Cell Migration Research Based on Organ-on-Chip-Related Approaches
Abstract
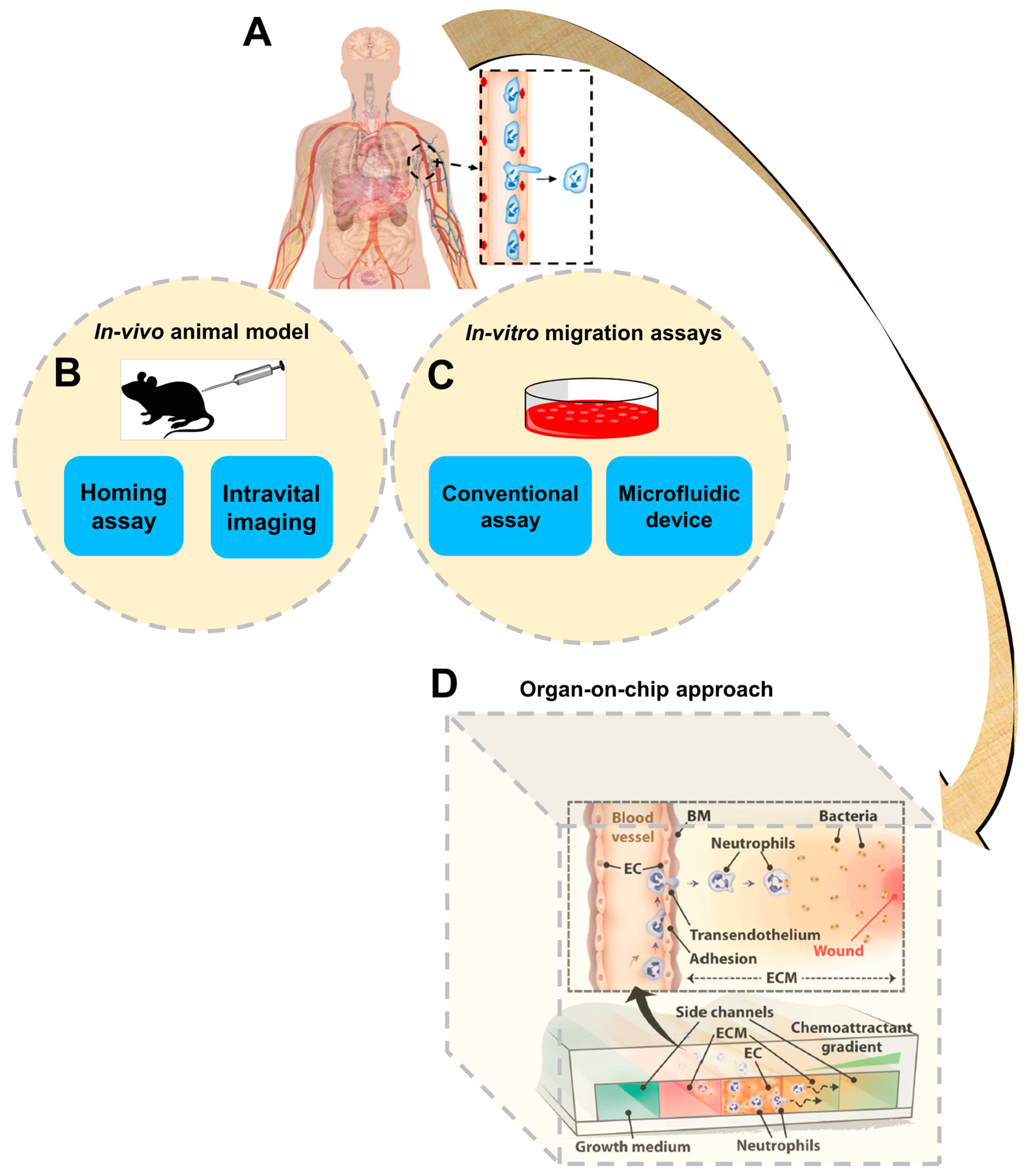
:1. Introduction
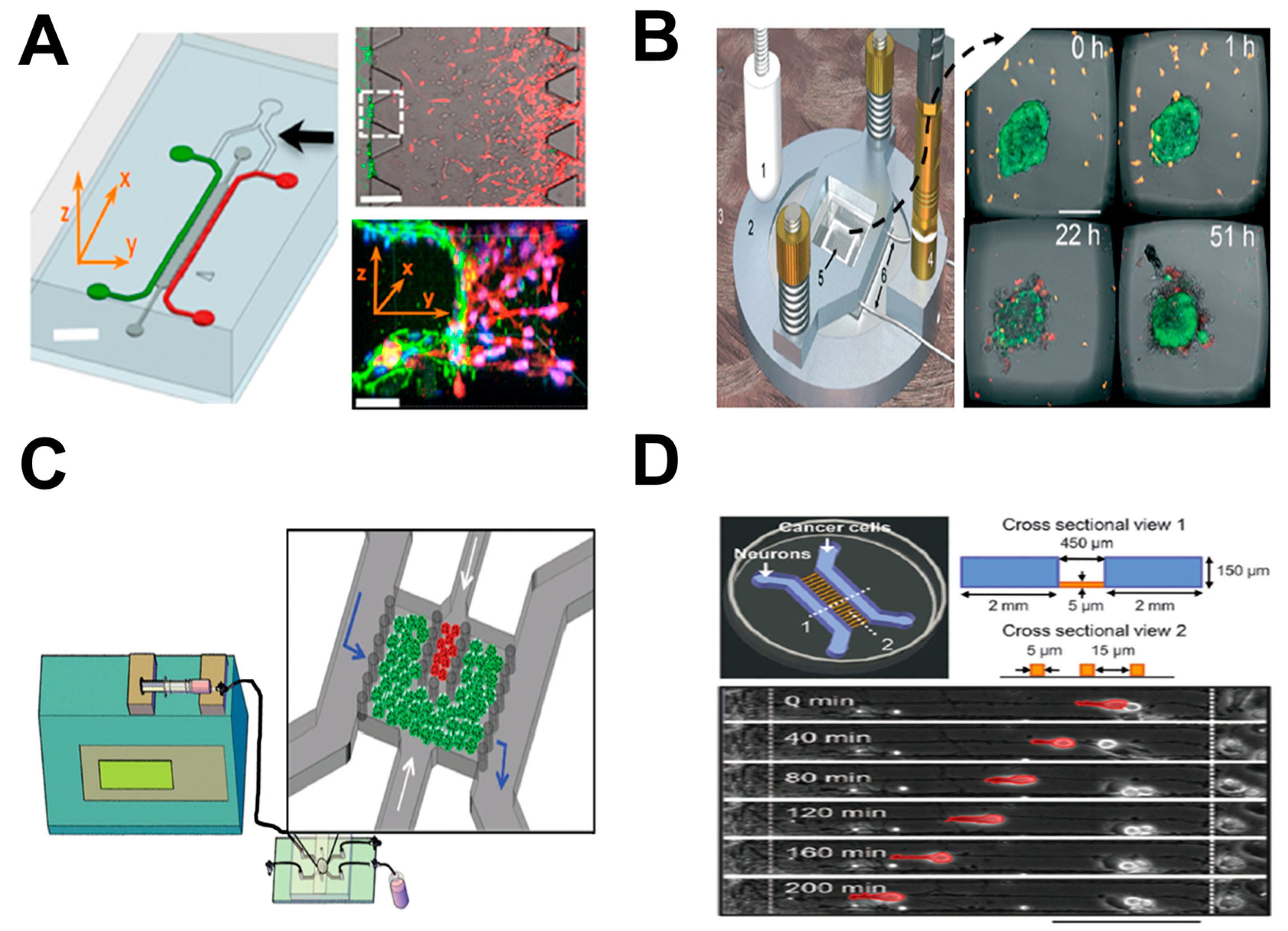
2. Tumor-on-Chip
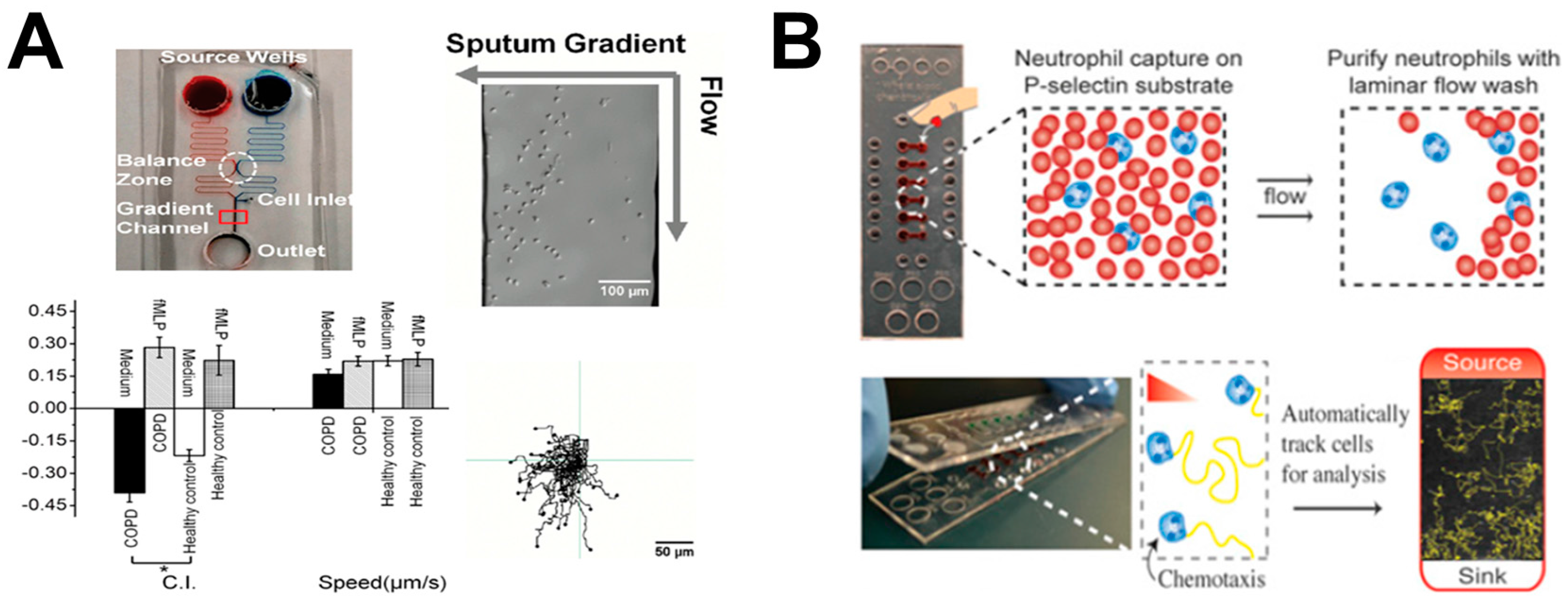
3. Lung-on-Chip
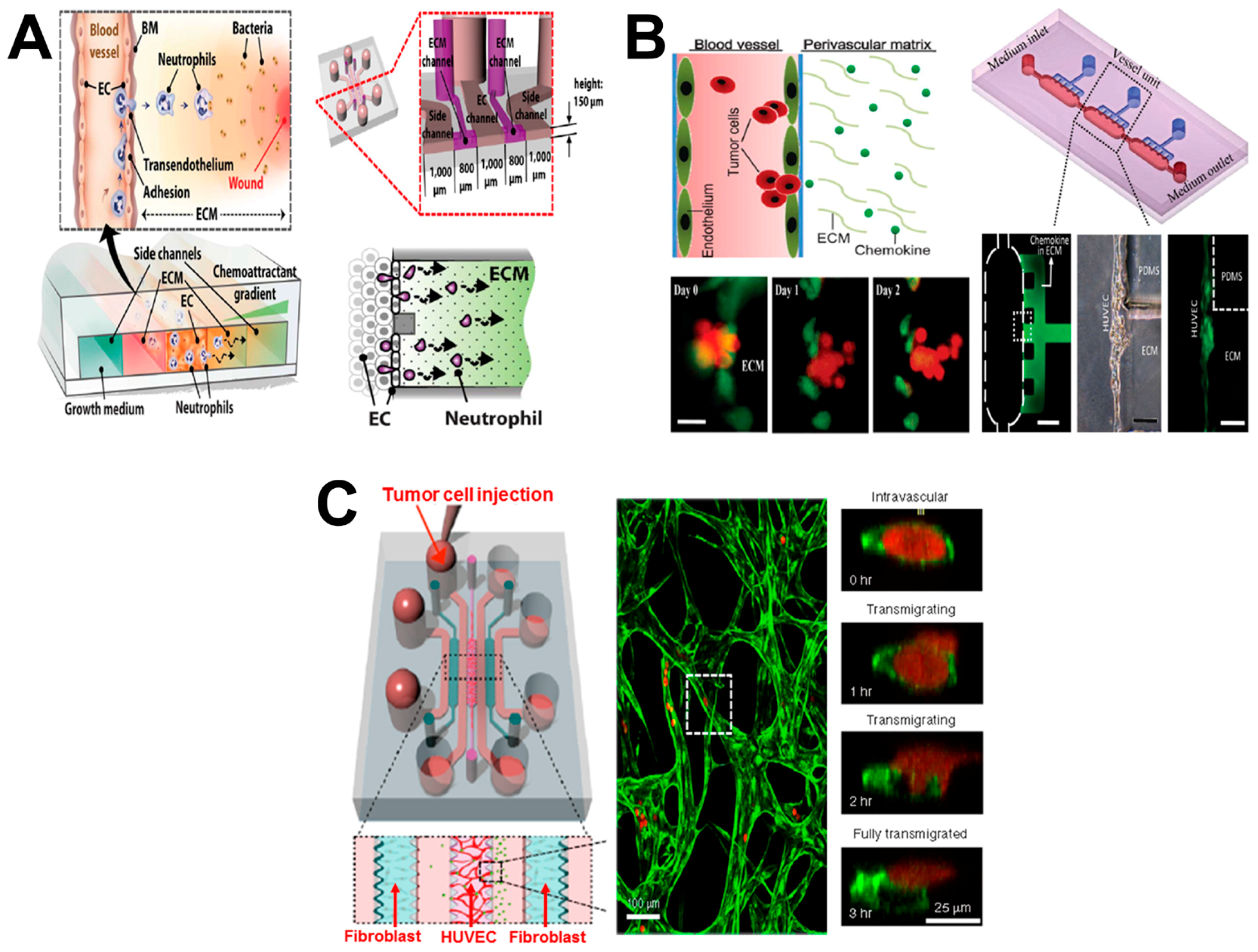
4. Vessel-on-Chip
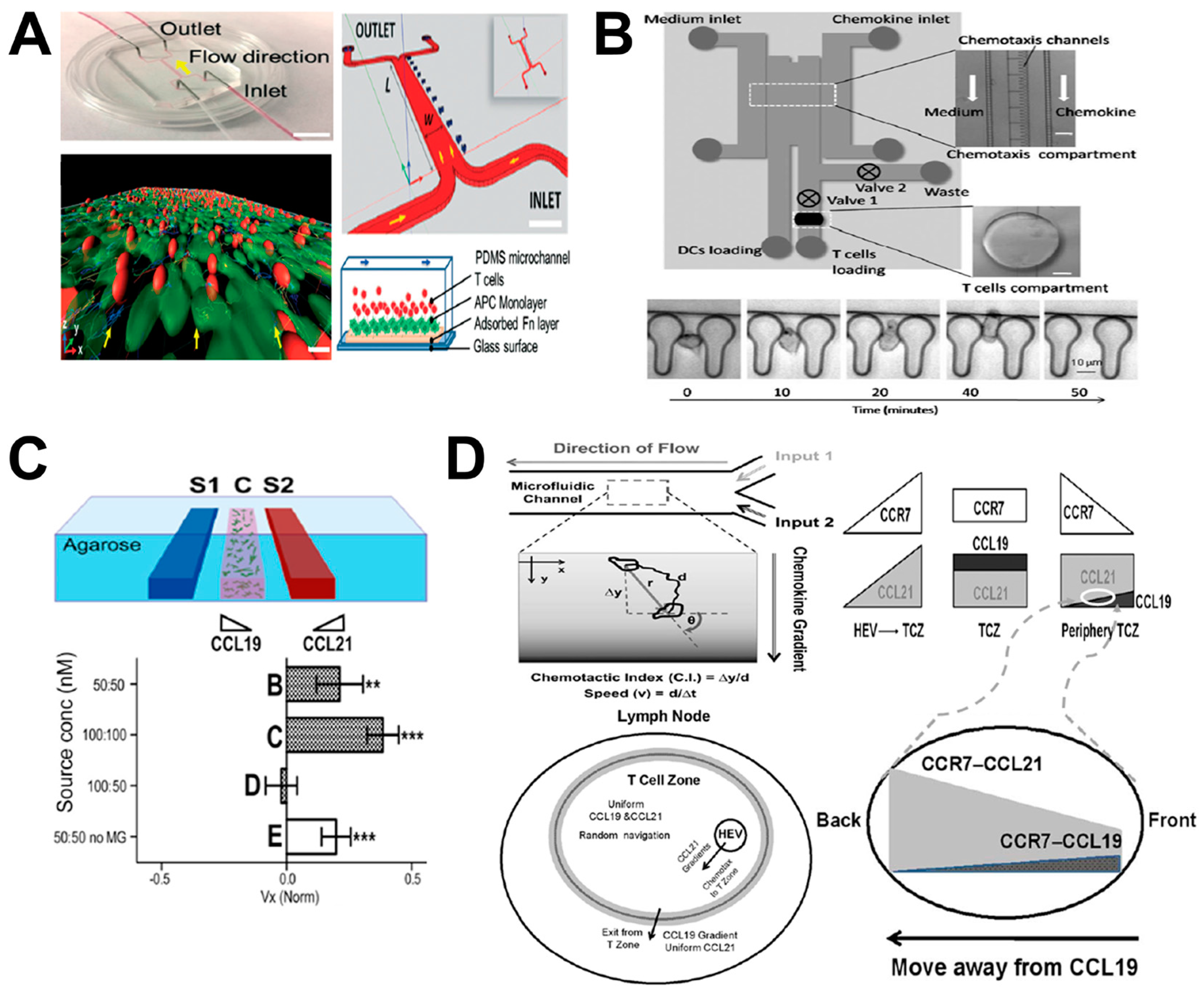
5. Lymph Node (LN)-on-Chip
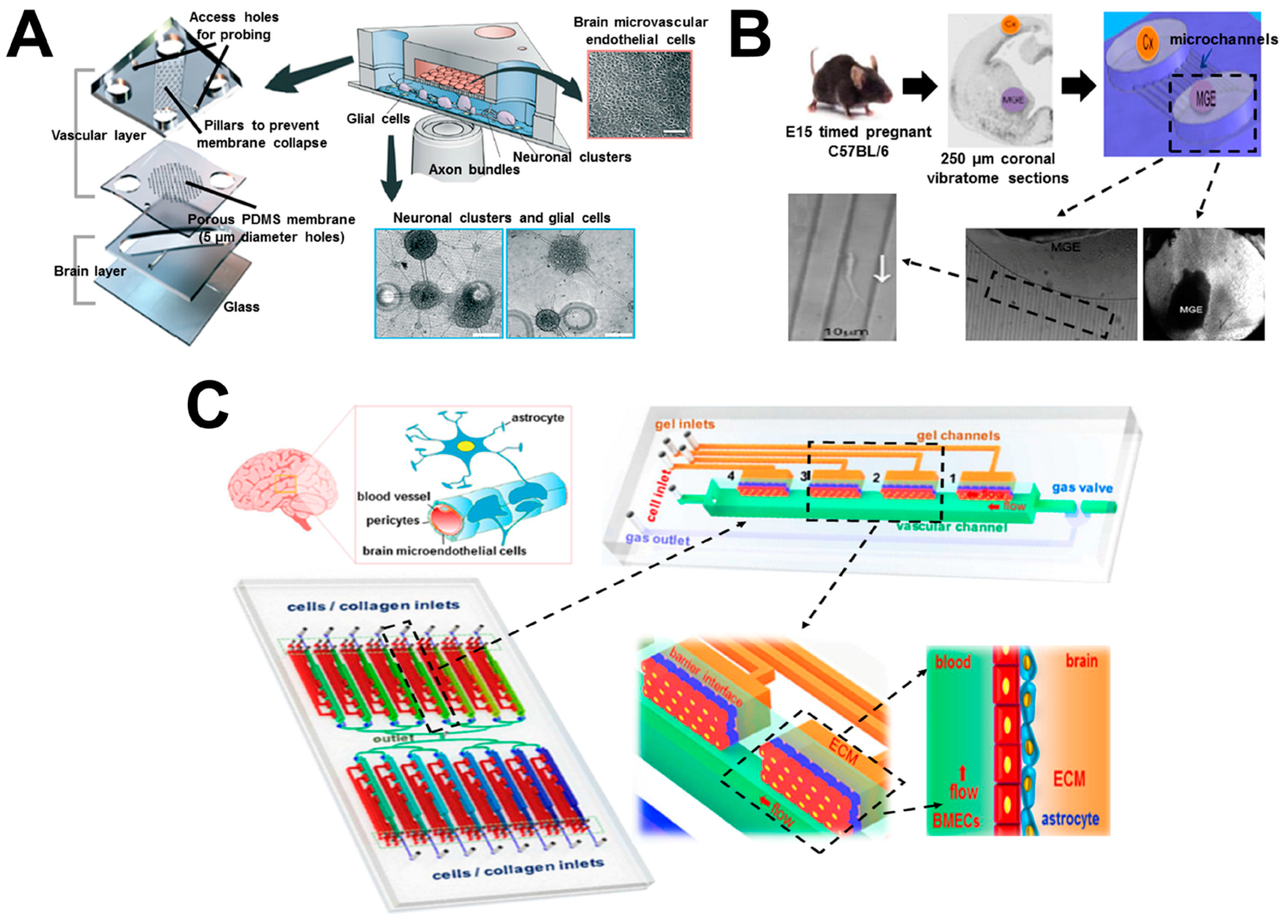
6. Brain-on-Chip
7. Conclusions and Outlook
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| 3D | three dimensional |
| ACC | adenoid cystic carcinoma |
| aHPBTs | activated human peripheral blood T cells |
| APC | antigen presenting cell |
| BBB | blood–brain barrier |
| BME | basement membrane extract |
| CAF-μTP | cancer–activated fibroblast microtissues |
| CNS | central nervous system |
| COPD | chronic obstructive pulmonary disease |
| DC | dendritic cell |
| DRG | dorsal root ganglion |
| EC | endothelial cell |
| ECM | extracellular matrices |
| FB | fibroblast |
| FGF-2 | fibroblast growth factor 2 |
| fMLP | N-formyl-methionyl-leucyl-phenylalanine |
| FN | fibronectin |
| hBMECs | human brain microvascular endothelial cells |
| HCC | hepatocellular carcinoma |
| HEV | high endothelial venule |
| hNPC | human fetal neural progenitor cell |
| hNSC | human neural stem cell |
| HUVEC | human umbilical vein endothelial cell |
| ICAM-1 | intercellular adhesion molecule 1 |
| iDC | immature DC |
| IL-8 | interleukin-8 |
| LN | lymph node |
| LPS | lipopolysaccharide |
| mDC | mature DC |
| MGE | medial ganglionic eminence |
| NF-μTP | normal fibroblast microtissues |
| NK | natural killer |
| NPC | neural progenitor cell |
| PDMS | polydimethylsiloxane |
| pMHC | peptide-major histocompatibility complex |
| PNI | perineural invasion |
| TCR | T cell receptor |
| TCZ | T cell zone |
| TEI | transendothelial invasion |
| TEM | transendothelial migration |
| TNF-α | tumor necrosis factor alpha |
| TVM | transvascular migration |
| μTP | microtissues |
| μVNs | microvascular networks |
References
- Dupre, L.; Houmadi, R.; Tang, C.; Rey-Barroso, J. T lymphocyte migration: An action movie starring the actin and associated actors. Front. Immunol. 2015, 6, 586. [Google Scholar] [CrossRef] [PubMed]
- Behar, T.N.; Schaffner, A.E.; Colton, C.A.; Somogyi, R.; Olah, Z.; Lehel, C.; Barker, J.L. Gaba-induced chemokinesis and NGF-induced chemotaxis of embryonic spinal cord neurons. J. Neurosci. 1994, 14, 29. [Google Scholar] [PubMed]
- Brinkman, C.; Peske, J.; Engelhard, V. Peripheral tissue homing receptor control of naïve, effector, and memory CD8 t cell localization in lymphoid and non-lymphoid tissues. Front. Immunol. 2013, 4. [Google Scholar] [CrossRef]
