The Mode of Action of Cyclo(l-Ala-l-Pro) in Inhibiting Aflatoxin Production of Aspergillus flavus
Abstract
:1. Introduction
2. Results
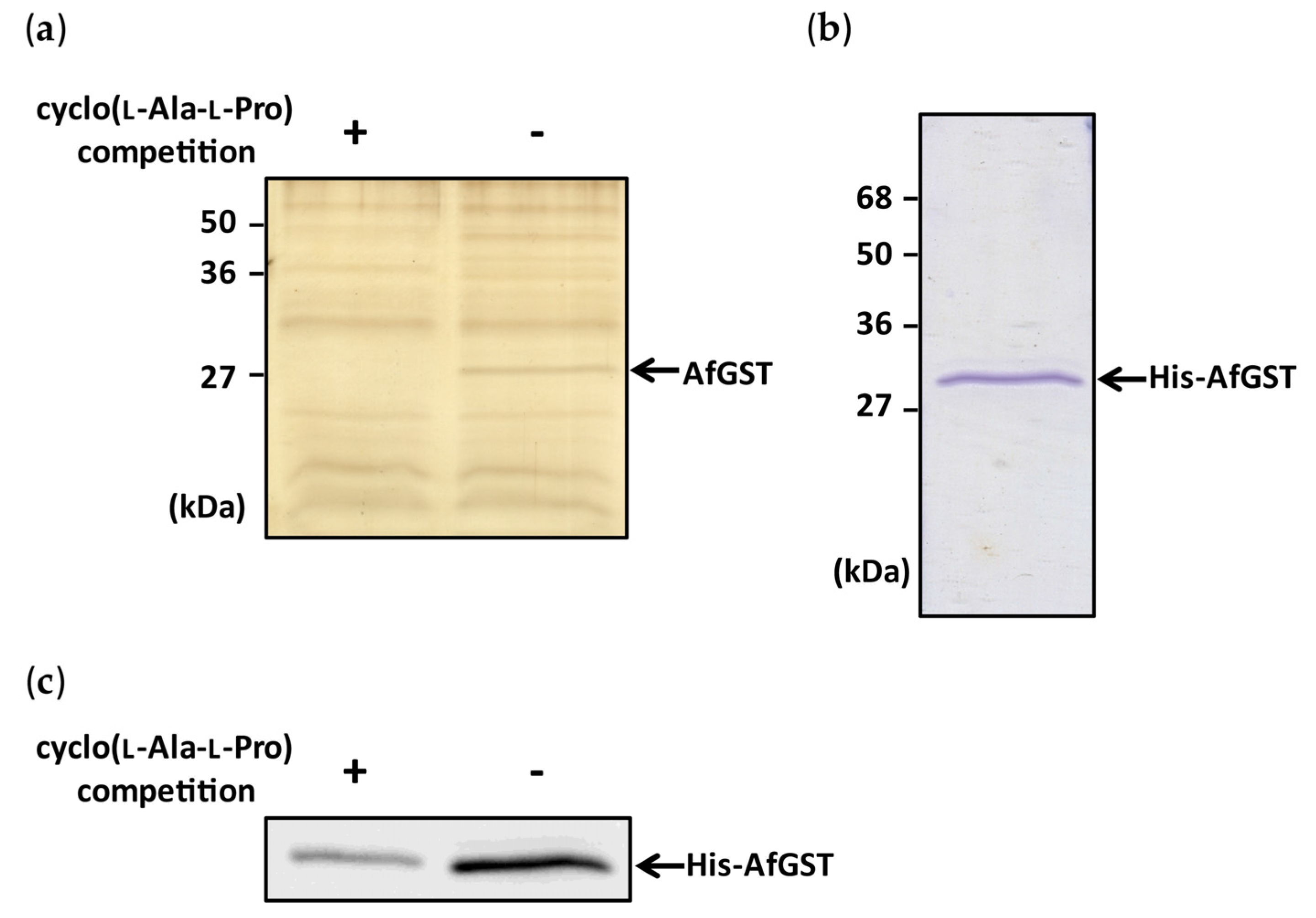
2.1. Identification of Cyclo(l-Ala-l-Pro) Binding Protein
2.2. Time Course of AfGST Expression
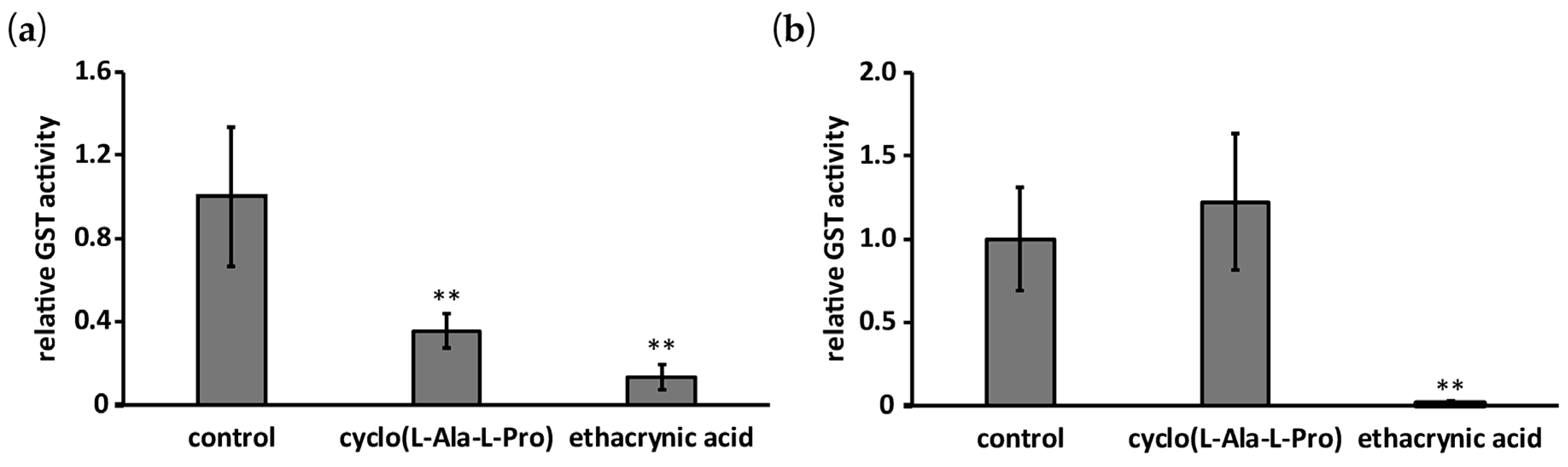
2.3. Effect of Cyclo(l-Ala-l-Pro) on GST Activity
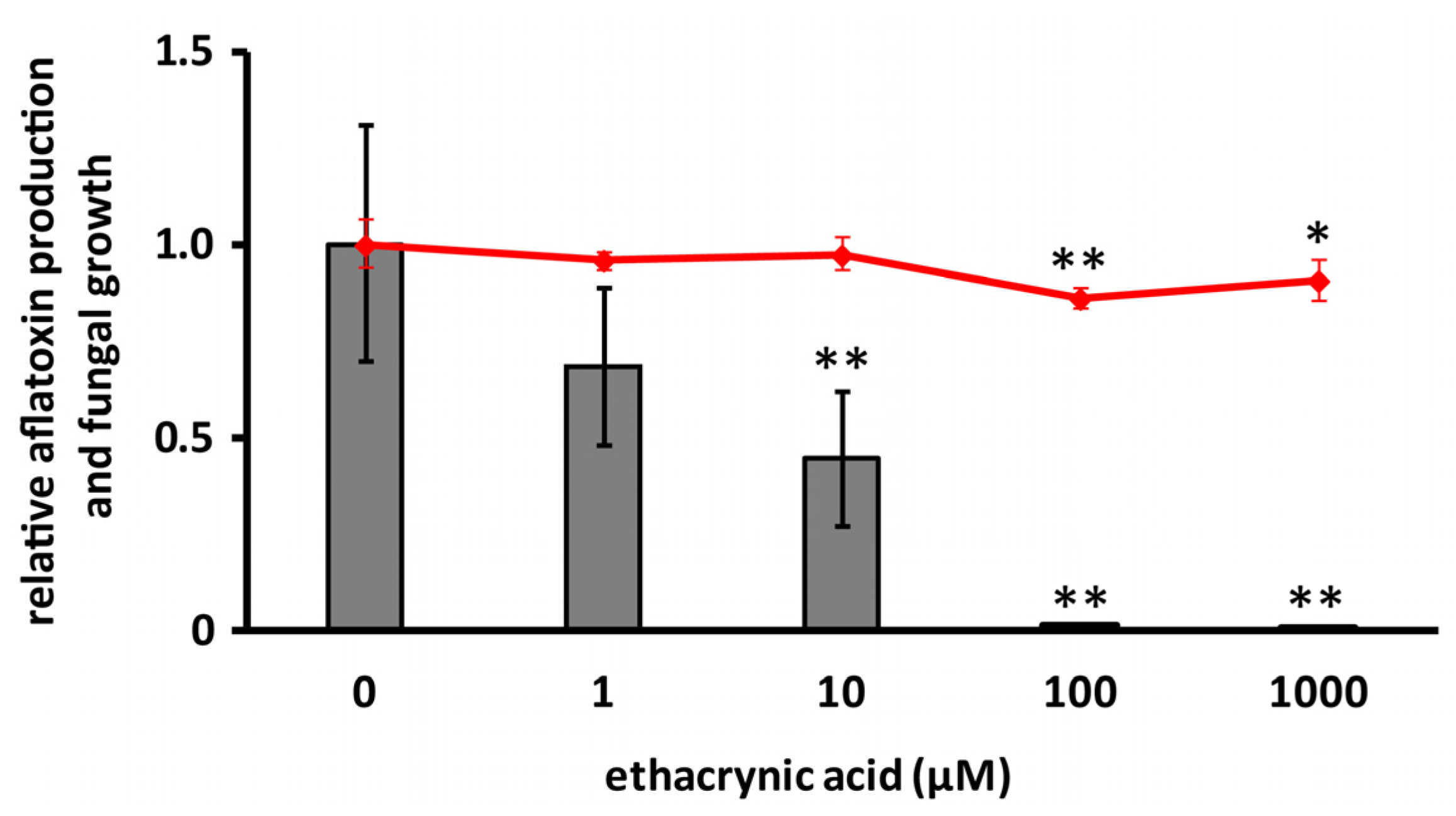
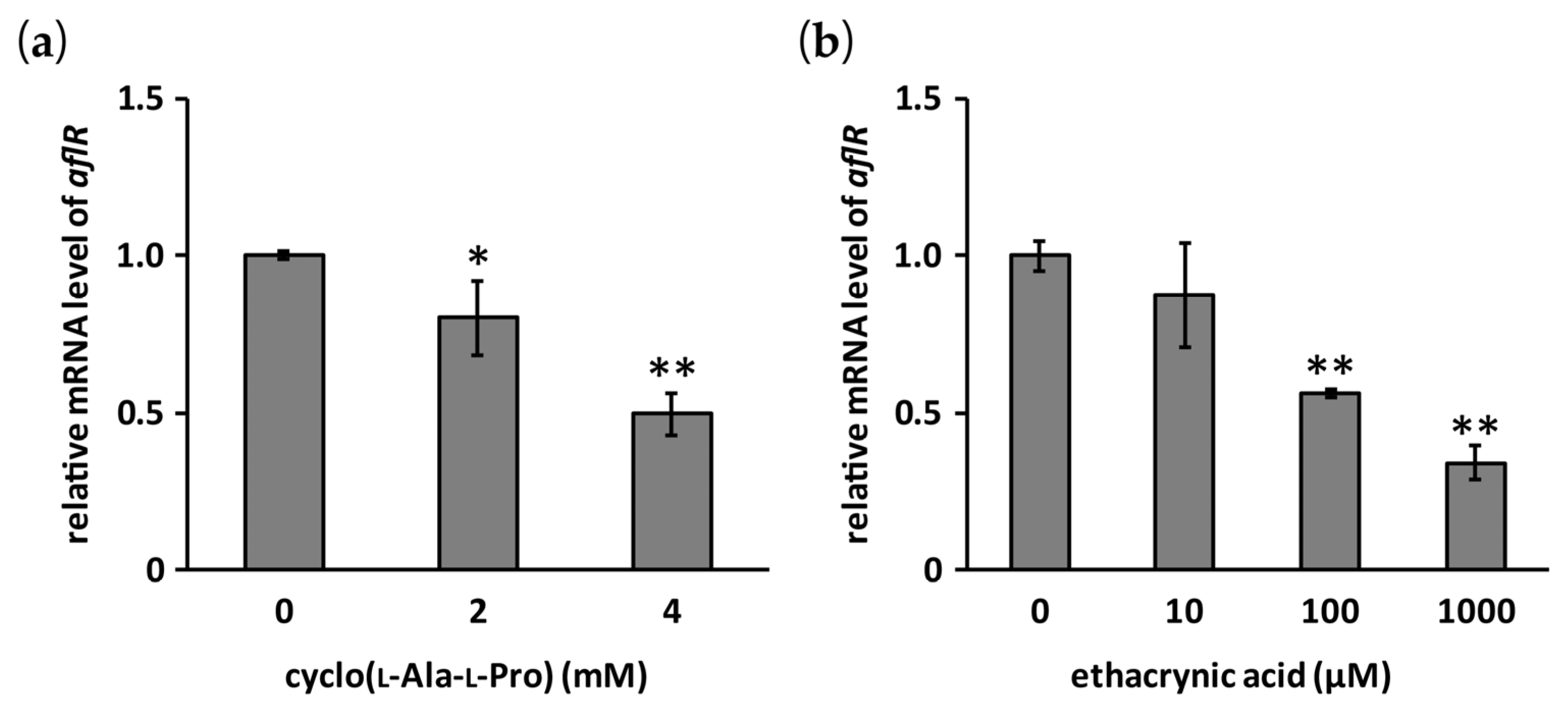
2.4. Effects of Cyclo(l-Ala-l-Pro) and Ethacrynic Acid on the mRNA Level of aflR
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Chemicals
5.2. Strains and Culture Conditions
5.3. Analyses of Aflatoxin Production and Fungal Growth
5.4. Preparation of Cyclo(l-Ala-l-Pro)-Immobilized Sepharose Beads
5.5. Detection of Cyclo(l-Ala-l-Pro)-Binding Protein
5.6. Binding Assay with Recombinant AfGST
5.7. RT-qPCR Analysis
5.8. GST Activity of His-AfGST and Schistoma Japonicum GST
5.9. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Strosnider, H.; Azziz-Baumgartner, E.; Banziger, M.; Bhat, R.V.; Breiman, R.; Brune, M.N.; DeCock, K.; Dilley, A.; Groopman, J.; Hell, K.; et al. Workgroup report: Public health strategies for reducing aflatoxin exposure in developing countries. Environ. Health Perspect. 2006, 114, 1898–1903. [Google Scholar] [CrossRef] [PubMed]
- Wu, F. Global impacts of aflatoxin in maize: Trade and human health. World Mycotoxin J. 2015, 8, 137–142. [Google Scholar] [CrossRef]
