Molecular Surveillance of Viral Processes Using Silicon Nitride Membranes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Viral Complexes
2.2. In vitro Transcription Reactions
2.3. Functionalization of SiN Microchips
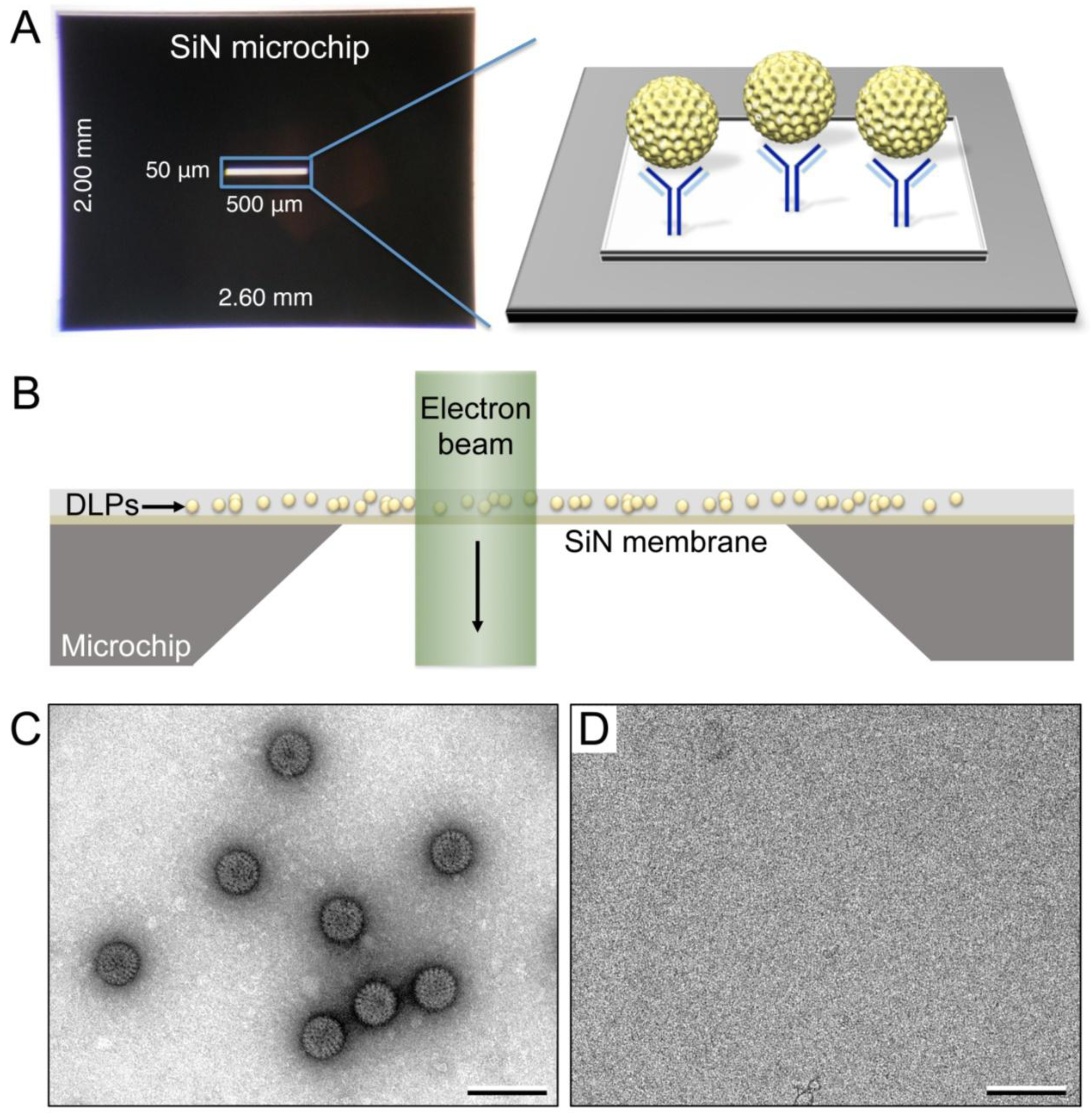
2.4. Recruitment of Viral Complexes to Microchips
2.5. EM Imaging and Computing
3. Results and Discussion
3.1. Functionalized Microchips Recruit Viral Complexes
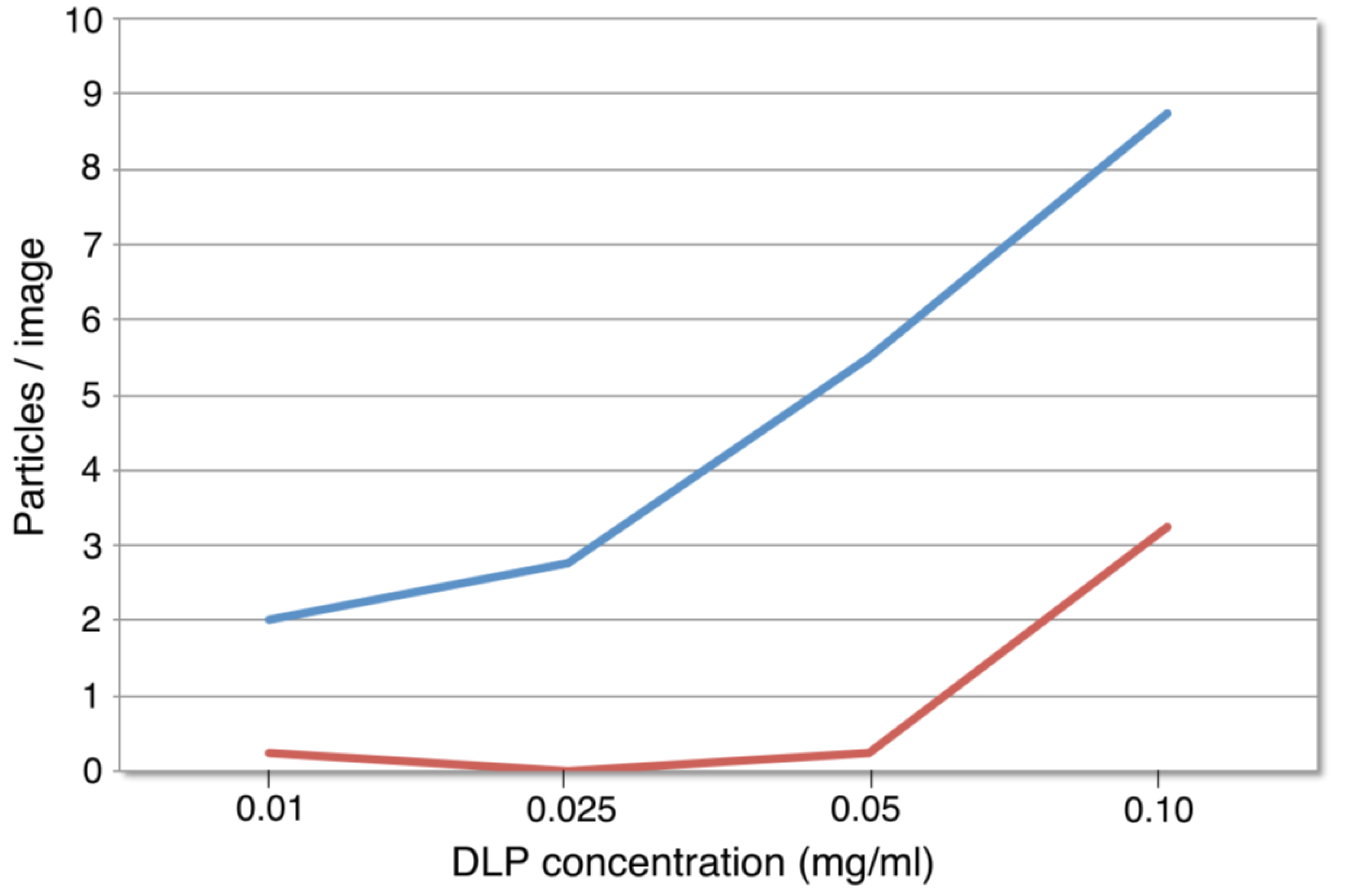
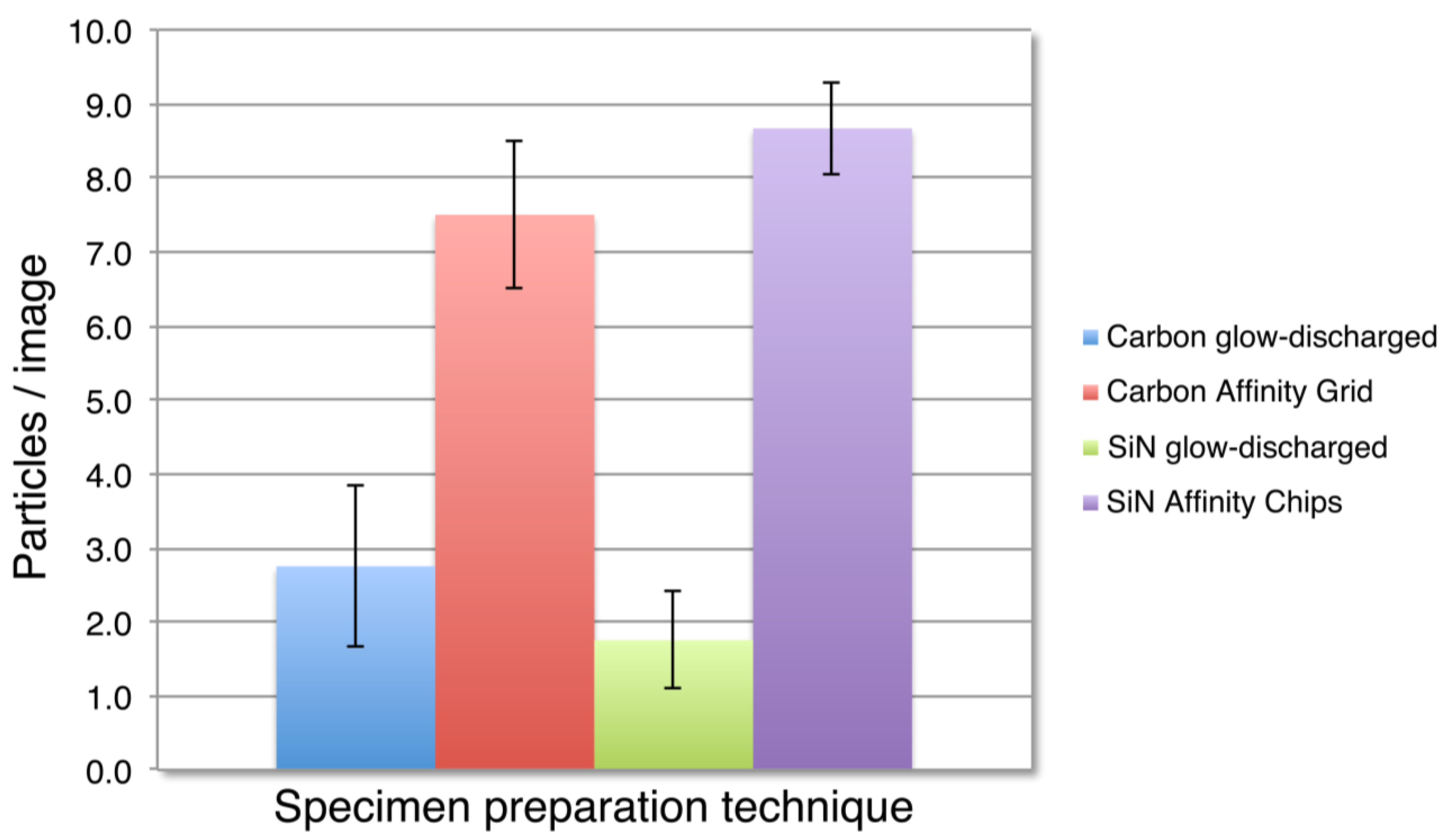
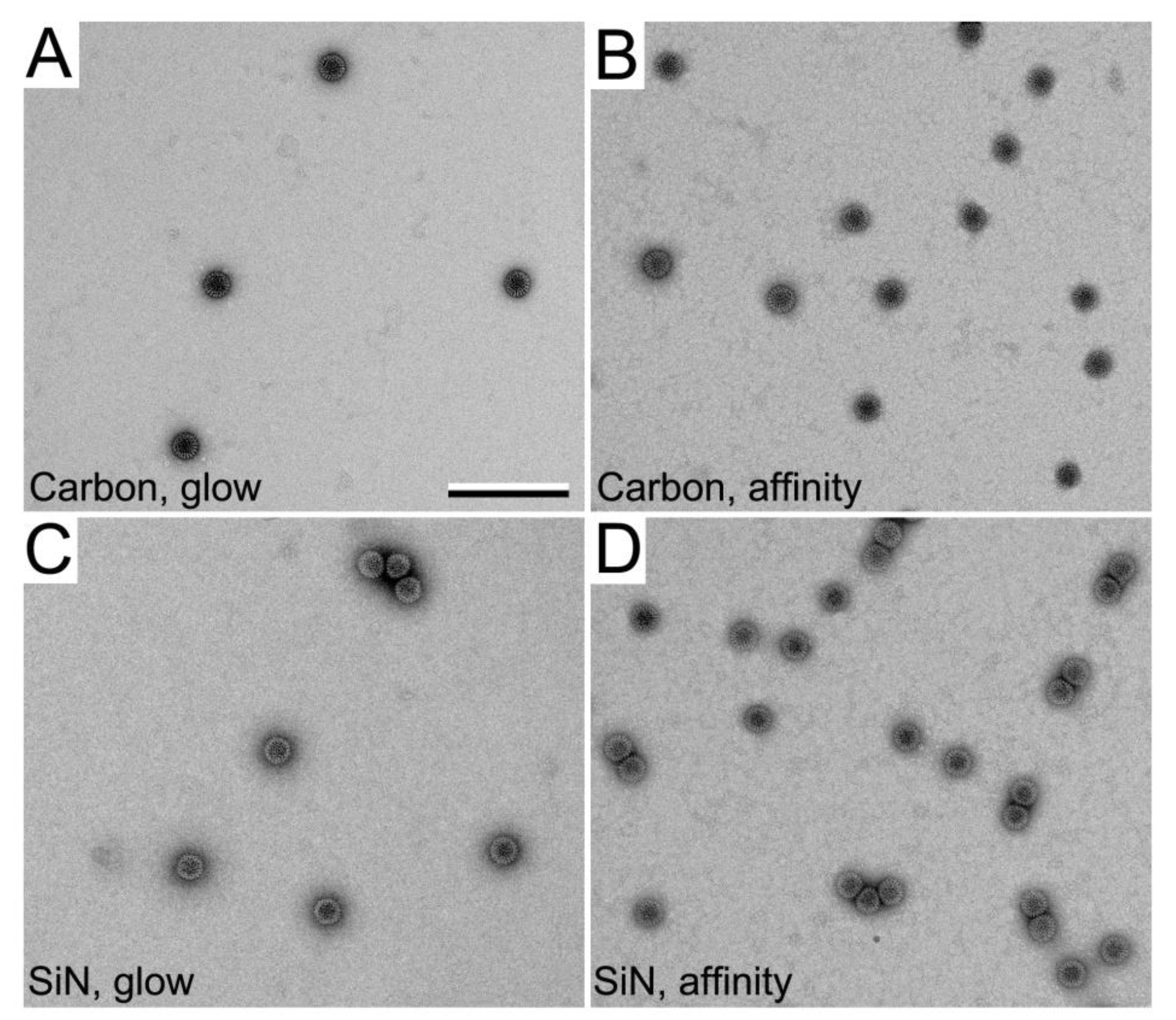
3.2. SiN versus Conventional Carbon Substrates
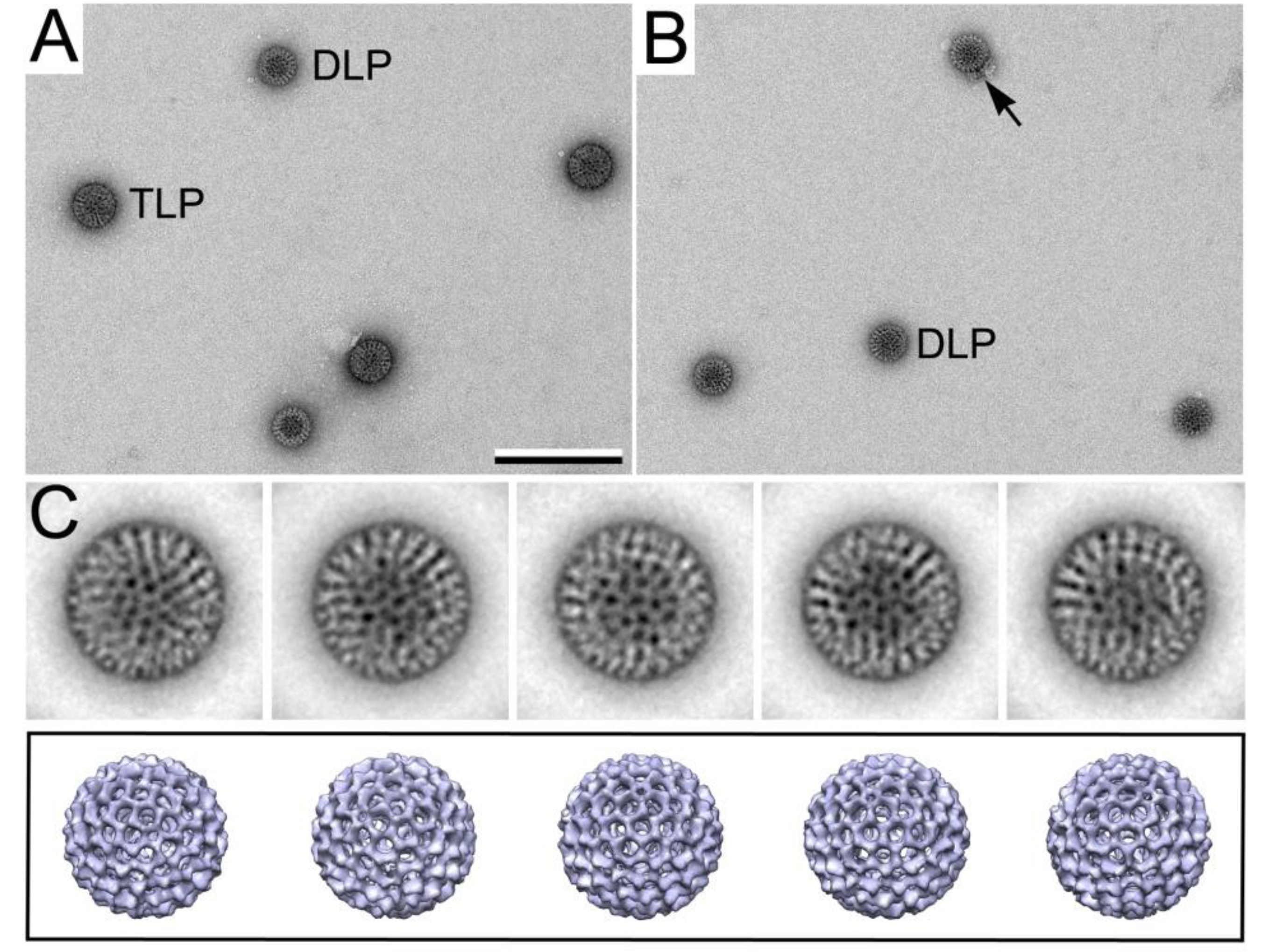
3.3. Monitoring Viral Mechanisms
3.3.1. Converting TLPs to DLPs
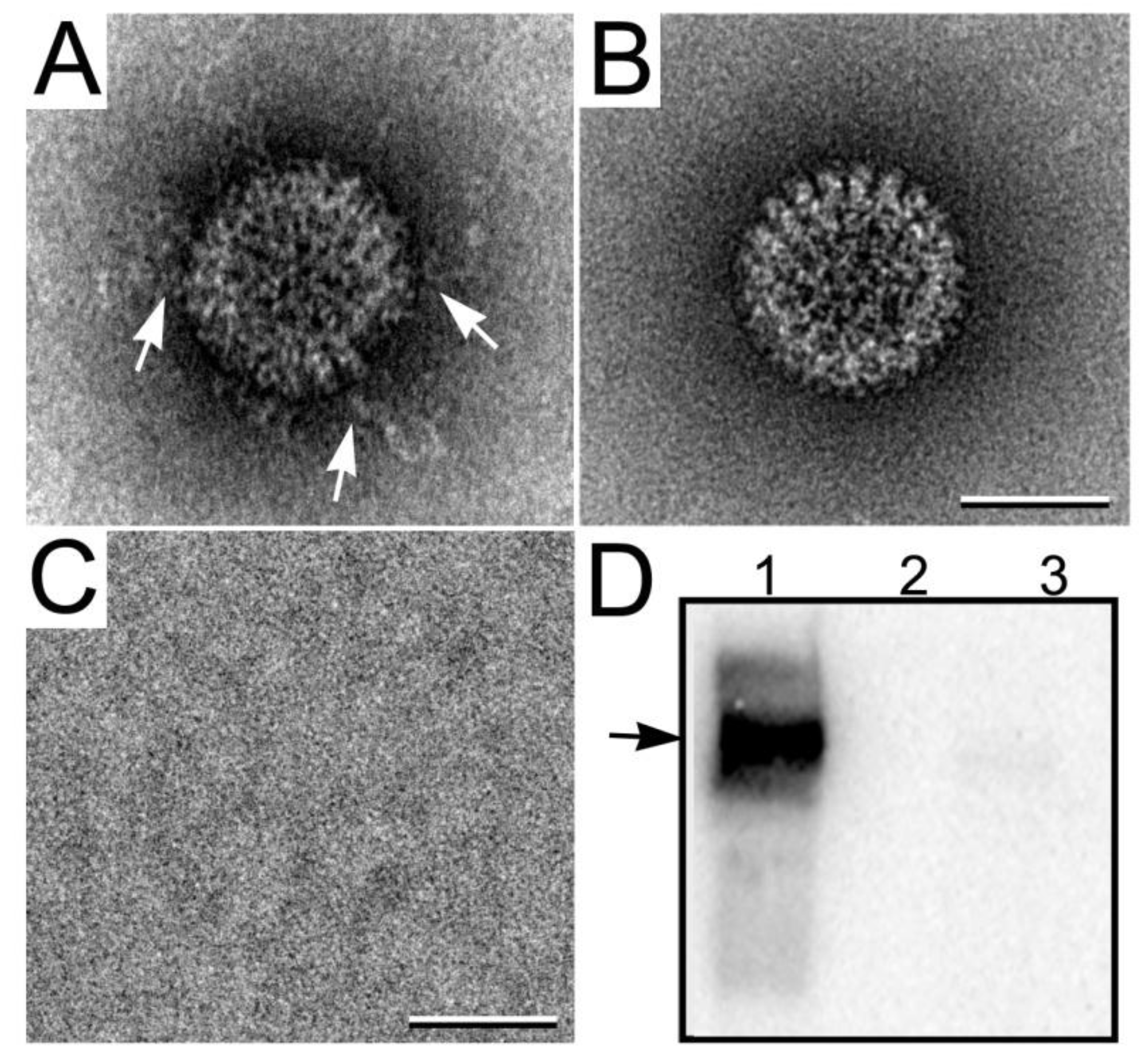
3.3.2. Viewing RNA Transcript Production
4. Conclusions
Acknowledgments
References
