Methods of Micropatterning and Manipulation of Cells for Biomedical Applications
Abstract
:1. Introduction
2. Techniques and Methods
2.1. Physical Cell Patterning
2.1.1. Inkjet Cell Printing
2.1.2. Optical and Optoelectronic Tweezers
2.1.3. Laser-Based Cell Patterning
2.1.4. Acoustic Force Patterning
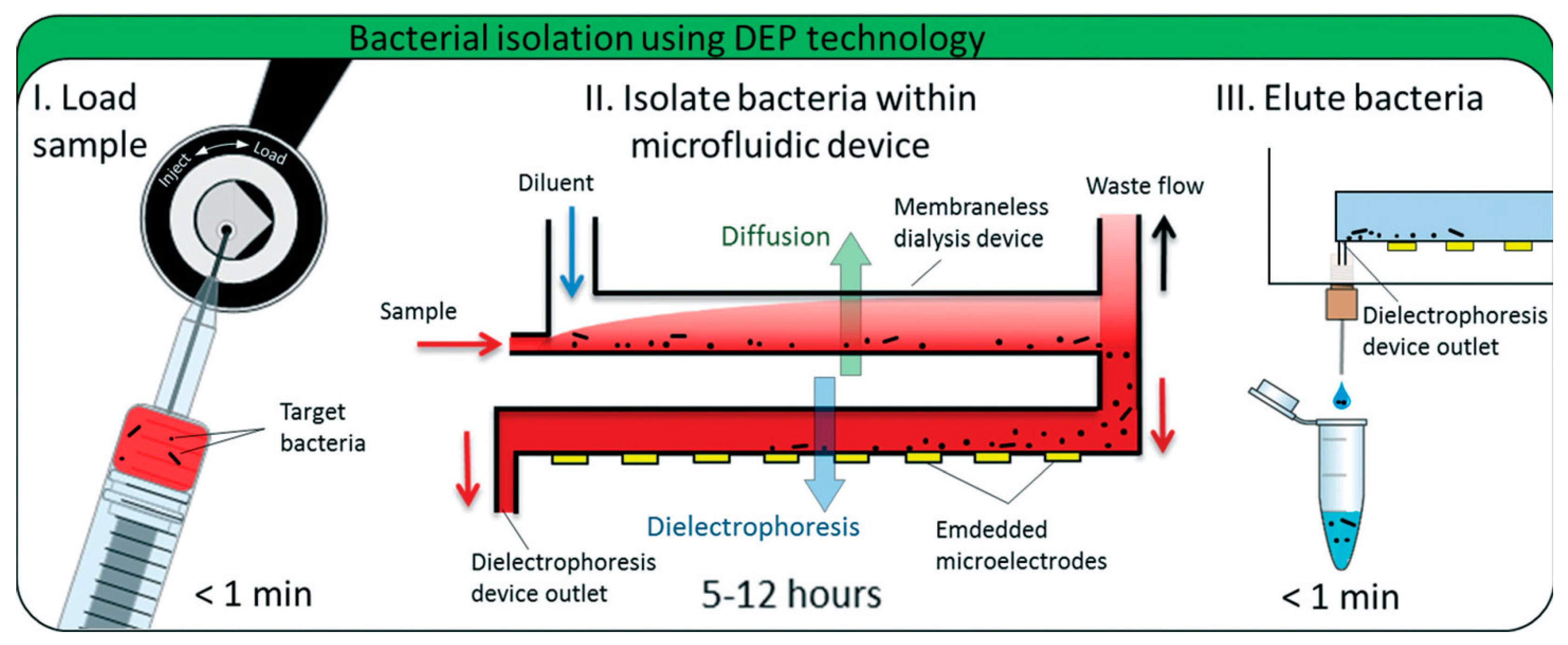
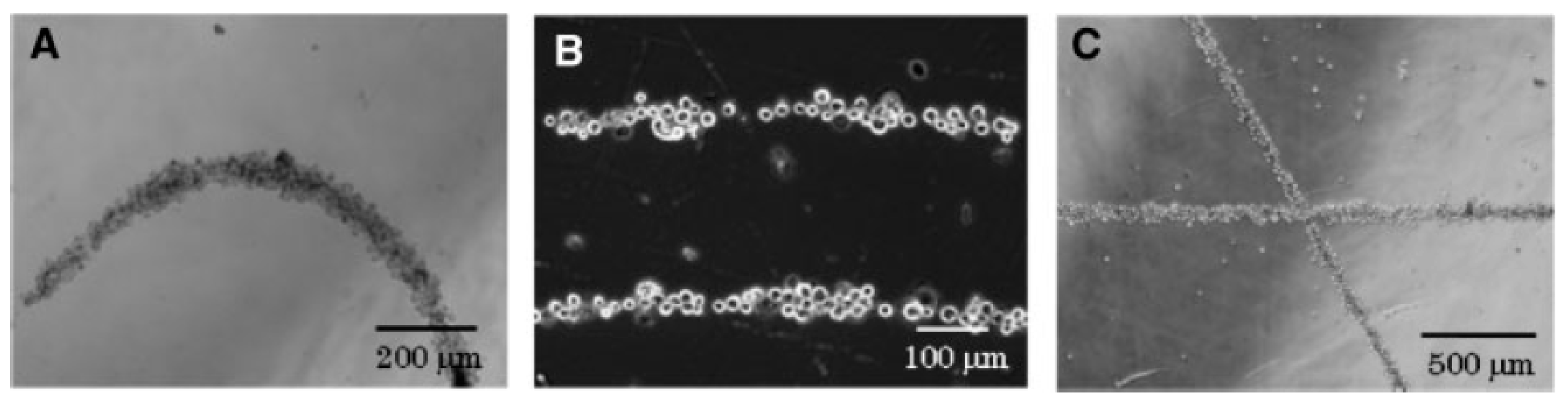
2.1.5. Electrokinetic Forces (Dielectrophoresis)
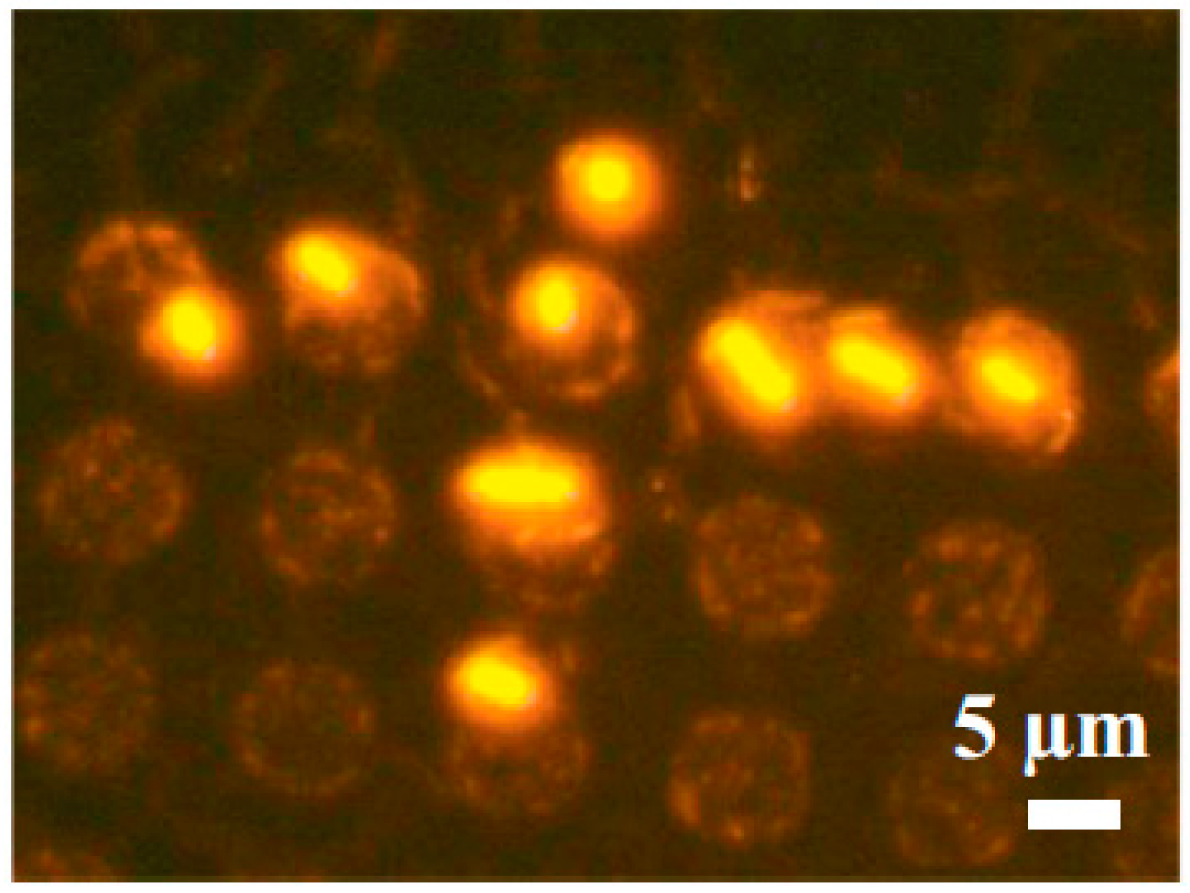
2.1.6. Magnetic Cell Manipulation
2.2. Chemical Patterning for Cells Assembly
Surface Chemistry Methodologies
2.3. Physical and Chemical Patterning
2.3.1. Microcontact Printing Overview
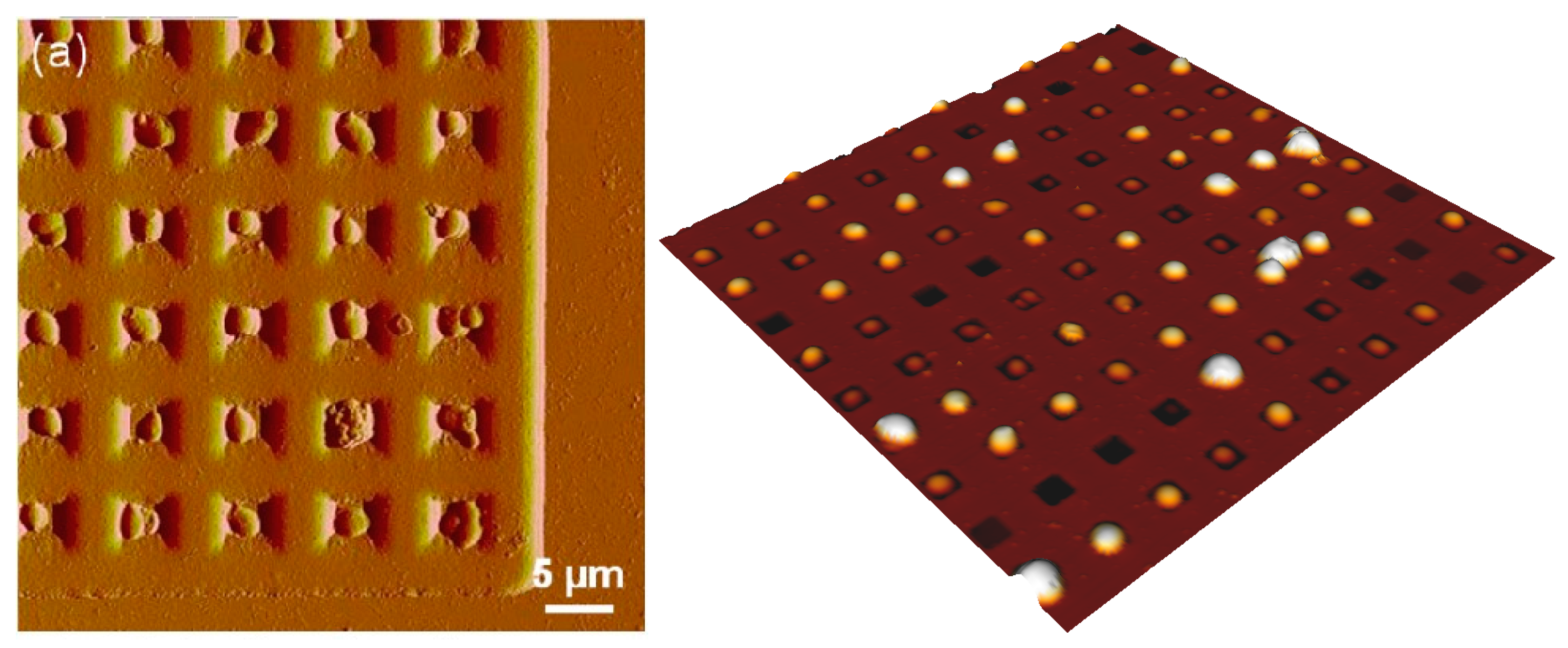
2.3.2. Microwells and Filtration
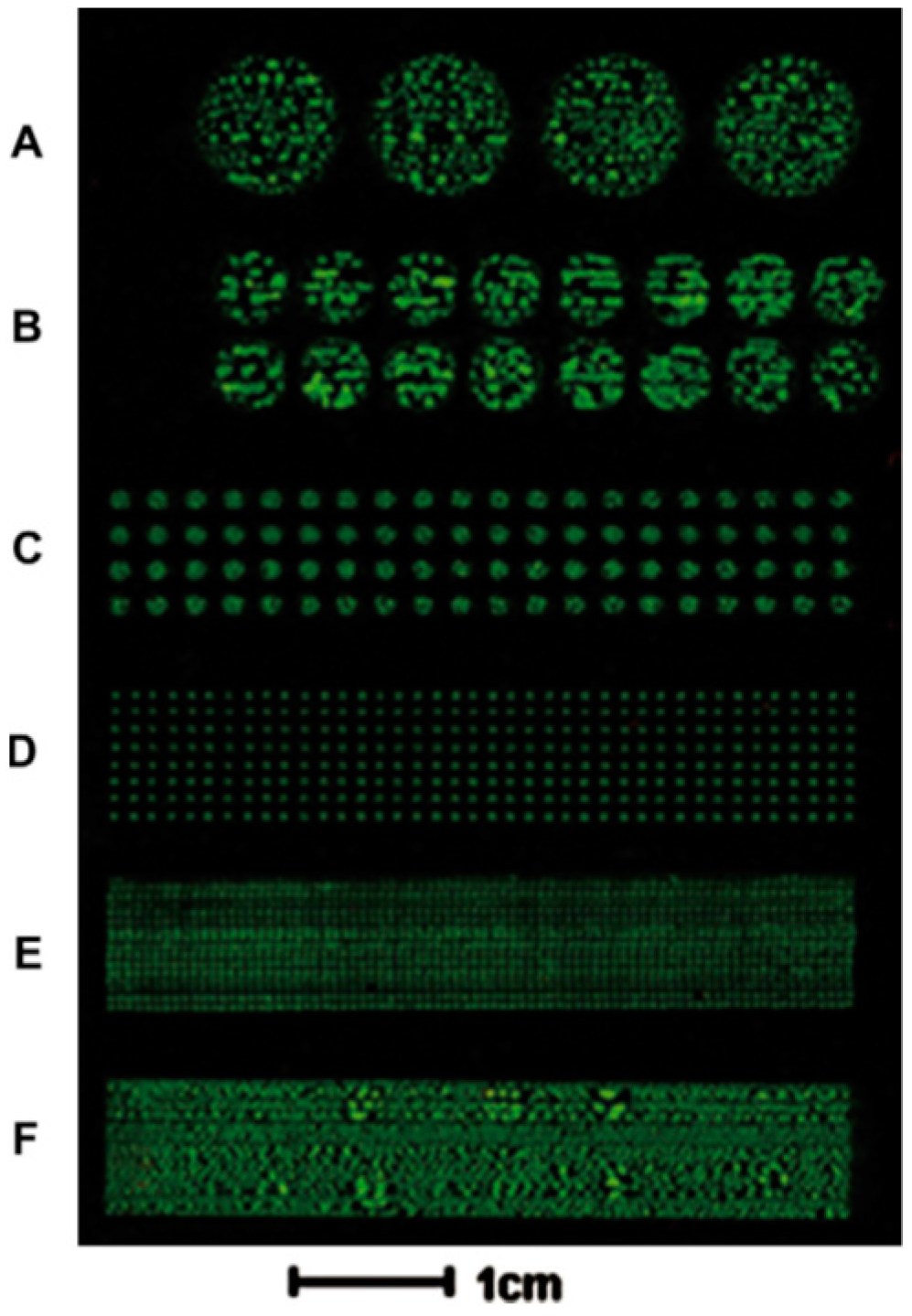
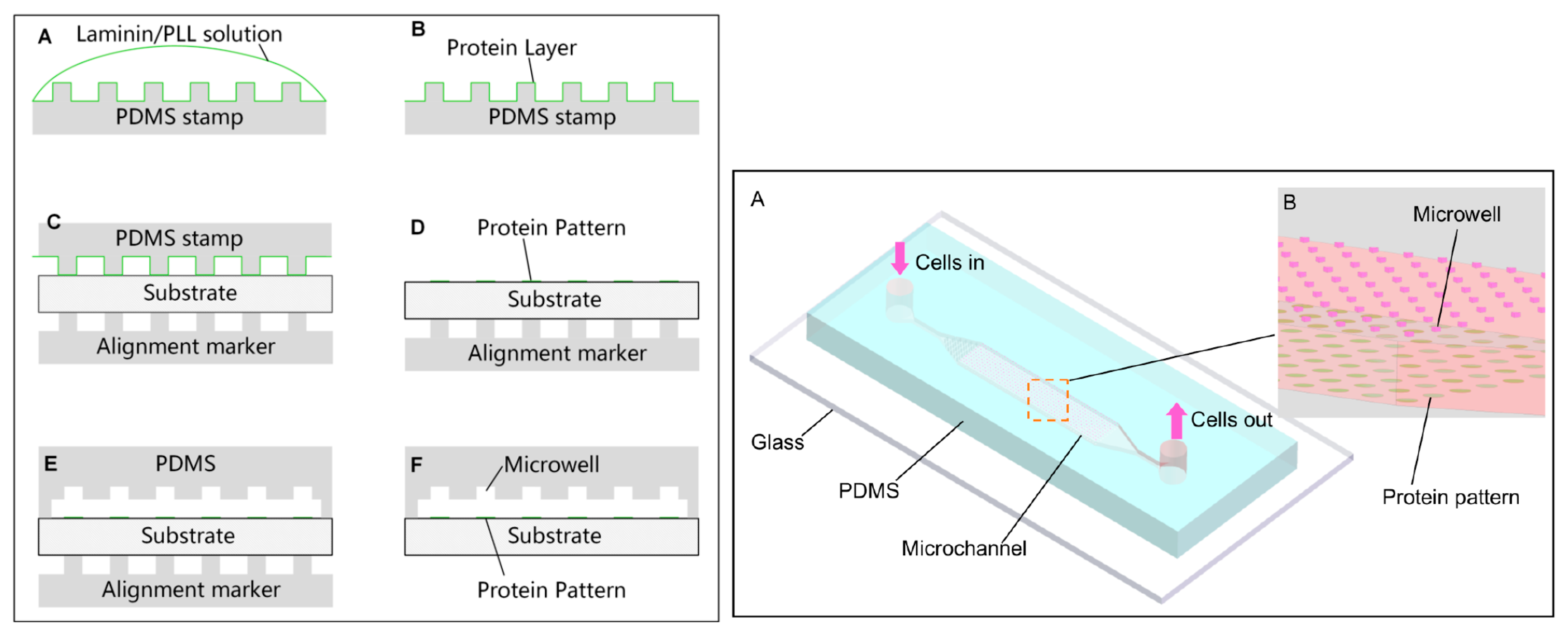
2.3.3. Cell Patterning in Microfluidic Devices Combined with Microcontact Printing
2.3.4. Deep UV Micropatterning
3. Perspective: Automatic Biophysical Measurements on Patterned Cells
4. Conclusions and Perspective
Acknowledgments
Conflicts of Interest
References
- Martinez-Rivas, A.; González-Quijano, G.K. Micro and nanoengineering advances for the development and commercialization of organ-on-chips. Biol. Eng. Med. 2017, 2, 2. [Google Scholar] [CrossRef]
- Shirure, V.S.; George, S.C. Design considerations to minimize the impact of drug absorption in polymer-based organ-on-a-chip platforms. Lab Chip 2017, 17, 681–690. [Google Scholar] [CrossRef] [PubMed]
- Ware, B.R.; Berger, D.R.; Khetani, S.R. Prediction of Drug-Induced Liver Injury in Micropatterned Co-cultures Containing iPSC-Derived Human Hepatocytes. Toxicol. Sci. 2015, 145, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Griep, L.M.; Wolbers, F.; de Wagenaar, B.; ter Braak, P.M.; Weksler, B.B.; Romero, I.A.; Couraud, P.O.; Vermes, I.; van der Meer, A.D.; van den Berg, A. BBB on chip: Microfluidic platform to mechanically and biochemically modulate blood-brain barrier function. Biomed. Microdevices 2013, 15, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Cao, Y.; DiPietro, L.A.; Liang, J. Dynamic cellular finite-element method for modelling large-scale cell migration and proliferation under the control of mechanical and biochemical cues: A study of re-epithelialization. J. R. Soc. Interface 2017, 14, 20160959. [Google Scholar] [CrossRef] [PubMed]
- Phan, D.T.; Bender, R.H.F.; Andrejecsk, J.W.; Sobrino, A.; Hachey, S.J.; George, S.C.; Hughes, C.C. Blood–brain barrier-on-a-chip: Microphysiological systems that capture the complexity of the blood–central nervous system interface. Exp. Biol. Med. 2017, 242, 1669–1678. [Google Scholar] [CrossRef] [PubMed]
- Huh, D.; Matthews, B.D.; Mammoto, A.; Montoya-Zavala, M.; Hsin, H.Y.; Ingber, D.E. Reconstituting Organ-Level Lung Functions on a Chip. Science 2010, 328, 1662–1668. [Google Scholar] [CrossRef] [PubMed]
- Beißner, N.; Lorenz, T.; Reichl, S. Chapter 11: Organ on chip. In Microsystems for Pharmatechnology: Manipulation of Fluids, Particles, Droplets, and Cells; Dietzel, A., Ed.; Springer: Cham, Switzerland, 2016; pp. 299–339. [Google Scholar] [CrossRef]
- Städler, B.; Falconnet, D.; Pfeiffer, I.; Höök, F.; Vörös, J. Micropatterning of DNA-Tagged Vesicles. Langmuir 2004, 20, 11348–11354. [Google Scholar] [CrossRef] [PubMed]