- Liu, Y.; Chang, C.C.H.; Marsh, G.M.; Wu, F. Population attributable risk of aflatoxin-related liver cancer: Systematic review and meta-analysis. Eur. J. Cancer 2012, 48, 2125–2136. [Google Scholar] [CrossRef] [PubMed]
- Abbas, H.K.; Accinelli, C.; Shier, W.T. Biological control of aflatoxin contamination in U.S. crops and the use of bioplastic formulations of Aspergillus flavus biocontrol strains to optimize application strategies. J. Agric. Food Chem. 2017. [Google Scholar] [CrossRef] [PubMed]
- Abbas, H.K.; Wilkinson, J.R.; Zablotowicz, R.M.; Accinelli, C.; Abel, C.A.; Bruns, H.A.; Weaver, M.A. Ecology of Aspergillus flavus, regulation of aflatoxin production, and management strategies to reduce aflatoxin contamination of corn. Toxin Rev. 2009, 28, 142–153. [Google Scholar] [CrossRef]
- Sakuda, S.; Yoshinari, T.; Furukawa, T.; Jermnak, U.; Takagi, K.; Iimura, K.; Yamamoto, T.; Suzuki, M.; Nagasawa, H. Search for aflatoxin and trichothecene production inhibitors and analysis of their modes of action. Biosci. Biotech. Biochem. 2016, 80, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Holmes, R.A.; Boston, R.S.; Payne, G.A. Diverse inhibitors of aflatoxin biosynthesis. Appl. Microbiol. Biotechnol. 2008, 78, 559–572. [Google Scholar] [CrossRef] [PubMed]
- Sakuda, S.; Prabowo, D.F.; Takagi, K.; Shiomi, K.; Mori, M.; Ōmura, S.; Nagasawa, H. Inhibitory effects of respiration inhibitors on aflatoxin production. Toxins 2014, 6, 1193–1200. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, T.; Iimura, K.; Kimura, T.; Yamamoto, T.; Sakuda, S. Inhibitory activities of alkyl syringates and related compounds on aflatoxin production. Toxins 2016, 8, 177. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, T.; Sakamoto, N.; Suzuki, M.; Kimura, M.; Nagasawa, H.; Sakuda, S. Precocene II, a trichothecene production inhibitor, binds to voltage-dependent anion channel and increases the superoxide level in mitochondria of Fusarium graminearum. PLoS ONE 2015, 10, e0135031. [Google Scholar] [CrossRef] [PubMed]
- Yan, P.S.; Song, Y.; Sakuno, E.; Nakajima, H.; Nakagawa, H.; Yabe, K. Cyclo(l-Leucyl-l-Prolyl) produced by Achromobacter xylosoxidans inhibits flatoxin production by Aspergillus parasiticus. Appl. Environ. Microbiol. 2004, 70, 7466–7473. [Google Scholar] [CrossRef] [PubMed]
- Jermnak, U.; Chinaphuti, A.; Poapolathep, A.; Kawai, R.; Nagasawa, H.; Sakuda, S. Prevention of aflatoxin contamination by a soil bacterium of Stenotrophomonas sp. that produces aflatoxin production inhibitors. Microbiology 2013, 159, 902–912. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Chang, P.K.; Ehrlich, K.C.; Cary, J.W.; Bhatnagar, D.; Cleveland, T.E.; Payne, G.A.; Linz, J.E.; Woloshuk, C.P.; Bennett, J.W. Clustered pathways genes in aflatoxin biosynthesis. Appl. Environ. Microbiol. 2004, 70, 1253–1262. [Google Scholar] [CrossRef] [PubMed]
- Kawatani, M.; Okumura, H.; Honda, K.; Kanoh, N.; Muroi, M.; Dohmae, N.; Takami, M.; Kitagawa, M.; Futamura, Y.; Imoto, M.; et al. The identification of an osteoclastogenesis inhibitor through the inhibition of glyoxalase I. Proc. Natl. Acad. Sci. USA 2009, 105, 11691–11696. [Google Scholar] [CrossRef] [PubMed]