- Pratt, E.D.; Huang, C.; Hawkins, B.G.; Gleghorn, J.P.; Kirby, B.J. Rare Cell Capture in Microfluidic Devices. Chem. Eng. Sci. 2011, 66, 1508–1522. [Google Scholar] [CrossRef]
- Mernier, G.; Majocchi, S.; Mermod, N.; Renaud, P. In situ evaluation of single-cell lysis by cytosol extraction observation through fluorescence decay and dielectrophoretic trapping time. Sensor Actuat. B 2012, 166, 907–912. [Google Scholar] [CrossRef]
- Maguire, T.J.; Novik, E.; Chao, P.; Barminko, J.; Nahmias, Y.; Yarmush, M.L.; Cheng, K.C. Design and application of microfluidic systems for in vitro pharmacokinetic evaluation of drug candidates. Curr. Drug. MeTable 2009, 10, 1192–1199. [Google Scholar] [CrossRef]
- Akagi, J.; Khoshmanesh, K.; Evans, B.; Hall, C.J.; Crosier, K.E.; Cooper, J.M.; Crosier, P.S.; Wlodkowic, D. Miniaturized Embryo Array for Automated Trapping, Immobilization and Microperfusion of Zebrafish Embryos. PLoS One 2012, 7. [Google Scholar] [CrossRef]
