Physiol. Rev. 1996, 76, 949–966. [Google Scholar] [PubMed]
- Sandvig, K.; van Deurs, B. Delivery into cells: Lessons learned from plant and bacterial toxins. Gene Ther. 2005, 12, 865–872. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Shimizu, T.; Watarai, M.; Kobayashi, M.; Kano, S.; Hamabata, T.; Takeda, Y.; Yamasaki, S. Genome analysis of a novel Shiga toxin 1 (Stx1)-converting phage which is closely related to Stx2-converting phages but not to other Stx1-converting phages. J. Bacteriol. 2003, 185, 3966–3971. [Google Scholar] [CrossRef] [PubMed]
- Lamb, F.I.; Roberts, L.M.; Lord, J.M. Nucleotide sequence of cloned cDNA coding for preproricin. Eur. J. Biochem. 1985, 148, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Frigerio, L.; Vitale, A.; Lord, J.M.; Ceriotti, A.; Roberts, L.M. Free ricin A chain, proricin, and native toxin have different cellular fates when expressed in tobacco protoplasts. J. Biol. Chem. 1998, 273, 14194–14199. [Google Scholar] [CrossRef] [PubMed]
- Bolognesi, A.; Bortolotti, M.; Maiello, S.; Battelli, M.G.; Polito, L. Ribosome-inactivating proteins from plants: A historical overview. Molecules 2016, 21, 1627. [Google Scholar] [CrossRef] [PubMed]
- Fagerquist, C.K.; Sultan, O. Top-down proteomic identification of furin-cleaved alpha-subunit of Shiga toxin 2 from Escherichia coli O157:H7 using MALDI-TOF-TOF-MS/MS. J. Biomed. Biotechnol. 2010, 2010, 123460. [Google Scholar] [CrossRef] [PubMed]
- Nataro, J.P.; Kaper, J.B. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 1998, 11, 142–201. [Google Scholar] [PubMed]
- Manning, S.D.; Motiwala, A.S.; Springman, A.C.; Qi, W.; Lacher, D.W.; Ouellette, L.M.; Mladonicky, J.M.; Somsel, P.; Rudrik, J.T.; Dietrich, S.E.; et al. Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks. Proc. Natl. Acad. Sci. USA 2008, 105, 4868–4873. [Google Scholar] [CrossRef] [PubMed]
- Basu, D.; Li, X.P.; Kahn, J.N.; May, K.L.; Kahn, P.C.; Tumer, N.E. The A1 subunit of Shiga toxin 2 has higher affinity for ribosomes and higher catalytic activity than the A1 subunit of Shiga toxin 1. Infect. Immun. 2016, 84, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Chiou, J.C.; Li, X.P.; Remacha, M.; Ballesta, J.P.; Tumer, N.E. The ribosomal stalk is required for ribosome binding, depurination of the rRNA and cytotoxicity of ricin A chain in Saccharomyces cerevisiae. Mol. Microbiol. 2008, 70, 1441–1452. [Google Scholar] [CrossRef] [PubMed]
- Chiou, J.C.; Li, X.P.; Remacha, M.; Ballesta, J.P.; Tumer, N.E. Shiga toxin 1 is more dependent on the P proteins of the ribosomal stalk for depurination activity than Shiga toxin 2. Int. J. Biochem. Cell Biol. 2011, 43, 1792–1801. [Google Scholar] [CrossRef] [PubMed]
- McCluskey, A.J.; Poon, G.M.; Bolewska-Pedyczak, E.; Srikumar, T.; Jeram, S.M.; Raught, B.; Gariepy, J. The catalytic subunit of Shiga-like toxin 1 interacts with ribosomal stalk proteins and is inhibited by their conserved C-terminal domain. J. Mol. Biol. 2008, 378, 375–386. [Google Scholar] [CrossRef] [PubMed]
- McCluskey, A.J.; Bolewska-Pedyczak, E.; Jarvik, N.; Chen, G.; Sidhu, S.S.; Gariepy, J. Charged and hydrophobic surfaces on the A chain of Shiga-like toxin 1 recognize the C-terminal domain of ribosomal stalk proteins. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Li, X.P.; Kahn, P.C.; Kahn, J.N.; Grela, P.; Tumer, N.E. Arginine residues on the opposite side of the active site stimulate the catalysis of ribosome depurination by ricin A chain by interacting with the P-protein stalk. J. Biol. Chem. 2013, 288, 30270–30284. [Google Scholar] [CrossRef] [PubMed]
- Wahl, M.C.; Moller, W. Structure and function of the acidic ribosomal stalk proteins. Curr. Protein Pept. Sci. 2002, 3, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Gonzalo, P.; Reboud, J.P. The puzzling lateral flexible stalk of the ribosome. Biol. Cell 2003, 95, 179–193. [Google Scholar] [CrossRef]
- Ballesta, J.P.; Remacha, M. The large ribosomal subunit stalk as a regulatory element of the eukaryotic translational machinery. Prog. Nucleic Acid Res. Mol. Biol. 1996, 55, 157–193. [Google Scholar] [PubMed]
- Tchorzewski, M. The acidic ribosomal P proteins. Int. J. Biochem. Cell Biol. 2002, 34, 911–915. [Google Scholar] [CrossRef]