- Estevam-Alves, R.; Ferreira, P.H.D.; Coatrini, A.C.; Oliveira, O.N.; Fontana, C.R.; Mendonca, C.R. Femtosecond Laser Patterning of the Biopolymer Chitosan for Biofilm Formation. Int. J. Mol. Sci. 2016, 17, 1243. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Mei, Y.; Thurmer, D.J.; Coric, E.; Schmidt, O.G. Rolled-up transparent microtubes as two-dimensionally confined culture scaffolds of individual yeast cells. Lab Chip 2009, 9, 263–268. [Google Scholar] [CrossRef] [PubMed]
- Credi, C.; De Marco, C.; Molena, E.; Pla Roca, M.; Samitier Martí, J.; Marques, J.; Fernàndez-Busquets, X.; Levi, M.; Turri, S. Heparin micropatterning onto fouling-release perfluoropolyether-based polymers via photobiotin activation. Colloids Surf. B Biointerfaces 2016, 146, 250–259. [Google Scholar] [CrossRef] [PubMed]
- Gascoyne, P.; Mahidol, C.; Ruchirawat, M.; Satayavivad, J.; Watcharasit, P.; Becker, F.F. Microsample preparation by dielectrophoresis : Isolation of malaria. Lab Chip 2002, 2, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Lee, H.J.; Jin Son, K.; Koh, W.-G. Fabrication of hydrogel-micropatterned nanofibers for highly sensitive microarray-based immunosensors having additional enzyme-based sensing capability. J. Mater. Chem. 2011, 21, 4476–4483. [Google Scholar] [CrossRef]
- Formosa, C.; Pillet, F.; Schiavone, M.; Duval, R.E.; Ressier, L.; Dague, E. Generation of living cell arrays for atomic force microscopy studies. Nat. Protoc. 2015, 10, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Imamura, Y.; Mukohara, T.; Shimono, Y.; Funakoshi, Y.; Chayahara, N.; Toyoda, M.; Kiyota, N.; Takao, S.; Kono, S.; Nakatsura, T.; et al. Comparison of 2D- and 3D-culture models as drug-testing platforms in breast cancer. Oncol. Rep. 2015, 33, 1837–1843. [Google Scholar] [CrossRef] [PubMed]
- Langer, R.; Tirrell, D.A. Designing materials for biology and medicine. Nature 2004, 428, 487–492. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Rivas, A.; González-Quijano, G.K. Capítulo 8: Nanobiosensores con aplicaciones en biomedicina. In Nanobiotecnología: Fundamentos y Perspectivas; Ramón-Gallegos, E., Ed.; Editorial Académica Española: Saarbrücken, Deutschland, Alemania, 2016; p. 371. ISBN 978-3-8417-5270-3. [Google Scholar]
- Yusof, A.; Keegan, H.; Spillane, C.D.; Sheils, O.M.; Martin, C.M.; O’Leary, J.J.; Zengerle, R.; Koltay, P. Inkjet-like printing of single-cells. Lab Chip 2011, 11, 2447–2454. [Google Scholar] [CrossRef] [PubMed]
- Matsusaki, M.; Sakaue, K.; Kadowaki, K.; Akashi, M. Three-Dimensional Human Tissue Chips Fabricated by Rapid and Automatic Inkjet Cell Printing. Adv. Healthc. Mater. 2013, 2, 534–539. [Google Scholar] [CrossRef] [PubMed]
- Hynes, W.F.; Doty, N.J.; Zarembinski, T.I.; Schwartz, M.P.; Toepke, M.W.; Murphy, W.L.; Atzet, S.K.; Clark, R.; Melendez, J.A.; Cady, N.C. Micropatterning of 3D Microenvironments for Living Biosensor Applications. Biosensors 2014, 4, 28–44. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Krishnamoorthy, S.; Song, H.; Zhang, Z.; Xu, C. Ligament flow during drop-on-demand inkjet printing of bioink containing living cells. J. Appl. Phys. 2017, 121, 124904. [Google Scholar] [CrossRef]
- Mecozzi, L.; Gennari, O.; Rega, R.; Battista, L.; Ferraro, P.; Grilli, S. Simple and Rapid Bioink Jet Printing for Multiscale Cell Adhesion Islands. Macromol. Biosci. 2017, 17. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Lu, J.; Chen, H.; Huang, L.; Cai, J.; Xu, Z. Application of inkjet printing technique for biological material delivery and antimicrobial assays. Anal. Biochem. 2011, 410, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Srimongkon, T.; Mandai, S.; Enomae, T. Application of Biomaterials and Inkjet Printing to Develop Bacterial Culture System. Adv. Mater. Sci. Eng. 2015, 2015, e290790. [Google Scholar] [CrossRef]
