Rapid Fabrication of Electrophoretic Microfluidic Devices from Polyester, Adhesives and Gold Leaf
Abstract
:1. Introduction
2. Materials and Methods
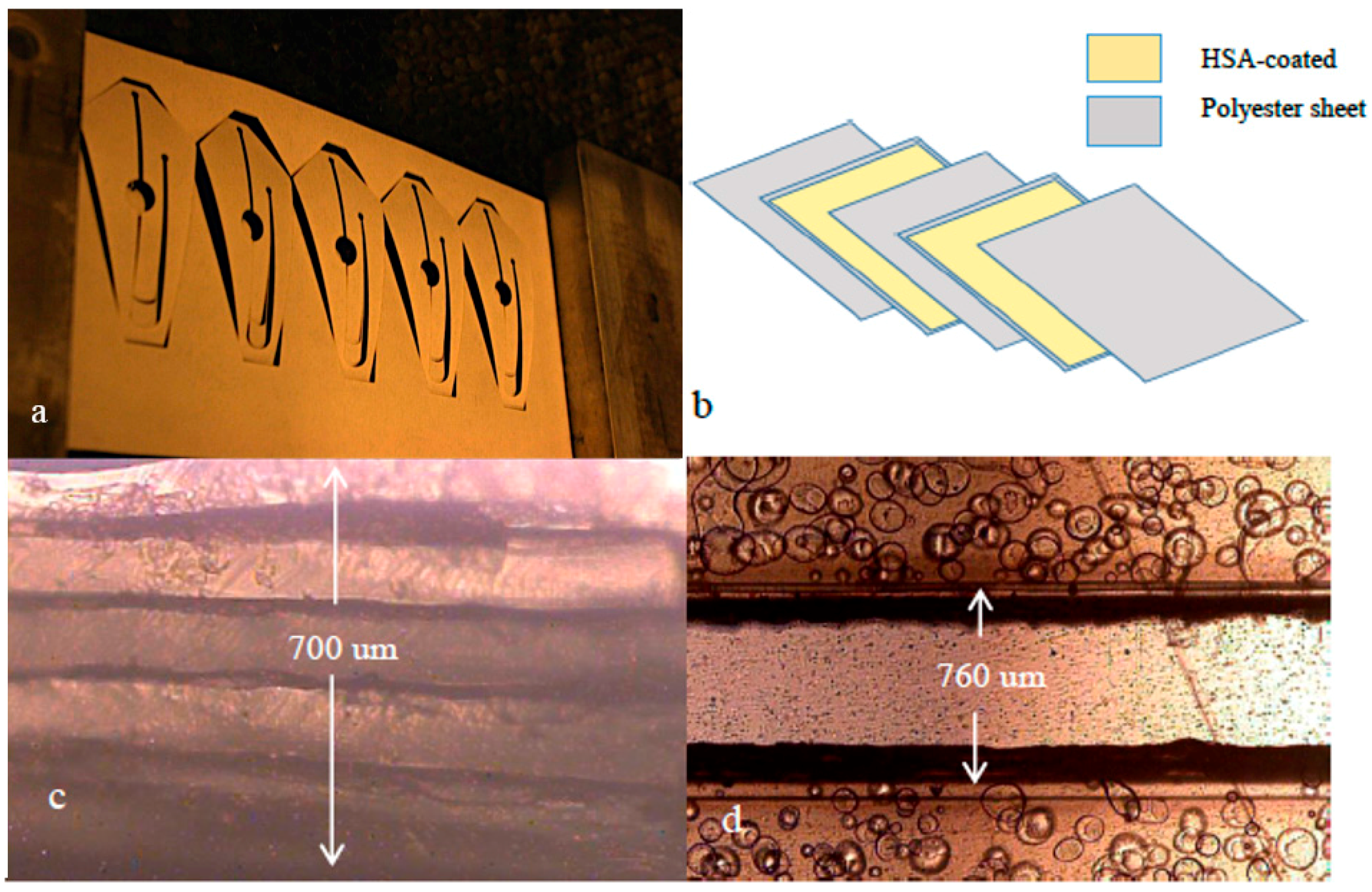
2.1. Device Fabrication
2.2. Reagents
3. Results
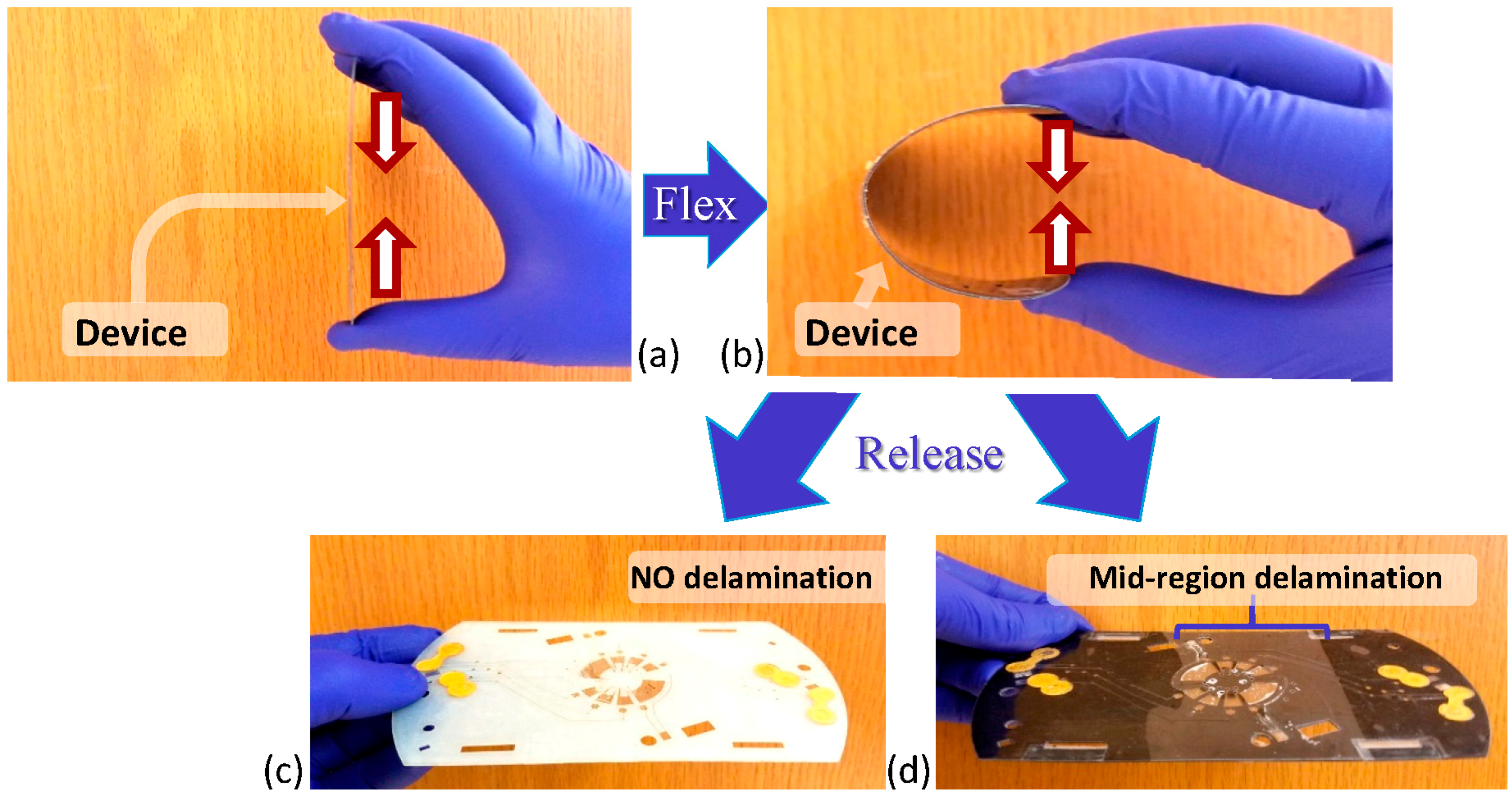
3.1. Fabrication
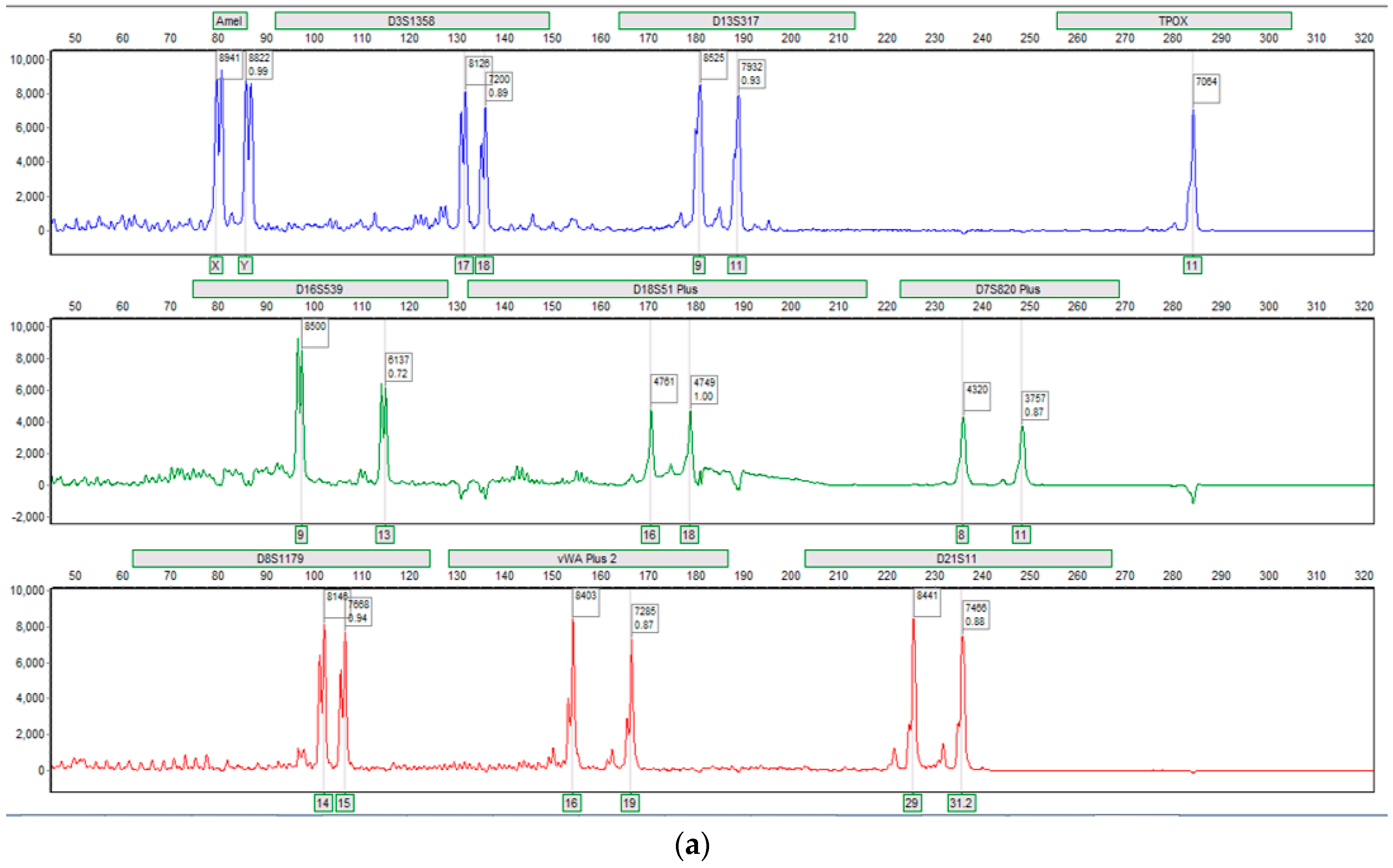
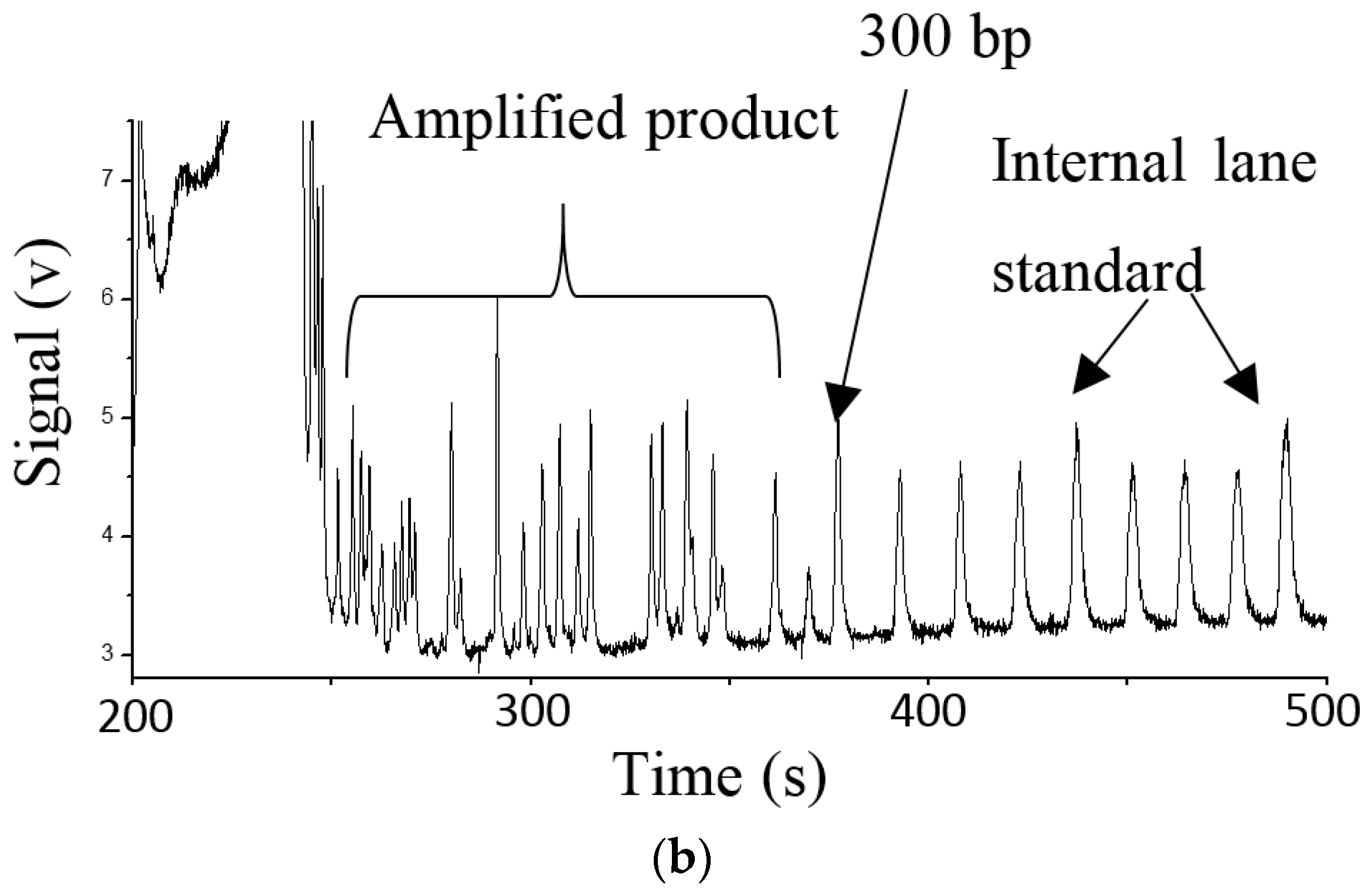
3.2. Fluidics and DNA Assays
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bruijns, B.; van Asten, A.; Tiggelaar, R.; Gardeniers, H. Microfluidic devices for forensic DNA analysis: A review. Biosensors 2016, 6, 41. [Google Scholar] [CrossRef] [PubMed]
- Whitesides, G.M. The origins and the future of microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Becker, H.; Gartner, C. Polymer microfabrication technologies for microfluidic systems. Anal. Bioanal. Chem. 2008, 390, 89–111. [Google Scholar] [CrossRef] [PubMed]
- Ren, K.; Zhou, J.; Wu, H. Materials for microfluidic fabrication. Acc. Chem. Res. 2013, 46, 2396–2406. [Google Scholar] [CrossRef] [PubMed]
