Investigation of the Mechanical Properties of the Human Osteosarcoma Cell at Different Cell Cycle Stages †
Abstract
:1. Introduction
2. Methods and Materials
2.1. Cell Culture
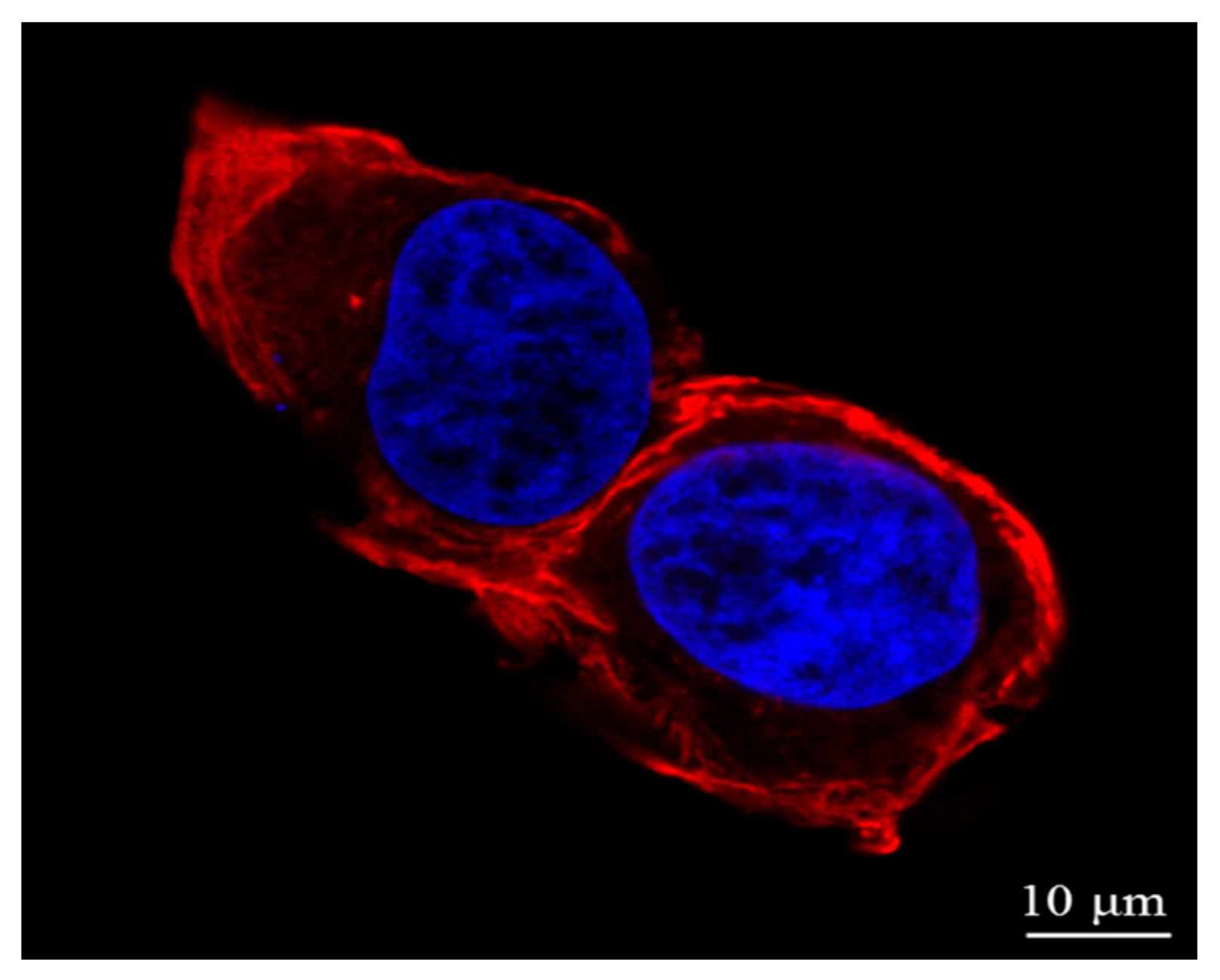
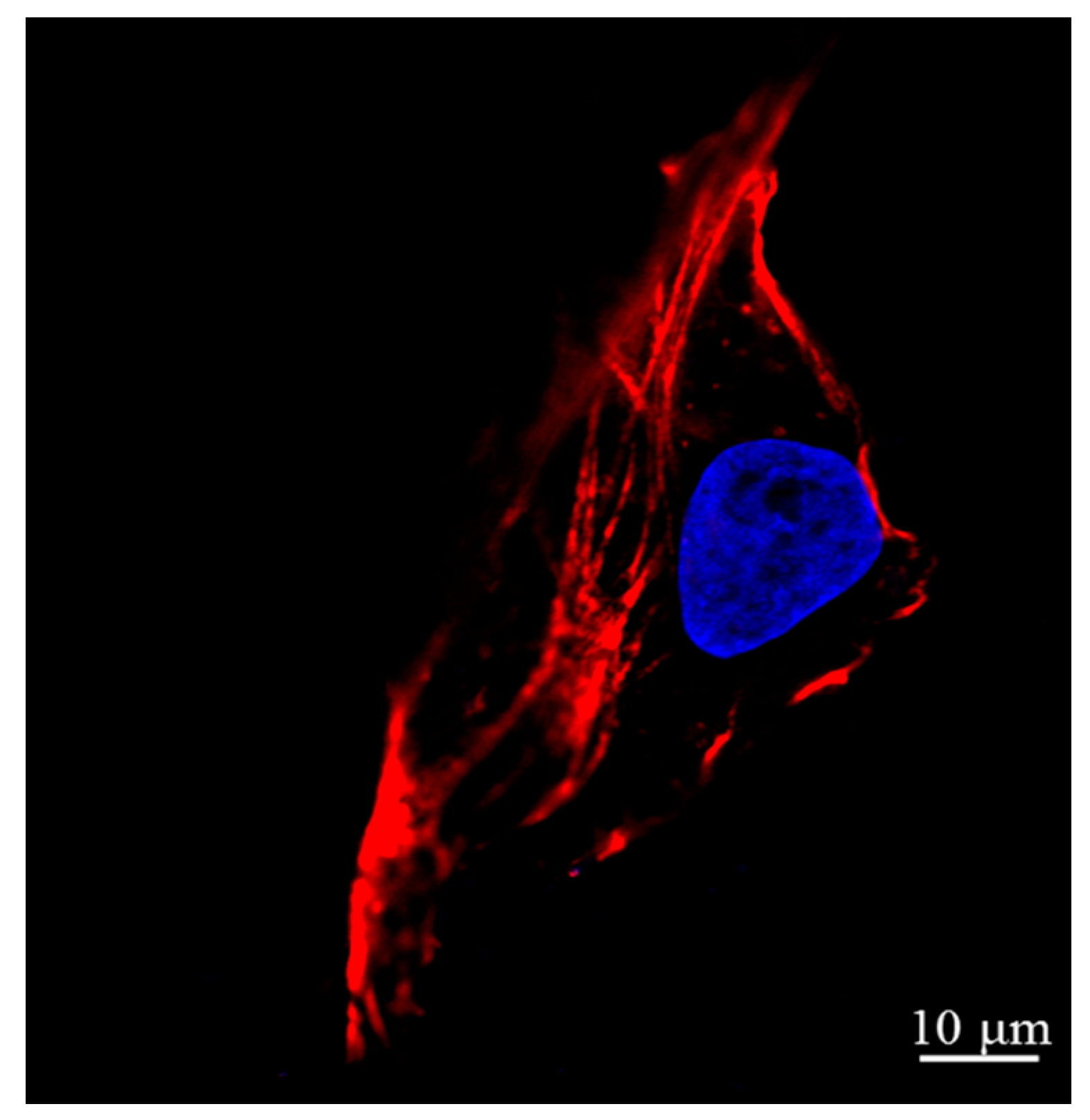