- Ozkan, M.; Pisanic, T.; Scheel, J.; Barlow, C.; Esener, S.; Bhatia, S.N. Electro-Optical Platform for the Manipulation of Live Cells. Langmuir 2003, 19, 1532–1538. [Google Scholar] [CrossRef]
- Kim, J.J.; Bong, K.W.; Reátegui, E.; Irimia, D.; Doyle, P.S. Porous microwells for geometry-selective, large-scale microparticle arrays. Nat. Mater. 2017, 16, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Chiou, P.Y.; Ohta, A.T.; Wu, M.C. Massively parallel manipulation of single cells and microparticles using optical images. Nature 2005, 436, 370–372. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Maltais, T.R.; Walter, T.M.; Wei, A.; Williams, S.J.; Wereley, S.T. Trapping and viability of swimming bacteria in an optoelectric trap. Lab Chip 2016, 16, 1039–1046. [Google Scholar] [CrossRef] [PubMed]
- Jing, P.; Wu, J.; Liu, G.W.; Keeler, E.G.; Pun, S.H.; Lin, L.Y. Photonic Crystal Optical Tweezers with High Efficiency for Live Biological Samples and Viability Characterization. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Schiele, N.R.; Corr, D.T.; Huang, Y.; Raof, N.A.; Xie, Y.; Chrisey, D.B. Laser-based direct-write techniques for cell printing. Biofabrication 2010, 2, 032001. [Google Scholar] [CrossRef] [PubMed]
- Phamduy, T.B.; Sweat, R.S.; Azimi, M.S.; Burow, M.E.; Murfee, W.L.; Chrisey, D.B. Printing cancer cells into intact microvascular networks: A model for investigating cancer cell dynamics during angiogenesis. Integr. Biol. 2015, 7, 1068–1078. [Google Scholar] [CrossRef] [PubMed]
- Nahmias, Y.; Odde, D.J. Micropatterning of living cells by laser-guided direct writing: application to fabrication of hepatic–endothelial sinusoid-like structures. Nat. Protoc. 2006, 1, 2288–2296. [Google Scholar] [CrossRef] [PubMed]
- Collins, D.J.; Devendran, C.; Ma, Z.; Ng, J.W.; Neild, A.; Ai, Y. Acoustic tweezers via sub–time-of-flight regime surface acoustic waves. Sci. Adv. 2016, 2, e1600089. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Lin, S.-C.S.; Kiraly, B.; Yue, H.; Li, S.; Chiang, I.-K.; Shi, J.; Benkovic, S.J.; Huang, T.J. On-chip manipulation of single microparticles, cells, and organisms using surface acoustic waves. Proc. Natl. Acad. Sci. USA 2012, 109, 11105–11109. [Google Scholar] [CrossRef] [PubMed]
- Naseer, S.M.; Manbachi, A.; Samandari, M.; Walch, P.; Gao, Y.; Zhang, Y.S.; Davoudi, F.; Wang, W.; Abrinia, K.; Cooper, J.M.; et al. Surface acoustic waves induced micropatterning of cells in gelatin methacryloyl (GelMA) hydrogels. Biofabrication 2017, 9, 015020. [Google Scholar] [CrossRef] [PubMed]
- Ai, Y.; Sanders, C.K.; Marrone, B.L. Separation of Escherichia coli Bacteria from Peripheral Blood Mononuclear Cells Using Standing Surface Acoustic Waves. Anal. Chem. 2013, 85, 9126–9134. [Google Scholar] [CrossRef] [PubMed]
- Hughes, M.P. Strategies for dielectrophoretic separation in laboratory-on-a-chip systems. Electrophoresis 2002, 23, 2569–2582. [Google Scholar] [CrossRef]
- Sadeghian, H.; Hojjat, Y.; Soleimani, M. Interdigitated electrode design and optimization for dielectrophoresis cell separation actuators. J. Electrost. 2017, 86, 41–49. [Google Scholar] [CrossRef]
- Ho, C.-T.; Lin, R.-Z.; Chang, W.-Y.; Chang, H.-Y.; Liu, C.-H. Rapid heterogeneous liver-cell on-chip patterning via the enhanced field-induced dielectrophoresis trap. Lab Chip 2006, 6, 724–734. [Google Scholar] [CrossRef] [PubMed]