- Cooper McDonald, J.; Whitesides, G.M. Poly(dimethylsiloxane) as a material for fabricating microfluidic devices. Acc. Chem. Res. 2002, 35, 491–499. [Google Scholar] [CrossRef]
- Thompson, B.L.; Ouyang, Y.; Duarte, G.R.M.; Carrilho, E.; Krauss, S.T.; Landers, J.P. Inexpensive, rapid protoyping of microfluidic devices using overhead transparencies and laser print, cut and laminate fabrication method. Nat. Protoc. 2015, 10, 875–886. [Google Scholar] [CrossRef] [PubMed]
- Thompson, B.L.; Birch, C.; Li, J.; Duvall, J.A.; Le Roux, D.S.; Nelson, D.A.; Tsuei, A.; Mill, D.L.; Krauss, S.T.; Root, B.E.; et al. Microfluidic enzymatic DNA extraction on a hybrid polyester-toner-PMMA device. Analyst 2016, 141, 4667–4675. [Google Scholar] [CrossRef] [PubMed]
- Duvall, J.A.; Le Roux, D.S.; Tsuei, A.; Thompson, B.L.; Birch, C.; Li, J.; Nelson, D.A.; Mills, D.L.; Ewing, M.M.; McLaren, R.S.; et al. A rotationally-driven