- Gurdon, J.B.; Bourillot, P.Y. Morphogen gradient interpretation. Nature 2001, 413, 797–803. [Google Scholar] [CrossRef] [PubMed]
- Nandagopal, S.; Wu, D.; Lin, F. Combinatorial guidance by ccr7 ligands for t lymphocytes migration in co-existing chemokine fields. PLoS ONE 2011, 6, e18183. [Google Scholar] [CrossRef]
- Askari, A.T.; Unzek, S.; Popovic, Z.B.; Goldman, C.K.; Forudi, F.; Kiedrowski, M.; Rovner, A.; Ellis, S.G.; Thomas, J.D.; DiCorleto, P.E. Effect of stromal-cell-derived factor 1 on stem-cell homing and tissue regeneration in ischaemic cardiomyopathy. Lancet 2003, 362, 697–703. [Google Scholar] [CrossRef]
- Maness, P.F.; Schachner, M. Neural recognition molecules of the immunoglobulin superfamily: Signaling transducers of axon guidance and neuronal migration. Nat. Neurosci. 2007, 10, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Müller, A.; Homey, B.; Soto, H.; Ge, N.; Catron, D.; Buchanan, M.E.; McClanahan, T.; Murphy, E.; Yuan, W.; Wagner, S.N. Involvement of chemokine receptors in breast cancer metastasis. Nature 2001, 410, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Butcher, E.C. T cell chemotaxis in a simple microfluidic device. Lab Chip 2006, 6, 1462–1469. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Baldessari, F.; Gyenge, C.C.; Sato, T.; Chambers, R.D.; Santiago, J.G.; Butcher, E.C. Lymphocyte electrotaxis in vitro and in vivo. J. Immunol. 2008, 181, 2465–2471. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lin, F. Microfluidic devices for studying chemotaxis and electrotaxis. Trends Cell Biol. 2011, 21, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Menon, S.; Beningo, K.A. Cancer cell invasion is enhanced by applied mechanical stimulation. PLoS ONE 2011, 6, e17277. [Google Scholar] [CrossRef] [PubMed]
- Diamond, M.S.; Springer, T.A. The dynamic regulation of integrin adhesiveness. Curr. Biol. 1994, 4, 506–517. [Google Scholar] [CrossRef]
- Johnston, B.; Butcher, E.C. Chemokines in rapid leukocyte adhesion triggering and migration. Semin. Immunol. 2002, 14, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, M.; Bianchi, E.; Fumagalli, L.; Pardi, R. Regulation of lymphocyte traffic by adhesion molecules. Inflamm. Res. 1999, 48, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Ward, E.J.; Marelli-Berg, F.M. Mechanisms of t cell organotropism. Cell. Mol. Life Sci. 2016, 73, 3009–3033. [Google Scholar] [CrossRef] [PubMed]
- Yum, K.; Hong, S.G.; Healy, K.E.; Lee, L.P. Physiologically relevant organs on chips. Biotechnol. J. 2014, 9, 16–27. [Google Scholar] [CrossRef] [PubMed]