2.2. Actin and Nucleic Acid Staining
2.3. Cell Imaging by Atomic Force Microscopy (AFM)
2.4. AFM Force Spectroscopy Analysis
3. Results and Discussion
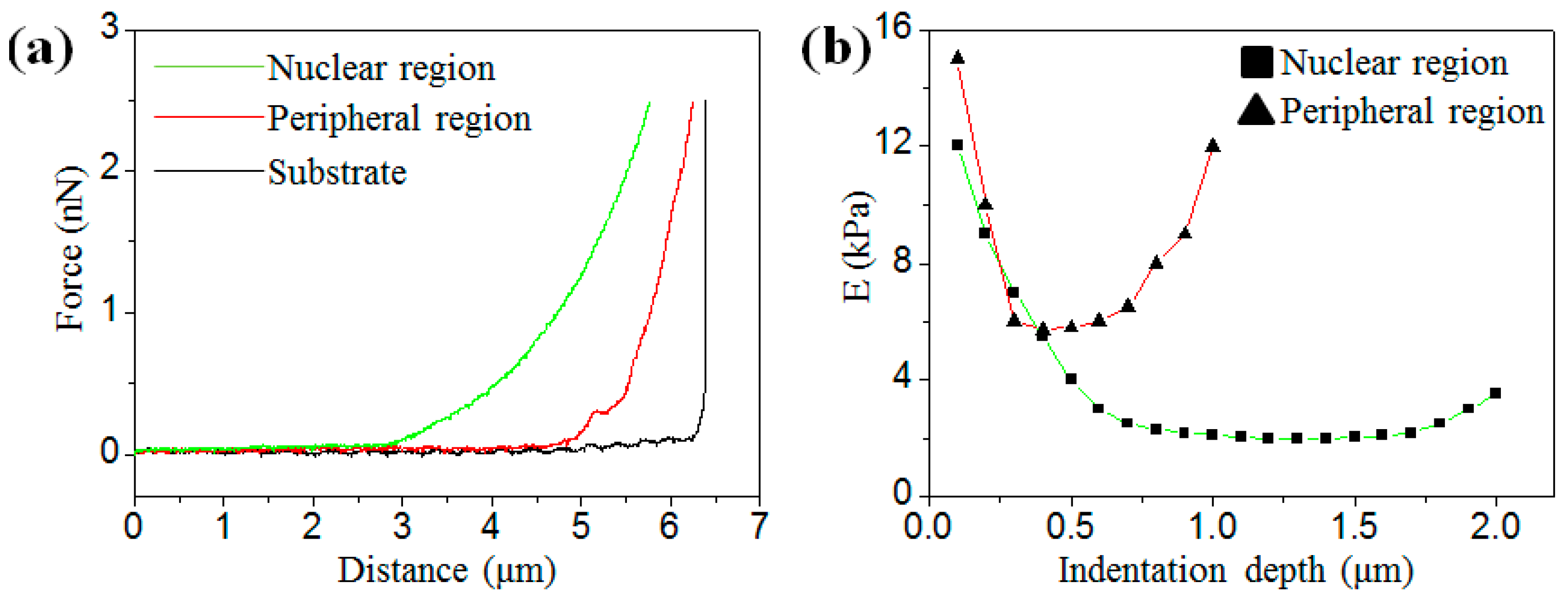
3.1. Analysis of Elastic Modulus under Different Indentation Depths
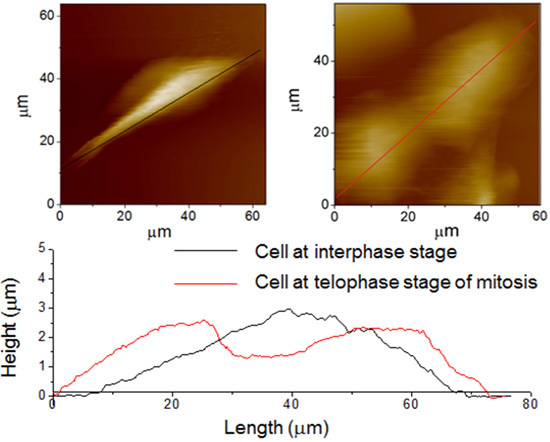