- Burns, C.; Geraghty, R.; Neville, C.; Murphy, A.; Kavanagh, K.; Doyle, S. Identification, cloning, and functional expression of three glutathione transferase genes from Aspergillus fumigatus. Fungal Genet. Biol. 2005, 42, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Sato, I.; Shimizu, M.; Hoshino, T.; Takaya, N. The glutathione system of Aspergillus nidulans involves a fungus-specific glutathione S-transferase. J. Biol. Chem. 2009, 284, 8042–8053. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.D.; Flanagan, J.U.; Jowsey, I.R. Glutathione transferases. Annu. Rev. Rharmacol. Toxicol. 2005, 45, 51–88. [Google Scholar] [CrossRef] [PubMed]
- Fraser, J.A.; Davis, M.A.; Hynes, M.J. A gene from Aspergillus nidulans with similarity to URE2 of Saccharomyces cerevisiae encodes a glutathione S-transferase which contributes to heavy metal and xenobiotic resistance. Appl. Environ. Microbiol. 2002, 68, 2802–2808. [Google Scholar] [CrossRef] [PubMed]
- Saxena, M.; Mukerji, K.G.; Raj, H.G. Positive correlation exists between glutathione S-trabsferase activity and aflatoxin formation in Aspergillus flavus. Biochem. J. 1988, 254, 567–570. [Google Scholar] [CrossRef] [PubMed]
- Saxena, M.; Allameh, A.; Mukerji, K.G.; Raj, H.G. Studies on glutathione S-transferases of A. flavus group in relation to aflatoxin production. J. Toxicol. Toxin Rev. 1989, 8, 319–328. [Google Scholar] [CrossRef]