- Ring, E.A.; de Jonge, N. Microfluidic system for transmission electron microscopy. Microsc. Microanal. 2010, 16, 622–629. [Google Scholar] [CrossRef]
- Klein, K.L.; Anderson, I.M.; de Jonge, N. Transmission electron microscopy with a liquid flow cell. J. Microsc. 2011, 242, 117–123. [Google Scholar] [CrossRef]
- Ramachandra, R.; Demers, H.; de Jonge, N. Atomic-resolution scanning transmission electron microscopy through 50-nm-thick silicon nitride membranes. Appl. Phys. Lett. 2011, 98. [Google Scholar] [CrossRef]
- Yuk, J.M.; Park, J.; Ercius, P.; Kim, K.; Hellebusch, D.J.; Crommie, M.F.; Lee, J.Y.; Zettl, A.; Alivisatos, A.P. High-resolution EM of colloidal nanocrystal growth using graphene liquid cells. Science 2012, 336, 61–64. [Google Scholar] [CrossRef]
- Taylor, K.A.; Glaeser, R.M. Retrospective on the early development of cryoelectron microscopy of macromolecules and a prospective on opportunities for the future. J. Struct. Biol. 2008, 163, 214–223. [Google Scholar] [CrossRef]
- Rhinow, D.; Kuhlbrandt, W. Electron cryo-microscopy of biological specimens on conductive titanium-silicon metal glass films. Ultramicroscopy 2008, 108, 698–705. [Google Scholar] [CrossRef]
- Yoshioka, C.; Carragher, B.; Potter, C.S. Cryomesh: A new substrate for cryo-electron microscopy. Microsc. Microanal. 2010, 16, 43–53. [Google Scholar] [CrossRef]
- Degen, K.; Dukes, M.; Tanner, J.R.; Kelly, D.F. The development of affinity capture devices—a nanoscale purification platform for biological in situ transmission electron microscopy. Rsc. Adv. 2012, 2, 2408–2412. [Google Scholar] [CrossRef]