- van den Broek, L.J.; Bergers, L.I.J.C.; Reijnders, C.M.A.; Gibbs, S. Progress and future prospectives in skin-on-chip development with emphasis on the use of different cell types and technical challenges. Stem Cell Rev. Rep. 2017, 13, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Seok, J.; Warren, H.S.; Cuenca, A.G.; Mindrinos, M.N.; Baker, H.V.; Xu, W.; Richards, D.R.; McDonald-Smith, G.P.; Gao, H.; Hennessy, L.; et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. USA 2013, 110, 3507–3512. [Google Scholar] [CrossRef] [PubMed]
- Mak, I.W.Y.; Evaniew, N.; Ghert, M. Lost in translation: Animal models and clinical trials in cancer treatment. Am. J. Transl. Res. 2014, 6, 114–118. [Google Scholar] [PubMed]
- Boyden, S. The chemotactic effect of mixtures of antibody and antigen on polymorphonuclear leucocytes. J. Exp. Med. 1962, 115, 453–466. [Google Scholar] [CrossRef] [PubMed]
- van der Meer, A.D.; Poot, A.A.; Duits, M.H.G.; Feijen, J.; Vermes, I. Microfluidic technology in vascular research. J. Biomed. Biotechnol. 2009, 2009, 823148. [Google Scholar] [CrossRef] [PubMed]
- Velve-Casquillas, G.; Le Berre, M.; Piel, M.; Tran, P.T. Microfluidic tools for cell biological research. Nano Today 2010, 5, 28–47. [Google Scholar] [CrossRef] [PubMed]
- Lin, F. Chapter 15—A microfluidics-based method for analyzing leukocyte migration to chemoattractant gradients. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 2009; Volume 461, pp. 333–347. [Google Scholar]
- Wu, J.; Wu, X.; Lin, F. Recent developments in microfluidics-based chemotaxis studies. Lab Chip 2013, 13, 2484–2499. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Hillier, C.; Komenda, P.; Lobato de Faria, R.; Levin, D.; Zhang, M.; Lin, F. A microfluidic platform for evaluating neutrophil chemotaxis induced by sputum from copd patients. PLoS ONE 2015, 10, e0126523. [Google Scholar] [CrossRef] [PubMed]
- Sackmann, E.K.-H.; Berthier, E.; Schwantes, E.A.; Fichtinger, P.S.; Evans, M.D.; Dziadzio, L.L.; Huttenlocher, A.; Mathur, S.K.; Beebe, D.J. Characterizing asthma from a drop of blood using neutrophil chemotaxis. Proc. Natl. Acad. Sci. USA 2014, 111, 5813–5818. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, S.N.; Ingber, D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014, 32, 760–772. [Google Scholar] [CrossRef] [PubMed]
- Zheng, F.; Fu, F.; Cheng, Y.; Wang, C.; Zhao, Y.; Gu, Z. Organ-on-a-chip systems: Microengineering to biomimic living systems. Small 2016, 12, 2253–2282. [Google Scholar] [CrossRef] [PubMed]
- Khademhosseini, A. Micro and Nanoengineering of the Cell Microenvironment: Technologies and Applications; Artech House: Norwood, MA, USA, 2008. [Google Scholar]
- Lee, P.J.; Hung, P.J.; Lee, L.P. An artificial liver sinusoid with a microfluidic endothelial-like barrier for primary hepatocyte culture. Biotechnol. Bioeng. 2007, 97, 1340–1346. [Google Scholar] [CrossRef] [PubMed]