- Ziglari, T.; Allameh, A.; Razzaghi-Abyaneh, M.; Khosravi, A.R.; Yadegari, M.H. Comparison of glutathione S-transferase activity and concentration in aflatoxin-producing and their non-toxigenic counterpart isolates. Mycopathologia 2008, 166, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Reverberi, M.; Zjalic, S.; Ricelli, A.; Fabbri, A.A.; Fanelli, C. Oxidant/antioxidant balance in Aspergillus parasiticus affects aflatoxin biosynthesis. Mycotoxin Res. 2006, 22, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Duran, R.M.; Cary, J.W.; Calvo, A.M. Production of cyclopiazonic acid, aflatrem, and aflatoxin by Aspergillus flavus is regulated by veA, a gene necessary for sclerotial formation. Appl. Microbiol. Biotechnol. 2007, 73, 1158–1168. [Google Scholar] [CrossRef] [PubMed]
- Baidya, S.; Duran, R.M.; Lohmar, J.M.; Harris-Coward, P.Y.; Cary, J.W.; Hong, S.Y.; Roze, L.V.; Linz, J.E.; Calvo, A.M. VeA is associated with the response to oxidative stress in the aflatoxin-producer Aspergillus flavus. Eukaryot. Cell 2014, 1, 1095–1103. [Google Scholar] [CrossRef] [PubMed]
- Schultz, M.; Dutta, S.; Tew, K.D. Inhibitors of glutathione S-transferases as therapeutic agents. Adv. Drug Deliv. Rev. 1997, 26, 91–104. [Google Scholar] [CrossRef]
- Musdal, Y.; Hegazy, U.M.; Aksoy, Y.; Mannervik, B. FDA-approved drugs and other compounds tested as inhibitors of human glutathione transferase P1-1. Chem. Biol. Interact. 