- D’Amico, L.; Ajami, N.J.; Adachi, J.A.; Gascoyne, P.R.C.; Petrosino, J.F. Isolation and concentration of bacteria from blood using microfluidic membraneless dialysis and dielectrophoresis. Lab Chip 2017, 17, 1340–1348. [Google Scholar] [CrossRef] [PubMed]
- Ino, K.; Ito, A.; Honda, H. Cell patterning using magnetite nanoparticles and magnetic force. Biotechnol. Bioeng. 2007, 97, 1309–1317. [Google Scholar] [CrossRef] [PubMed]
- Souza, G.R.; Tseng, H.; Gage, J.A.; Mani, A.; Desai, P.; Leonard, F.; Liao, A.; Longo, M.; Refuerzo, J.S.; Godin, B. Magnetically Bioprinted Human Myometrial 3D Cell Rings as A Model for Uterine Contractility. Int. J. Mol. Sci. 2017, 18, 683. [Google Scholar] [CrossRef] [PubMed]
- Pivetal, J.; Royet, D.; Ciuta, G.; Frenea-Robin, M.; Haddour, N.; Dempsey, N.M.; Dumas-Bouchiat, F.; Simonet, P. Micro-magnet arrays for specific single bacterial cell positioning. J. Magn. Magn. Mater. 2015, 380, 72–77. [Google Scholar] [CrossRef]
- Wei, S.C.; Fattet, L.; Tsai, J.H.; Guo, Y.; Pai, V.H.; Majeski, H.E.; Chen, A.C.; Sah, R.L.; Taylor, S.S.; Engler, A.J.; et al. Matrix stiffness drives epithelial-mesenchymal transition and tumour metastasis through a TWIST1-G3BP2 mechanotransduction pathway. Nat. Cell Biol. 2015, 17, 678–688. [Google Scholar] [CrossRef] [PubMed]
- Kwon, M.; Godinho, S.A.; Chandhok, N.S.; Ganem, N.J.; Azioune, A.; Thery, M.; Pellman, D. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008, 22, 2189–2203. [Google Scholar] [CrossRef] [PubMed]
- Théry, M.; Racine, V.; Piel, M.; Pépin, A.; Dimitrov, A.; Chen, Y.; Sibarita, J.-B.; Bornens, M. Anisotropy of cell adhesive microenvironment governs cell internal organization and orientation of polarity. Proc. Natl. Acad. Sci. USA 2006, 103, 19771–19776. [Google Scholar] [CrossRef] [PubMed]
- Théry, M.; Racine, V.; Pépin, A.; Piel, M.; Chen, Y.; Sibarita, J.-B.; Bornens, M. The extracellular matrix guides the orientation of the cell division axis. Nat. Cell Biol. 2005, 7, 947–953. [Google Scholar] [CrossRef] [PubMed]
- Formosa, C.; Grare, M.; Jauvert, E.; Coutable, A.; Regnouf-de-Vains, J.B.; Mourer, M.; Duval, R.E.; Dague, E. Nanoscale analysis of the effects of antibiotics and CX1 on a Pseudomonas aeruginosa multidrug-resistant strain. Sci. Rep. 2012, 2. [Google Scholar] [CrossRef] [PubMed]
- Formosa, C.; Herold, M.; Vidaillac, C.; Duval, R.E.; Dague, E. Unravelling of a mechanism of resistance to colistin in Klebsiella pneumoniae using atomic force microscopy. J. Antimicrob. Chemother. 2015, 70, 2261–2270. [Google Scholar] [CrossRef] [PubMed]
- Camesano, T.A.; Natan, M.J.; Logan, B.E. Observation of changes in bacterial cell morphology using tapping mode atomic force microscopy. Langmuir 2000, 16, 4563–4572. [Google Scholar] [CrossRef]
- Emerson, R.J.; Camesano, T.A. Nanoscale investigation of pathogenic microbial adhesion to a biomaterial. Appl. Environ. Microbiol. 2004, 70, 6012–6022. [Google Scholar] [CrossRef] [PubMed]
- Cerf, A.; Cau, J.-C.; Vieu, C. Controlled assembly of bacteria on chemical patterns using soft lithography. Colloids Surf. B Biointerfaces 2008, 65, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Ressier, L.; Viallet, B.; Beduer, A.; Fabie, D.; Fabié, L.; Palleau, E.; Dague, E. Combining convective/capillary deposition and AFM oxidation lithography for close-packed directed assembly of colloids. Langmuir 2008, 24, 13254–13257. [Google Scholar] [CrossRef] [PubMed]