- Kozlov, Y.V.; Chernaia, M.M.; Fraser, M.E.; James, M.N. Purification and crystallization of Shiga toxin from Shigella dysenteriae. J. Mol. Biol. 1993, 232, 704–706. [Google Scholar] [CrossRef] [PubMed]
- Fraser, M.E.; Fujinaga, M.; Cherney, M.M.; Melton-Celsa, A.R.; Twiddy, E.M.; O’Brien, A.D.; James, M.N. Structure of Shiga toxin type 2 (Stx2) from Escherichia coli O157:H7. J. Biol. Chem. 2004, 279, 27511–27517. [Google Scholar] [CrossRef] [PubMed]
- Fraser, M.E.; Cherney, M.M.; Marcato, P.; Mulvey, G.L.; Armstrong, G.D.; James, M.N. Binding of adenine to Stx2, the protein toxin from Escherichia coli O157:H7. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2006, 62, 627–630. [Google Scholar] [CrossRef] [PubMed]
- Robertus, J.D. Toxin structure. Cancer Treat. Res. 1988, 37, 11–24. [Google Scholar] [PubMed]
- Katzin, B.J.; Collins, E.J.; Robertus, J.D. Structure of ricin A-chain at 2.5 Å. Proteins 1991, 10, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.W.; Tang, Y.S.; Sze, S.Y.; Zhu, Z.N.; Wong, K.B.; Shaw, P.C. Crystal structure of ribosome-inactivating protein ricin A chain in complex with the C-terminal peptide of the ribosomal stalk protein P2. Toxins 2016, 8. [Google Scholar] [CrossRef] [PubMed]
- Too, P.H.; Ma, M.K.; Mak, A.N.; Wong, Y.T.; Tung, C.K.; Zhu, G.; Au, S.W.; Wong, K.B.; Shaw, P.C. The C-terminal fragment of the ribosomal P protein complexed to trichosanthin reveals the interaction between the ribosome-inactivating protein and the ribosome. Nucleic Acids Res. 2009, 37, 602–610. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Zhu, Y.; Wang, C.; Niu, L.; Teng, M.; Li, X. Structural insights into the interaction of the ribosomal P stalk protein P2 with a type II ribosome-inactivating protein ricin. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Basu, D.; Kahn, J.N.; Li, X.P.; Tumer, N.E. Conserved arginines at the P-protein stalk binding site and the active site are critical for ribosome interactions of Shiga toxins but do not contribute to differences in the affinity of the A1 subunits for the ribosome. Infect. Immun. 2016, 84, 3290–3301. [Google Scholar] [CrossRef] [PubMed]
- Brigotti, M.; Carnicelli, D.; Alvergna, P.; Mazzaracchio, R.; Sperti, S.; Montanaro, L. The RNA-N-glycosidase activity of Shiga-like toxin I: Kinetic parameters of the native and activated toxin. Toxicon 1997, 35, 1431–1437. [Google Scholar] [CrossRef]
- Sturm, M.B.; Schramm, V.L. Detecting ricin: Sensitive luminescent assay for ricin A-chain ribosome depurination kinetics. Anal. Chem. 2009, 81, 2847–2853. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.Y.; Link, T.M.; Schramm, V.L. Ricin A-chain: Kinetics, mechanism, and RNA stem-loop inhibitors. Biochemistry 1998, 37, 11605–11613. [Google Scholar] [CrossRef] [PubMed]
- Wahome, P.G.; Robertus, J.D.; Mantis, N.J. Small-molecule inhibitors of ricin and Shiga toxins. Curr. Top. Microbiol. Immunol. 2012, 357, 179–207. [Google Scholar] [PubMed]
- Li, X.P.; Chiou, J.C.; Remacha, M.; Ballesta, J.P.; Tumer, N.E. A two-step binding model proposed for the electrostatic interactions of ricin A chain with ribosomes. Biochemistry 2009, 48, 3853–3863. [Google Scholar] [CrossRef] [PubMed]
- Donohue-Rolfe, A.; Acheson, D.W.; Kane, A.V.; Keusch, G.T. Purification of Shiga toxin and Shiga-like toxins I and II by receptor analog affinity chromatography with immobilized P1 glycoprotein and production of cross-reactive monoclonal antibodies. Infect. Immun. 1989, 57, 3888–3893. [Google Scholar] [PubMed]




© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.-P.; Tumer, N.E. Differences in Ribosome Binding and Sarcin/Ricin Loop Depurination by Shiga and Ricin Holotoxins. Toxins 2017, 9, 133. https://doi.org/10.3390/toxins9040133
Li X-P, Tumer NE. Differences in Ribosome Binding and Sarcin/Ricin Loop Depurination by Shiga and Ricin Holotoxins. Toxins. 2017; 9(4):133. https://doi.org/10.3390/toxins9040133
Chicago/Turabian StyleLi, Xiao-Ping, and Nilgun E. Tumer. 2017. "Differences in Ribosome Binding and Sarcin/Ricin Loop Depurination by Shiga and Ricin Holotoxins" Toxins 9, no. 4: 133. https://doi.org/10.3390/toxins9040133