- Gilmore, B.L.; Showalter, S.P.; Dukes, M.J.; Tanner, J.R.; Demmert, A.C.; McDonald, S.M.; Kelly, D.F. Visualizing viral assemblies in a nanoscale biosphere. Lab Chip 2013, 13, 216–219. [Google Scholar] [CrossRef]
- Trask, S.D.; McDonald, S.M.; Patton, J.T. Structural insights into the coupling of virion assembly and rotavirus replication. Nat. Rev. Microbiol. 2012, 10, 165–177. [Google Scholar] [CrossRef]
- Arnold, M.M.; Patton, J.T. Rotavirus antagonism of the innate immune response. Viruses 2009, 1, 1035–1056. [Google Scholar] [CrossRef]
- Ring, E.A.; Peckys, D.B.; Dukes, M.J.; Baudoin, J.P.; de Jonge, N. Silicon nitride windowsfor electron microscopy of whole cells. J. Microsc-Oxford 2011, 243, 273–283. [Google Scholar] [CrossRef]
- Ohi, M.; Li, Y.; Cheng, Y.; Walz, T. Negative staining and image classification—powerful tools in modern electron microscopy. Biol. Proced. Online 2004, 6, 23–34. [Google Scholar] [CrossRef]
- Frank, J.; Radermacher, M.; Penczek, P.; Zhu, J.; Li, Y.; Ladjadj, M.; Leith, A. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996, 116, 190–199. [Google Scholar] [CrossRef]
- Scheres, S.H. A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 2012, 415, 406–418. [Google Scholar] [CrossRef]
- Zhang, X.; Settembre, E.; Xu, C.; Dormitzer, P.R.; Bellamy, R.; Harrison, S.C.; Grigorieff, N. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc. Natl. Acad. Sci. USA 2008, 105, 1867–1872. [Google Scholar]
- Grigorieff Laboratory website. Available online: http://emlab.rose2.brandeis.edu/rotavirusdlp (accessed on 18 April 2012).
- Mellado, M.C.; Mena, J.A.; Lopes, A.; Ramirez, O.T.; Carrondo, M.J.; Palomares, L.A.; Alves, P.M. Impact of physicochemical parameters on in vitro assembly and disassembly kinetics of recombinant triple-layered rotavirus-like particles. Biotechnol. Bioeng. 2009, 104, 674–686. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Gilmore, B.L.; Tanner, J.R.; McKell, A.O.; Boudreaux, C.E.; Dukes, M.J.; McDonald, S.M.; Kelly, D.F. Molecular Surveillance of Viral Processes Using Silicon Nitride Membranes. Micromachines 2013, 4, 90-102. https://doi.org/10.3390/mi4010090
Gilmore BL, Tanner JR, McKell AO, Boudreaux CE, Dukes MJ, McDonald SM, Kelly DF. Molecular Surveillance of Viral Processes Using Silicon Nitride Membranes. Micromachines. 2013; 4(1):90-102. https://doi.org/10.3390/mi4010090
Chicago/Turabian StyleGilmore, Brian L., Justin R. Tanner, Allison O. McKell, Crystal E. Boudreaux, Madeline J. Dukes, Sarah M. McDonald, and Deborah F. Kelly. 2013. "Molecular Surveillance of Viral Processes Using Silicon Nitride Membranes" Micromachines 4, no. 1: 90-102. https://doi.org/10.3390/mi4010090