- Vernetti, L.A.; Senutovitch, N.; Boltz, R.; DeBiasio, R.; Ying Shun, T.; Gough, A.; Taylor, D.L. A human liver microphysiology platform for investigating physiology, drug safety, and disease models. Exp. Biol. Med. 2016, 241, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Jang, K.-J.; Mehr, A.P.; Hamilton, G.A.; McPartlin, L.A.; Chung, S.; Suh, K.-Y.; Ingber, D.E. Human kidney proximal tubule-on-a-chip for drug transport and nephrotoxicity assessment. Integr. Biol. 2013, 5, 1119–1129. [Google Scholar] [CrossRef] [PubMed]
- Wufuer, M.; Lee, G.; Hur, W.; Jeon, B.; Kim, B.J.; Choi, T.H.; Lee, S. Skin-on-a-chip model simulating inflammation, edema and drug-based treatment. Sci. Rep. 2016, 6, 37471. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Yan, J.-J.; Shin, Y.; Jeon, J.J.; Won, J.; Eun Jeong, H.; Kamm, R.D.; Kim, Y.-J.; Chung, S. A versatile assay for monitoring in vivo-like transendothelial migration of neutrophils. Lab Chip 2012, 12, 3861–3865. [Google Scholar] [CrossRef] [PubMed]
- Zervantonakis, I.K.; Hughes-Alford, S.K.; Charest, J.L.; Condeelis, J.S.; Gertler, F.B.; Kamm, R.D. Three-dimensional microfluidic model for tumor cell intravasation and endothelial barrier function. Proc. Natl. Acad. Sci. USA 2012, 109, 13515–13520. [Google Scholar] [CrossRef] [PubMed]
- Christakou, A.E.; Ohlin, M.; Onfelt, B.; Wiklund, M. Ultrasonic three-dimensional on-chip cell culture for dynamic studies of tumor immune surveillance by natural killer cells. Lab Chip 2015, 15, 3222–3231. [Google Scholar] [CrossRef] [PubMed]
- Gioiella, F.; Urciuolo, F.; Imparato, G.; Brancato, V.; Netti, P.A. An engineered breast cancer model on a chip to replicate ecm-activation in vitro during tumor progression. Adv. Healthc. Mater. 2016, 5, 3074–3084. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Li, J.; Wang, N.; Yang, X.; Hamada, Y.; Li, Q.; Zheng, W.; Jiang, X. An on-chip model for investigating the interaction between neurons and cancer cells. Integr. Biol. 2016, 8, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Huh, D.; Matthews, B.D.; Mammoto, A.; Montoya-Zavala, M.; Hsin, H.Y.; Ingber, D.E. Reconstituting organ-level lung functions on a chip. Science 2010, 328, 1662. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Wu, J.; Zhu, L.; Liu, Y.; Zhang, M.; Lin, F. An all-on-chip method for rapid neutrophil chemotaxis analysis directly from a drop of blood. J. Vis. Exp. JoVE 2017. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Liu, T.; Qin, J. A microfluidic-based device for study of transendothelial invasion of tumor aggregates in realtime. Lab Chip 2012, 12, 2837–2842. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.B.; Whisler, J.A.; Fröse, J.; Yu, C.; Shin, Y.; Kamm, R.D. On-chip human microvasculature assay for visualization and quantitation of tumor cell extravasation dynamics. Nat. Protoc. 2017, 12, 865. [Google Scholar] [CrossRef] [PubMed]