- Cerf, A.; Cau, J.-C.; Vieu, C.; Dague, E. Nanomechanical properties of dead or alive single-patterned bacteria. Langmuir 2009, 25, 5731–5736. [Google Scholar] [CrossRef] [PubMed]
- Jauvert, E.; Palleau, E.; Dague, E.; Ressier, L. Directed Assembly of Living Pseudomonas aeruginosa Bacteria on PEI Patterns Generated by Nanoxerography for Statistical AFM Bioexperiments. ACS Appl. Mater. Interfaces 2014, 6, 21230–21236. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Biebuyck, H.A.; Whitesides, G.M. Patterning Self-Assembled Monolayers: Applications in Materials Science. Langmuir 1994, 10, 1498–1511. [Google Scholar] [CrossRef]
- Thibault, C.; Severac, C.; Trevisiol, E.; Vieu, C. Microtransfer molding of hydrophobic dendrimer. Microelectron. Eng. 2006, 83, 1513–1516. [Google Scholar] [CrossRef]
- Hua, F.; Sun, Y.; Gaur, A.; Meitl, M.A.; Bilhaut, L.; Rotkina, L.; Wang, J.; Geil, P.; Shim, M.; Rogers, J.A.; et al. Polymer Imprint Lithography with Molecular-Scale Resolution. Nano. Lett. 2004, 4, 2467–2471. [Google Scholar] [CrossRef]
- Kane, R.S.; Takayama, S.; Ostuni, E.; Ingber, D.E.; Whitesides, G.M. Patterning proteins and cells using soft lithography. Biomaterials 1999, 20, 2363–2376. [Google Scholar] [CrossRef]
- Mrksich, M.; Whitesides, G.M. Using self-assembled monolayers to understand the interactions of man-made surfaces with proteins and cells. Annu. Rev. Biophys. Biomol. Struct. 1996, 25, 55–78. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Yan, L.; Altman, M.; Lässle, M.; Nugent, H.; Frankel, F.; Lauffenburger, D.A.; Whitesides, G.M.; Rich, A. Biological surface engineering: A simple system for cell pattern formation. Biomaterials 1999, 20, 1213–1220. [Google Scholar] [CrossRef]
- Ruiz, S.A.; Chen, C.S. Microcontact printing: A tool to pattern. Soft Matter 2007, 3, 168–177. [Google Scholar] [CrossRef]
- Long, M.; Sato, M.; Lim, C.T.; Wu, J.; Adachi, T.; Inoue, Y. Advances in Experiments and Modeling in Micro- and Nano-Biomechanics: A Mini Review. Cell. Mol. Bioeng. 2011, 4, 327–339. [Google Scholar] [CrossRef]
- Foncy, J.; Cau, J.-C.; Bartual-Murgui, C.; François, J.M.; Trévisiol, E.; Sévérac, C. Comparison of polyurethane and epoxy resist master mold for nanoscale soft lithography. Microelectron. Eng. 2013, 110, 183–187. [Google Scholar] [CrossRef]
- Wong, L.; Pegan, J.D.; Gabela-Zuniga, B.; Khine, M.; McCloskey, K.E. Leaf-inspired microcontact printing vascular patterns. Biofabrication 2017, 9, 021001. [Google Scholar] [CrossRef] [PubMed]
- Malaquin, L.; Kraus, T.; Schmid, H.; Delamarche, E.; Wolf, H. Controlled Particle Placement through Convective and Capillary Assembly. Langmuir 2007, 23, 11513–11521. [Google Scholar] [CrossRef] [PubMed]
- Fredonnet, J.; Foncy, J.; Lamarre, S.; Cau, J.-C.; Trévisiol, E.; Peyrade, J.-P.; François, J.M.; Séverac, C. Dynamic PDMS inking for DNA patterning by soft lithography. Microelectron. Eng. 2013, 111, 379–383. [Google Scholar] [CrossRef]
- Cayron, H.; Berteloite, B.; Vieu, C.; Paveau, V.; Cau, J.-C.; Cerf, A. Controlled deposition and multi-layer architecturing of single biomolecules using automated directed capillary assembly and nano-contact printing processes. Microelectron. Eng. 2015, 135, 1–6. [Google Scholar] [CrossRef]
- Delapierre, F.-D.; Mottet, G.; Taniga, V.; Boisselier, J.; Viovy, J.-L.; Malaquin, L. High throughput micropatterning of interspersed cell arrays using capillary assembly. Biofabrication 2017, 9, 015015. [Google Scholar] [CrossRef] [PubMed]