3.2. AFM Images of Cells at the Different Cell Cycle Stages
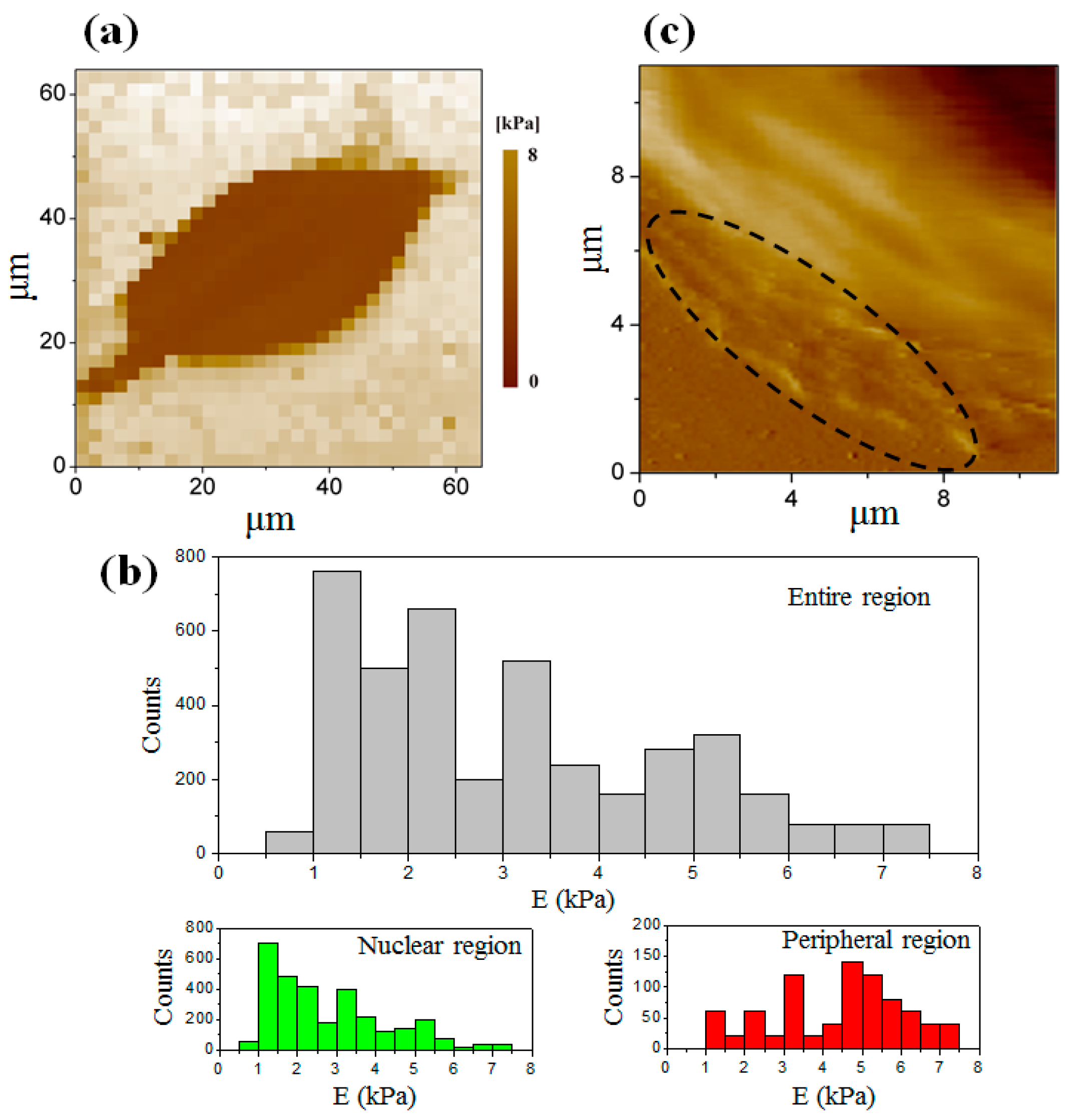
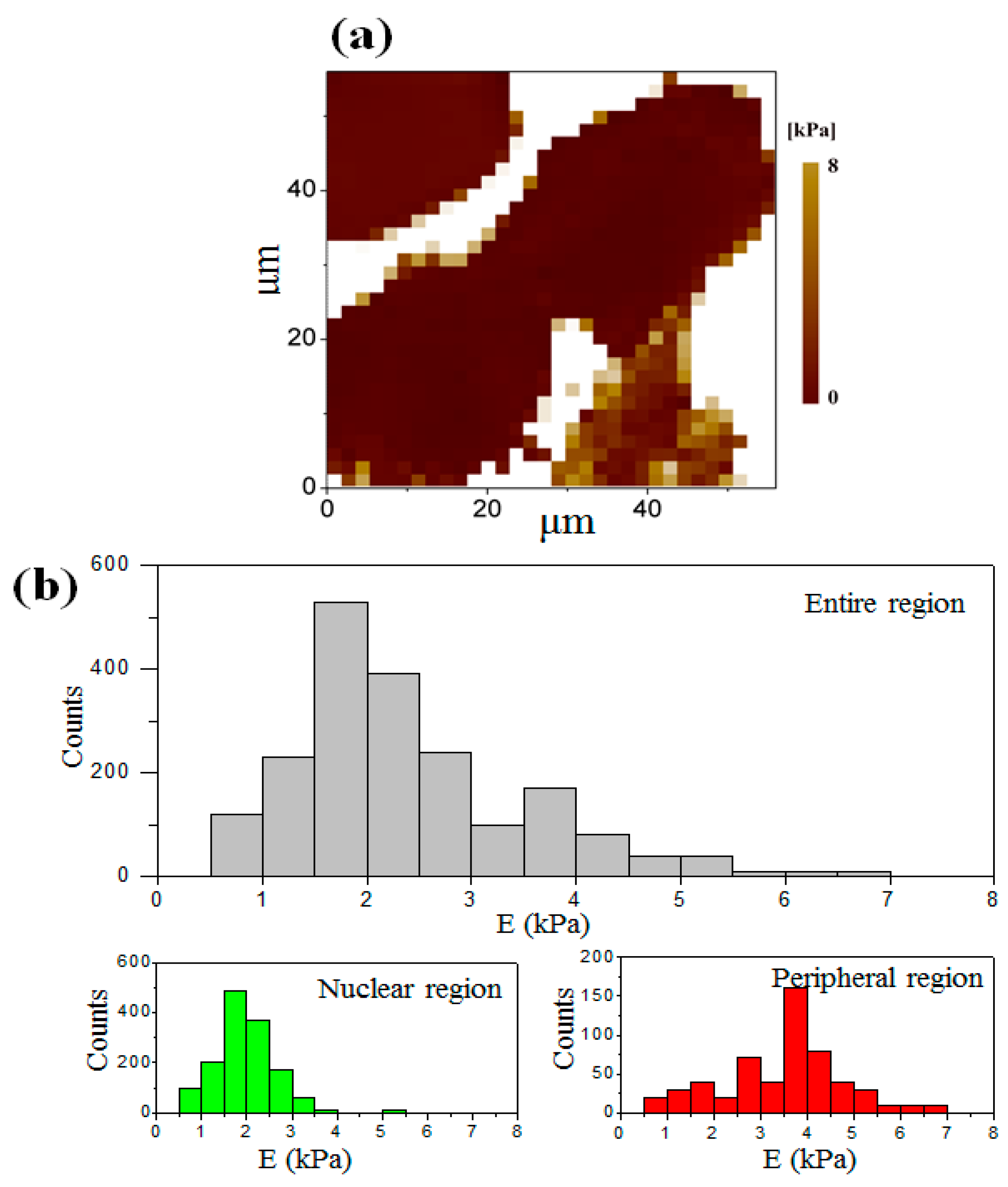
3.3. Elastic Modulus Map of the Cell at the Interphase
3.4. Elastic Modulus Map of the Cell at the Telophase of Mitosis