- Tourovskaia, A.; Fauver, M.; Kramer, G.; Simonson, S.; Neumann, T. Brief communication: Tissue-engineered microenvironment systems for modeling human vasculature. Exp. Biol. Med. 2014, 239, 1264–1271. [Google Scholar] [CrossRef] [PubMed]
- Moura Rosa, P.; Gopalakrishnan, N.; Ibrahim, H.; Haug, M.; Halaas, O. The intercell dynamics of t cells and dendritic cells in a lymph node-on-a-chip flow device. Lab Chip 2016, 16, 3728–3740. [Google Scholar] [CrossRef] [PubMed]
- Mitra, B.; Jindal, R.; Lee, S.; Dong, D.X.; Li, L.; Sharma, N.; Maguire, T.; Schloss, R.; Yarmush, M.L. Microdevice integrating innate and adaptive immune responses associated with antigen presentation by dendritic cells. RSC Adv. 2013, 3, 16002–16010. [Google Scholar] [CrossRef]
- Haessler, U.; Pisano, M.; Wu, M.; Swartz, M.A. Dendritic cell chemotaxis in 3D under defined chemokine gradients reveals differential response to ligands CCL21 and CCL19. Proc. Natl. Acad. Sci. USA 2011, 108, 5614–5619. [Google Scholar] [CrossRef] [PubMed]
- Ricart, B.G.; John, B.; Lee, D.; Hunter, C.A.; Hammer, D.A. Dendritic cells distinguish individual chemokine signals through CCR7 and CXCR4. J. Immunol. 2010, 186, 53. [Google Scholar] [CrossRef] [PubMed]
- Kilic, O.; Pamies, D.; Lavell, E.; Schiapparelli, P.; Feng, Y.; Hartung, T.; Bal-Price, A.; Hogberg, H.T.; Quinones-Hinojosa, A.; Guerrero-Cazares, H.; et al. Brain-on-a-chip model enables analysis of human neuronal differentiation and chemotaxis. Lab Chip 2016, 16, 4152–4162. [Google Scholar] [CrossRef] [PubMed]
- Nery, F.C.; da Hora, C.C.; Yaqub, U.; Zhang, X.; McCarthy, D.M.; Bhide, P.G.; Irimia, D.; Breakefield, X.O. New methods for investigation of neuronal migration in embryonic brain explants. J. Neurosci. Methods 2015, 239, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Keenan, T.M.; Grinager, J.R.; Procak, A.A.; Svendsen, C.N. In vitro localization of human neural stem cell neurogenesis by engineered FGF-2 gradients. Integr. Biol. 2012, 4, 1522–1531. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Li, Z.; Yu, Y.; Sizdahkhani, S.; Ho, W.S.; Yin, F.; Wang, L.; Zhu, G.; Zhang, M.; Jiang, L.; et al. A dynamic in vivo-like organotypic blood-brain barrier model to probe metastatic brain tumors. Sci. Rep. 2016, 6, 36670. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.X.; Bos, P.D.; Massagué, J. Metastasis: From dissemination to organ-specific colonization. Nat. Rev. Cancer 2009, 9, 274. [Google Scholar] [CrossRef] [PubMed]
- Steeg, P.S. Tumor metastasis: Mechanistic insights and clinical challenges. Nat. Med. 2006, 12, 895. [Google Scholar] [CrossRef] [PubMed]