- Lagraulet, A.; Foncy, J.; Berteloite, B.; Esteve, A.; Blatche, M.-C.; Malaquin, L.; Vieu, C. InnoStamp 40TM and InnoScan 1100ALTM: A complete automated platform for microstructured cell arrays. Nat. Methods 2015, 12. [Google Scholar] [CrossRef]
- McNulty, J.D.; Klann, T.; Sha, J.; Salick, M.; Knight, G.T.; Turng, L.-S.; Ashton, R.S. High-precision robotic microcontact printing (R-µCP) utilizing a vision guided selectively compliant articulated robotic arm. Lab Chip 2014, 14, 1923–1930. [Google Scholar] [CrossRef] [PubMed]
- Knight, G.T.; Klann, T.; McNulty, J.D.; Ashton, R.S. Fabricating Complex Culture Substrates Using Robotic Microcontact Printing (R-µCP) and Sequential Nucleophilic Substitution. J. Vis. Exp. JoVE 2014. [Google Scholar] [CrossRef] [PubMed]
- Ricoult, S.G.; Sanati Nezhad, A.; Knapp-Mohammady, M.; Kennedy, T.E.; Juncker, D. Humidified Microcontact Printing of Proteins: Universal Patterning of Proteins on Both Low and High Energy Surfaces. Langmuir 2014, 30, 12002–12010. [Google Scholar] [CrossRef] [PubMed]
- Polio, S.R.; Smith, M.L. Patterned Hydrogels for Simplified Measurement of Cell Traction Forces. Methods Cell Biol. 2014, 121, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Vedula, S.R.K.; Hirata, H.; Nai, M.H.; Brugués, A.; Toyama, Y.; Trepat, X.; Lim, C.T.; Ladoux, B. Epithelial bridges maintain tissue integrity during collective cell migration. Nat. Mater. 2014, 13, 87–96. [Google Scholar] [CrossRef] [PubMed]
- El Kirat, K.; Burton, I.; Dupres, V.; Dufrene, Y.F. Sample preparation procedures for biological atomic force microscopy. J. Microsc. 2005, 218, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Kailas, L.; Ratcliffe, E.C.; Hayhurst, E.J.; Walker, M.G.; Foster, S.J.; Hobbs, J.K. Immobilizing live bacteria for AFM imaging of cellular processes. Ultramicroscopy 2009, 109, 775–780. [Google Scholar] [CrossRef] [PubMed]
- Dague, E.; Jauvert, E.; Laplatine, L.; Viallet, B.; Thibault, C.; Ressier, L. Assembly of live micro-organisms on microstructured PDMS stamps by convective/capillary deposition for AFM bio-experiments. Nanotechnology 2011, 22, 395102. [Google Scholar] [CrossRef] [PubMed]
- Rhee, S.W.; Taylor, A.M.; Tu, C.H.; Cribbs, D.H.; Cotman, C.W.; Jeon, N.L. Patterned cell culture inside microfluidic devices. Lab Chip 2005, 5, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Torisawa, Y.; Mosadegh, B.; Luker, G.D.; Morell, M.; O’Shea, K.S.; Takayama, S. Microfluidic hydrodynamic cellular patterning for systematic formation of co-culture spheroids. Integr. Biol. 2009, 1, 649–654. [Google Scholar] [CrossRef] [PubMed]
- Tu, C.; Huang, B.; Zhou, J.; Liang, Y.; Tian, J.; Ji, L.; Liang, X.; Ye, X. A Microfluidic Chip for Cell Patterning Utilizing Paired Microwells and Protein Patterns. Micromachines 2017, 8. [Google Scholar] [CrossRef]
- Azioune, A.; Carpi, N.; Tseng, Q.; Théry, M.; Piel, M. Chapter 8—Protein Micropatterns: A Direct Printing Protocol Using Deep UVs. In Methods in Cell Biology; Cassimeris, L., Tran, P., Eds.; Academic Press: Cambridge, MA, USA, 2010; Volume 97, pp. 133–146. ISBN 978-0-12-381349-7. [Google Scholar]
- Carpi, N.; Piel, M.; Azioune, A.; Cuvelier, D.; Fink, J. Micropatterning on silicon elastomer (PDMS) with deep UVs. Protoc. Exch. 2011. [Google Scholar] [CrossRef]
- Carpi, N.; Piel, M.; Azioune, A.; Fink, J. Micropatterning on glass with deep UV. Protoc. Exch. 2011. [Google Scholar] [CrossRef]
- Alvéole. Available online: http://www.alveolelab.com/primo.html (accessed on 20 November 2017).