2013, 205, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Takayama, H.; Moriya, T.; Kanoh, N. Preparation of photo-cross-linked small molecule affinity matrices for affinity selection of protein targets for biologically active small molecules. In Chemical Genomics and Proteomics Reviews and Protocols; Zanders, E.D., Ed.; Humana Press: New York, NJ, USA, 2012; pp. 75–83. [Google Scholar]
- ASPERGILLUS GENOME PROJECT of the Broad Institute. Available online: http://www.broadinstitute.org/fungal-genome-initiative/aspergillus-genome-projects (accessed on 25 October 2015).
- Habdous, M.; Vincent-Viry, M.; Visvikis, S.; Siest, G. Rapid spectrophotometric method for serum glutathione S-transferases activity. Clin. Chim. Acta 2002, 326, 131–142. [Google Scholar] [CrossRef]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iimura, K.; Furukawa, T.; Yamamoto, T.; Negishi, L.; Suzuki, M.; Sakuda, S. The Mode of Action of Cyclo(l-Ala-l-Pro) in Inhibiting Aflatoxin Production of Aspergillus flavus. Toxins 2017, 9, 219. https://doi.org/10.3390/toxins9070219
Iimura K, Furukawa T, Yamamoto T, Negishi L, Suzuki M, Sakuda S. The Mode of Action of Cyclo(l-Ala-l-Pro) in Inhibiting Aflatoxin Production of Aspergillus flavus. Toxins. 2017; 9(7):219. https://doi.org/10.3390/toxins9070219
Chicago/Turabian StyleIimura, Kurin, Tomohiro Furukawa, Toshiyoshi Yamamoto, Lumi Negishi, Michio Suzuki, and Shohei Sakuda. 2017. "The Mode of Action of Cyclo(l-Ala-l-Pro) in Inhibiting Aflatoxin Production of Aspergillus flavus" Toxins 9, no. 7: 219. https://doi.org/10.3390/toxins9070219