- Friedl, P.; Alexander, S. Cancer invasion and the microenvironment: Plasticity and reciprocity. Cell 2011, 147, 992–1009. [Google Scholar] [CrossRef] [PubMed]
- Calvo, F.; Sahai, E. Cell communication networks in cancer invasion. Curr. Opin. Cell Biol. 2011, 23, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Roussos, E.T.; Condeelis, J.S.; Patsialou, A. Chemotaxis in cancer. Nat. Rev. Cancer 2011, 11, 573–587. [Google Scholar] [CrossRef] [PubMed]
- Gupta, G.P.; Massagué, J. Cancer metastasis: Building a framework. Cell 2006, 127, 679–695. [Google Scholar] [CrossRef] [PubMed]
- Pavesi, A.; Tan, A.T.; Koh, S.; Chia, A.; Colombo, M.; Antonecchia, E.; Miccolis, C.; Ceccarello, E.; Adriani, G.; Raimondi, M.T.; et al. A 3D microfluidic model for preclinical evaluation of tcr-engineered t cells against solid tumors. JCI Insight 2017, 2. [Google Scholar] [CrossRef] [PubMed]
- Bapat, A.A.; Hostetter, G.; Von Hoff, D.D.; Han, H. Perineural invasion and associated pain in pancreatic cancer. Nat. Rev. Cancer 2011, 11, 695–707. [Google Scholar] [CrossRef] [PubMed]
- Kolaczkowska, E.; Kubes, P. Neutrophil recruitment and function in health and inflammation. Nat. Rev. Immunol. 2013, 13, 159. [Google Scholar] [CrossRef] [PubMed]
- Fahy, J.V. Eosinophilic and neutrophilic inflammation in asthma: Insights from clinical studies. Proc. Am. Thorac. Soc. 2009, 6, 256–259. [Google Scholar] [CrossRef] [PubMed]
- Amulic, B.; Cazalet, C.; Hayes, G.L.; Metzler, K.D.; Zychlinsky, A. Neutrophil function: From mechanisms to disease. Annu. Rev. Immunol. 2012, 30, 459–489. [Google Scholar] [CrossRef] [PubMed]
- Turato, G.; Zuin, R.; Saetta, M. Pathogenesis and pathology of copd. Respiration 2001, 68, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, C.; Yoneda, T.; Yoshikawa, M.; Fu, A.; Tokayama, T.; Tsukaguchi, K.; Narita, N. Airway inflammation in copd assessed by sputum levels of lnterleukin-8. Chest 1997, 112, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Middleton, J.; Patterson, A.M.; Gardner, L.; Schmutz, C.; Ashton, B.A. Leukocyte extravasation: Chemokine transport and presentation by the endothelium. Blood 2002, 100, 3853–3860. [Google Scholar] [CrossRef] [PubMed]
- Woodfin, A.; Voisin, M.-B.; Imhof, B.A.; Dejana, E.; Engelhardt, B.; Nourshargh, S. Endothelial cell activation leads to neutrophil transmigration as supported by the sequential roles of ICAM-2, JAM-A, and PECAM-1. Blood 2009, 113, 6246–6257. [Google Scholar] [CrossRef] [PubMed]
- Miles, F.L.; Pruitt, F.L.; van Golen, K.L.; Cooper, C.R. Stepping out of the flow: Capillary extravasation in cancer metastasis. Clin. Exp. Metastasis 2008, 25, 305–324. [Google Scholar] [CrossRef] [PubMed]
- Randolph, G.J.; Angeli, V.; Swartz, M.A. Dendritic-cell trafficking to lymph nodes through lymphatic vessels. Nat. Rev. Immunol. 2005, 5, 617. [Google Scholar] [CrossRef] [PubMed]