- Hulshof, F.F.B.; Zhao, Y.; Vasilevich, A.; Beijer, N.R.M.; de Boer, M.; Papenburg, B.J.; van Blitterswijk, C.; Stamatialis, D.; de Boer, J. NanoTopoChip: High-throughput nanotopographical cell instruction. Acta Biomater. 2017, 62, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Liu, L.; Xi, N.; Wang, Y. Biological Applications of a Nanomanipulator Based on AFM: In situ visualization and quantification of cellular behaviors at the single-molecule level. IEEE Nanotechnol. Mag. 2015, 9, 25–35. [Google Scholar] [CrossRef]
- Fortier, H.; Variola, F.; Wang, C.; Zou, S. AFM force indentation analysis on leukemia cells. Anal. Methods 2016, 8, 4421–4431. [Google Scholar] [CrossRef]
- Minelli, E.; Ciasca, G.; Sassun, T.E.; Antonelli, M.; Palmieri, V.; Papi, M.; Maulucci, G.; Santoro, A.; Giangaspero, F.; Delfini, R.; et al. A fully-automated neural network analysis of AFM force-distance curves for cancer tissue diagnosis. Appl. Phys. Lett. 2017, 111, 143701. [Google Scholar] [CrossRef]
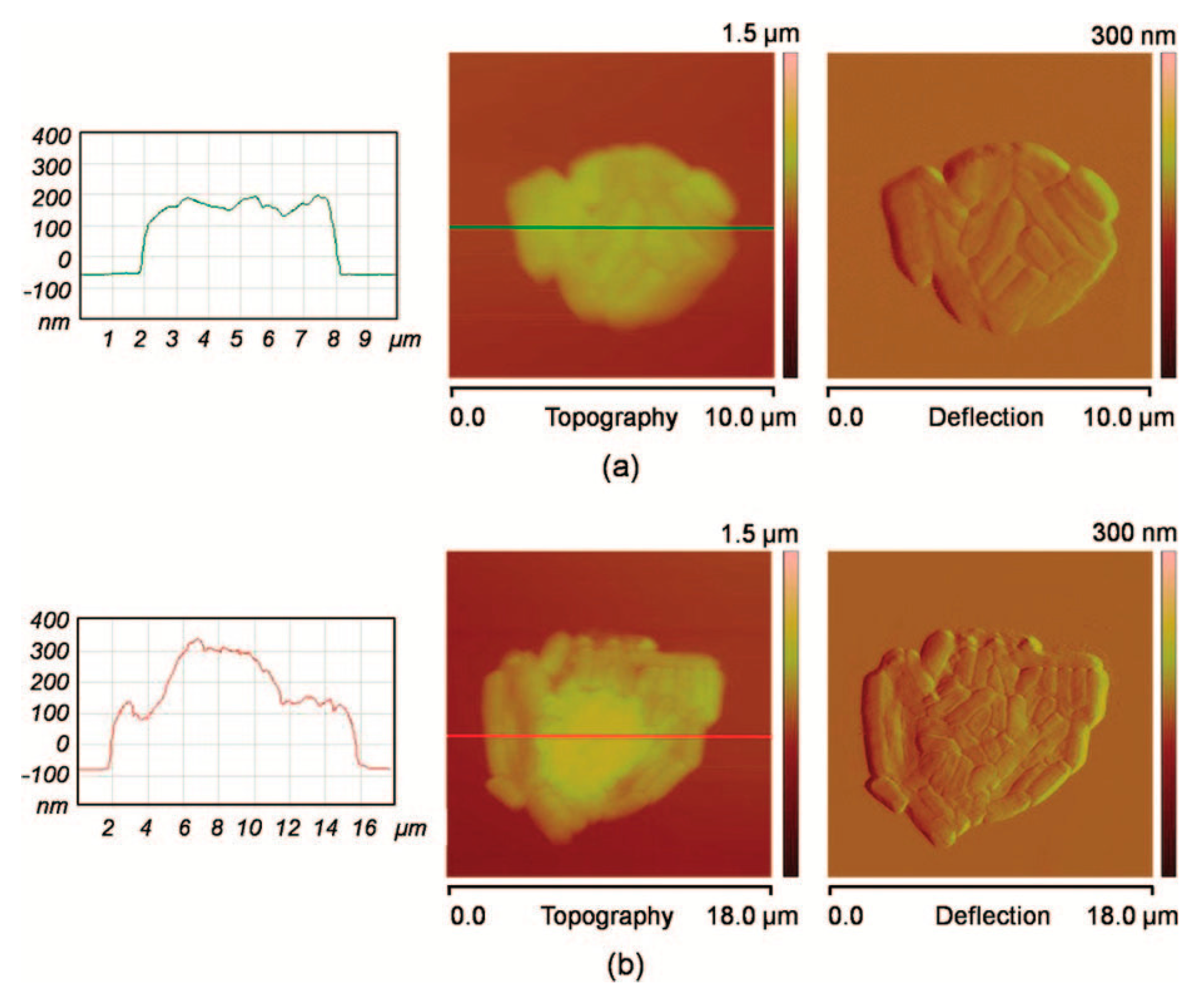

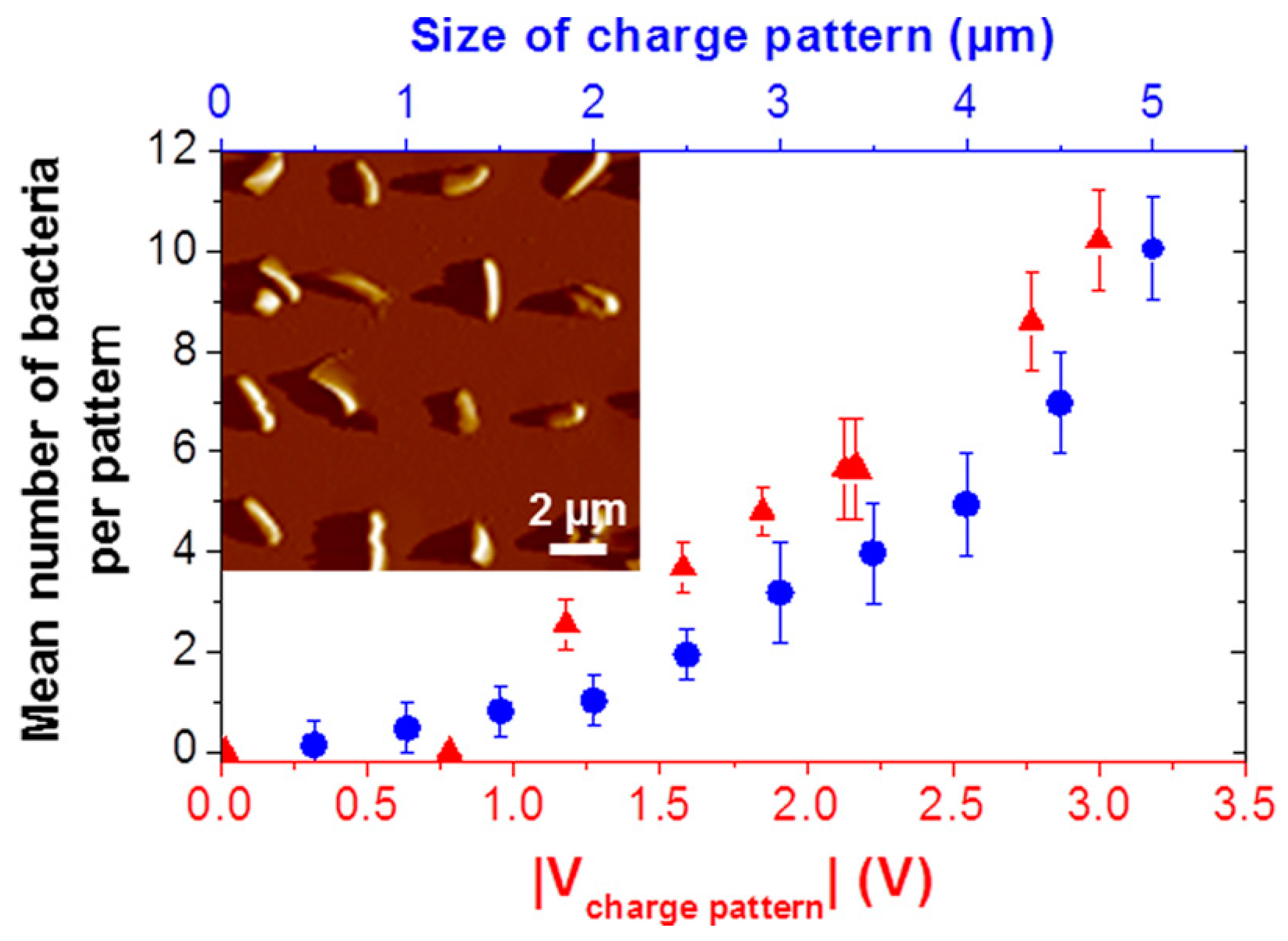
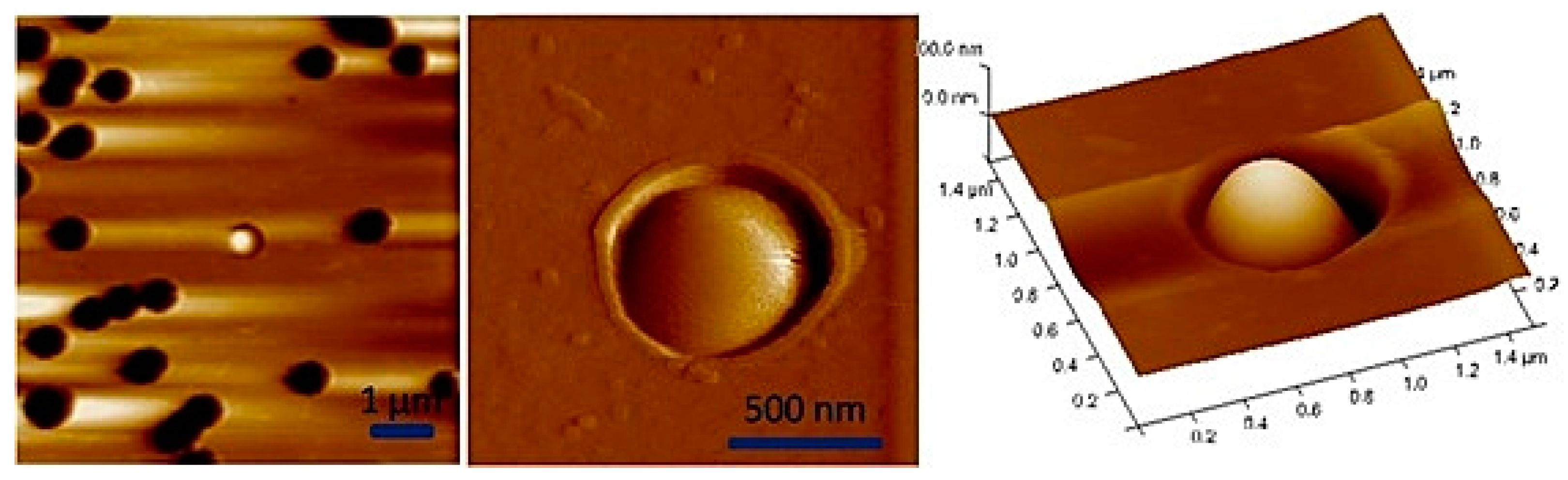
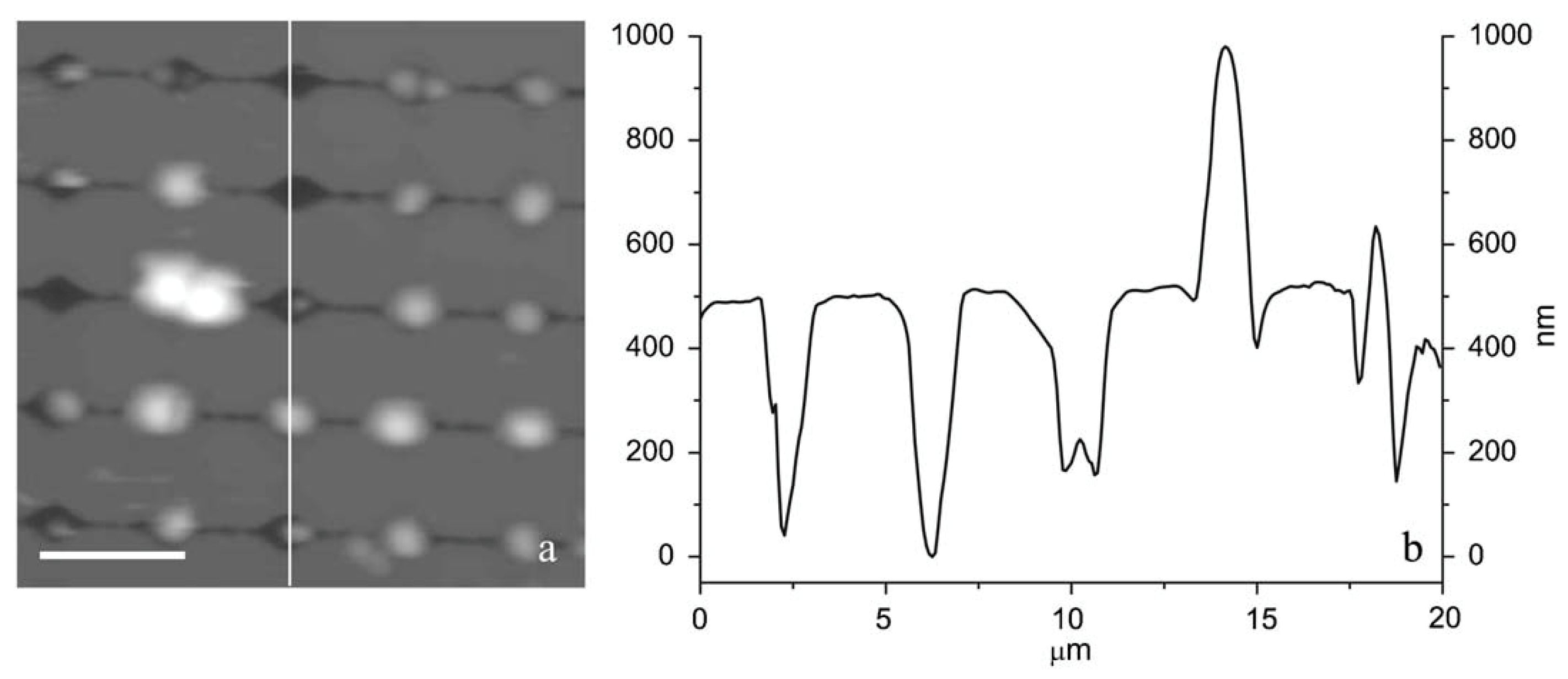
| Technique | Advantage | Disadvantage |
|---|---|---|
| Inkjet printing (Physical) | Moderate cost Good controllability | Droplet formation Requires an external power source |
| Optical and optoelectronic cell trapping (Physical) | Remote and large-scale manipulation Highly specific due to the intrinsic charge and dielectric properties of cells | Thermal effects and photodamage in cells Requires an external power source |
| Laser-based cell patterning (Physical) | Cells and any particles can be manipulated | Large instrumentation Complex set-up |
| Acoustic patterning (Physical) | No significant heat generation and no effects on cell viability | Requires an external power source, piezoelectric surface, and electrode fabrication. |
| Dielectrophoresis (Physical) | Combine electrokinetic forces with hydrodynamic effects High-resolution patterning Large-scale parallel manipulation | Requires an external power source Dielectric force decreases due to the separation distance of electrodes |