polyethylene terephthalate microdevice with integrated reagent mixing for multiplexed PCR amplification of DNA. Anal. Methods 2016, 8, 7331–7340. [Google Scholar] [CrossRef]
- Thompson, B.L.; Birch, C.; Nelson, D.A.; Li, J.; DuVall, J.A.; Le Roux, D.S.; Tsuei, A.; Mills, D.L.; Root, B.E.; Landers, J.P. A centrifugal microfluidic device with integrated gold leaf electrodes for the electrophoretic separation of DNA. Lab Chip 2016, 16, 4569–4580. [Google Scholar] [CrossRef] [PubMed]
- Duvall, J.A.; Cabaniss, S.T.; Angelotti, M.L.; Moore, J.H.; Abhyankar, M.; Shukla, N.; Mils, D.L.; Kessel, B.G.; Garner, G.T.; Swami, N.S.; et al. Rapid detection of Clostridium difficile via magnetic bead aggregation in cost-effective polyester microdevices with cell phone image analysis. Analyst 2016, 141, 5637–5645. [Google Scholar] [CrossRef] [PubMed]
- Krauss, S.T.; Remcho, T.P.; Lipe, S.M.; Aranda, R.; Maynard, H.P.; Sukla, N.; Li, J.; Tontarski, R.E.; Landers, J.P. Objective Method for Presumptive Field-Testing of Illicit Drug Possession Using Centrifugal Microdevices and Smartphone Analysis. Anal. Chem. 2016, 88, 8689–8697. [Google Scholar] [CrossRef] [PubMed]
- Thompson, B.L.; Gilbert, R.J.; Meiia, M.; Shukla, N.; Haverstick, D.M.; Garner, G.T.; Landers, J.P. Hematocrit analysis through the use of an inexpensive centrifugal polyester-toner device with finger-to-chip blood loading capability. Anal. Chim. Acta 2016, 924, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Krauss, S.T.; Remcho, T.P.; Monazami, E.; Thompson, B.L.; Reinke, P.; Begley, M.R.; Landers, J.P. Inkjet printing on transparency films for reagent storage with polyester–toner microdevices. Anal. Methods 2016, 8, 7061–7068. [Google Scholar] [CrossRef]
- Reedy, C.R.; Hagan, K.A.; Marchiarullo, D.J.; Dewald, A.H.; Barron, A.; Bienvenue, J.M.; Landers, J.P. A modular microfluidic system for deoxyribonucleic acid identification by shorttandem repeat analysis. Anal. Chim. Acta 2011, 687, 150–158. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Birch, C.; DuVall, J.A.; Le Roux, D.; Thompson, B.L.; Tsuei, A.-C.; Li, J.; Nelson, D.A.; Mills, D.L.; Landers, J.P.; Root, B.E. Rapid Fabrication of Electrophoretic Microfluidic Devices from Polyester, Adhesives and Gold Leaf. Micromachines 2017, 8, 17. https://doi.org/10.3390/mi8010017
Birch C, DuVall JA, Le Roux D, Thompson BL, Tsuei A-C, Li J, Nelson DA, Mills DL, Landers JP, Root BE. Rapid Fabrication of Electrophoretic Microfluidic Devices from Polyester, Adhesives and Gold Leaf. Micromachines. 2017; 8(1):17. https://doi.org/10.3390/mi8010017
Chicago/Turabian StyleBirch, Christopher, Jacquelyn A. DuVall, Delphine Le Roux, Brandon L. Thompson, An-Chi Tsuei, Jingyi Li, Daniel A. Nelson, Daniel L. Mills, James P. Landers, and Brian E. Root. 2017. "Rapid Fabrication of Electrophoretic Microfluidic Devices from Polyester, Adhesives and Gold Leaf" Micromachines 8, no. 1: 17. https://doi.org/10.3390/mi8010017