- Banchereau, J.; Steinman, R.M. Dendritic cells and the control of immunity. Nature 1998, 392, 245. [Google Scholar] [CrossRef] [PubMed]
- Lanzavecchia, A.; Sallusto, F. Dynamics of t lymphocyte responses: Intermediates, effectors, and memory cells. Science 2000, 290, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Quaratino, S.; Duddy, L.P.; Londei, M. Fully competent dendritic cells as inducers of t cell anergy in autoimmunity. Proc. Natl. Acad. Sci. USA 2000, 97, 10911–10916. [Google Scholar] [CrossRef] [PubMed]
- Sallusto, F.; Palermo, B.; Lenig, D.; Miettinen, M.; Matikainen, S.; Julkunen, I.; Forster, R.; Burgstahler, R.; Lipp, M.; Lanzavecchia, A. Distinct patterns and kinetics of chemokine production regulate dendritic cell function. Eur. J. Immunol. 1999, 29, 1617–1625. [Google Scholar] [CrossRef]
- Kim, C.H.; Broxmeyer, H.E. Chemokines: Signal lamps for trafficking of t and b cells for development and effector function. J. Leukoc. Biol. 1999, 65, 6–15. [Google Scholar] [PubMed]
- Wang, Y.; Li, G.; Stanco, A.; Long, J.E.; Crawford, D.; Potter, G.B.; Pleasure, S.J.; Behrens, T.; Rubenstein, J.L. Cxcr4 and cxcr7 have distinct functions in regulating interneuron migration. Neuron 2011, 69, 61–76. [Google Scholar] [CrossRef] [PubMed]
- Stumm, R.K.; Rummel, J.; Junker, V.; Culmsee, C.; Pfeiffer, M.; Krieglstein, J.; Höllt, V.; Schulz, S. A dual role for the SDF-1/CXCR4 chemokine receptor system in adult brain: Isoform-selective regulation of SDF-1 expression modulates CXCR4-dependent neuronal plasticity and cerebral leukocyte recruitment after focal ischemia. J. Neurosci. 2002, 22, 5865–5878. [Google Scholar] [PubMed]
- Zagzag, D.; Esencay, M.; Mendez, O.; Yee, H.; Smirnova, I.; Huang, Y.; Chiriboga, L.; Lukyanov, E.; Liu, M.; Newcomb, E.W. Hypoxia-and vascular endothelial growth factor-induced stromal cell-derived factor-1α/CXCR4 expression in glioblastomas: One plausible explanation of scherer’s structures. Am. J. Pathol. 2008, 173, 545–560. [Google Scholar] [CrossRef] [PubMed]
- Neufurth, M.; Wang, X.; Wang, S.; Steffen, R.; Ackermann, M.; Haep, N.D.; Schröder, H.C.; Müller, W.E.G. 3D printing of hybrid biomaterials for bone tissue engineering: Calcium-polyphosphate microparticles encapsulated by polycaprolactone. Acta Biomater. 2017. [Google Scholar] [CrossRef] [PubMed]
- Hixon, K.R.; Melvin, A.M.; Lin, A.Y.; Hall, A.F.; Sell, S.A. Cryogel scaffolds from patient-specific 3D-printed molds for personalized tissue-engineered bone regeneration in pediatric cleft-craniofacial defects. J. Biomater. Appl. 2017. [Google Scholar] [CrossRef] [PubMed]