| Magnetic techniques (Physical) | Remote manipulation High-resolution patterning, No stress behavior on cells | Magnets and labelling cells with magnetic particles are required |
| Surface chemistry methodologies (Chemical) | High precision and recognition by receptor or specific functional groups between the surface and cells | Pretreated surface is required The surface chemistry could modify the functionality of cells |
| Microcontact printing (Physicochemical) | Low cost, rapid prototyping | Difficulty in controlling the ink and the surface robustness |
| Microwells and filtration (Physicochemical) | Minimize the surface chemistry and conservation of cell functionality | Time consuming placing numerous cells inside microwells |
| DUV patterning (Physicochemical) | It does not require expensive facilities | The resolution depends on the photomask design and patterning substrate. |
| Cell patterning in microfluidic devices combined with microcontact printing (Physicochemical) | Study 3D culture cells and specialized biomedical microdevices | Requires specialized facilities, integration of techniques |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martinez-Rivas, A.; González-Quijano, G.K.; Proa-Coronado, S.; Séverac, C.; Dague, E. Methods of Micropatterning and Manipulation of Cells for Biomedical Applications. Micromachines 2017, 8, 347. https://doi.org/10.3390/mi8120347
Martinez-Rivas A, González-Quijano GK, Proa-Coronado S, Séverac C, Dague E. Methods of Micropatterning and Manipulation of Cells for Biomedical Applications. Micromachines. 2017; 8(12):347. https://doi.org/10.3390/mi8120347
Chicago/Turabian StyleMartinez-Rivas, Adrian, Génesis K. González-Quijano, Sergio Proa-Coronado, Childérick Séverac, and Etienne Dague. 2017. "Methods of Micropatterning and Manipulation of Cells for Biomedical Applications" Micromachines 8, no. 12: 347. https://doi.org/10.3390/mi8120347