- Gregor, A.; Filová, E.; Novák, M.; Kronek, J.; Chlup, H.; Buzgo, M.; Blahnová, V.; Lukášová, V.; Bartoš, M.; Nečas, A. Designing of pla scaffolds for bone tissue replacement fabricated by ordinary commercial 3D printer. J. Biol. Eng. 2017, 11, 31. [Google Scholar] [CrossRef] [PubMed]
- Domansky, K.; Leslie, D.C.; McKinney, J.; Fraser, J.P.; Sliz, J.D.; Hamkins-Indik, T.; Hamilton, G.A.; Bahinski, A.; Ingber, D.E. Clear castable polyurethane elastomer for fabrication of microfluidic devices. Lab Chip 2013, 13, 3956–3964. [Google Scholar] [CrossRef] [PubMed]
| Organ Type | Comments | Ref. |
|---|---|---|
| Tumor-on-chip | Investigating endothelial barrier function during tumor cell intravasation; | [36] |
| Investigating immune surveillance of natural killer (NK) cells for tumors; | [37] | |
| Investigating ECM activation during tumor progression; | [38] | |
| Investigating the interactions between neurons and cancer cells during tumor perineural invasion. | [39] | |
| Lung-on-chip | Investigating bacteria or inflammatory cytokine induced cell migration; | [40] |
| Investigating neutrophil chemotaxis with clinical samples for diagnosis of chronic obstructive pulmonary disease (COPD); | [26] | |
| Rapid analysis of neutrophil chemotaxis; | [41] | |
| Investigating neutrophil chemotaxis with clinical samples for asthma detection. | [27] | |
| Vessel-on-chip | Investigating neutrophil transendothelial migration (TEM) during inflammatory process; | [35] |
| Investigating tumor transendothelial invasion (TEI); | [42] | |
| Investigating tumor cell extravasation; | [43] | |
| Investigating angiogenesis. | [44] | |
| Lymph node-on-chip | Investigating the interaction between T cells and dendritic cells (DCs); | [45] |
| Evaluating DC chemotaxis and DC–T cell interaction; | [46] | |
| Investigating differential chemotaxis of DCs to CCL21 and CCL19; | [47] | |
| Studying the guidance of CCR7 ligands for T cell migration in Lymph Nodes (LNs); | [5] | |
| Investigating differential chemotaxis of DCs through CCR7 and CXCR4 signaling. | [48] | |
| Brain-on-chip | Investigating neuronal differentiation and chemotaxis; | [49] |
| Investigating neuronal migration; | [50] | |
| Investigating human neural stem cell (hNSC) neurogenesis; | [51] | |
| Investigating brain tumor metastasis. | [52] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, X.; Levin, D.; Lin, F. Cell Migration Research Based on Organ-on-Chip-Related Approaches. Micromachines 2017, 8, 324. https://doi.org/10.3390/mi8110324
Ren X, Levin D, Lin F. Cell Migration Research Based on Organ-on-Chip-Related Approaches. Micromachines. 2017; 8(11):324. https://doi.org/10.3390/mi8110324
Chicago/Turabian StyleRen, Xiaoou, David Levin, and Francis Lin. 2017. "Cell Migration Research Based on Organ-on-Chip-Related Approaches" Micromachines 8, no. 11: 324. https://doi.org/10.3390/mi8110324