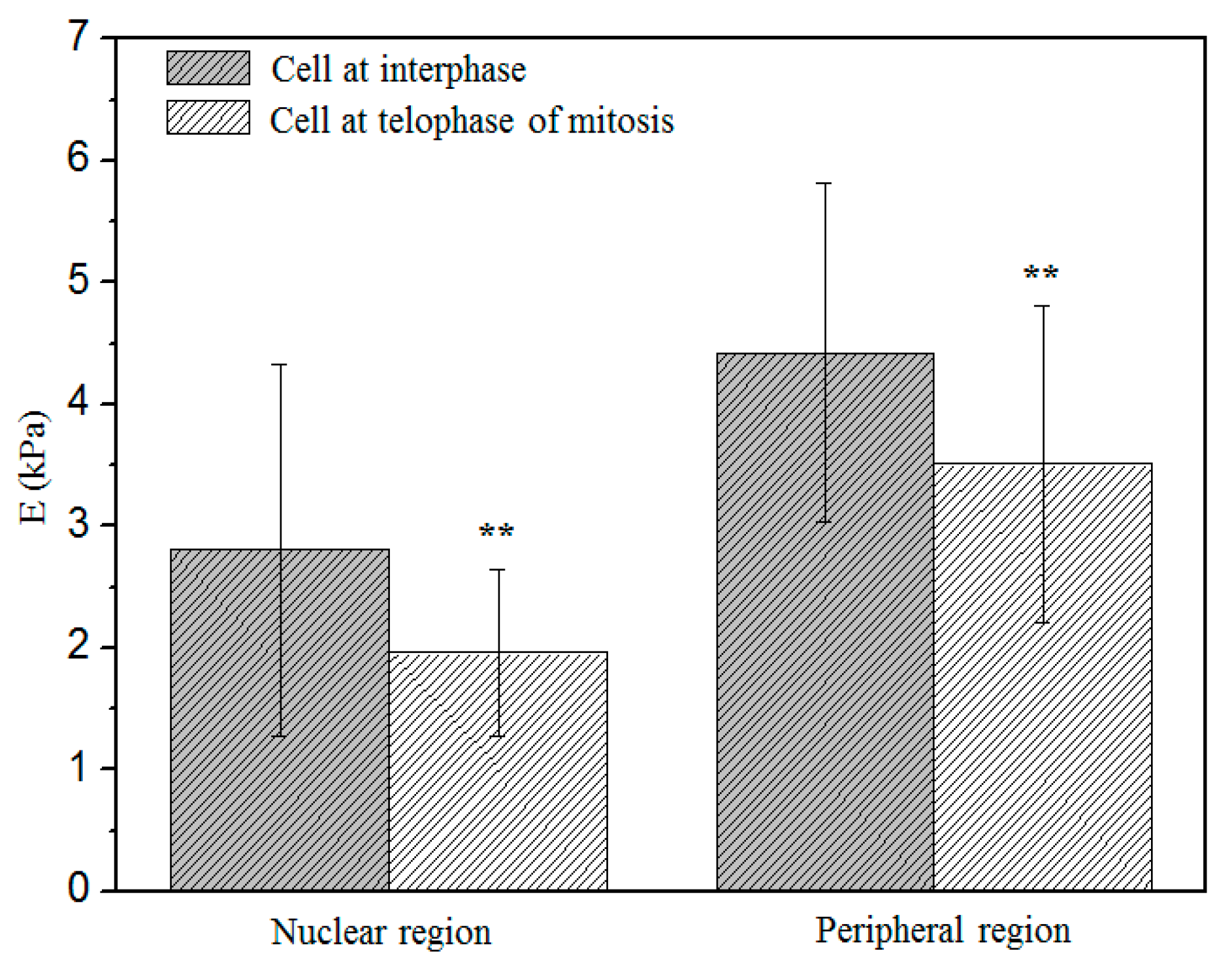
3.5. Comparison of the Elastic Modulus between Two Stages
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bao, G.; Suresh, S. Cell and molecular mechanics of biological materials. Nat. Mater. 2003, 2, 2715–2797. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Bao, G.; Wang, N. Cell mechanics: mechanical response, cell adhesion, and molecular deformation. Annu. Rev. Biomed. Eng. 2000, 2, 189–226. [Google Scholar] [CrossRef] [PubMed]
- Good, D.W.; Stewart, G.D.; Hammer, S. Elasticity as a biomarker for prostate cancer: A systematic review. BJU Int. 2014, 113, 523–534. [Google Scholar] [CrossRef] [PubMed]
- Lekka, M. Atomic force microscopy: A tip for diagnosing cancer. Nat. Nanotechnol. 2012, 7, 691–692. [Google Scholar] [CrossRef] [PubMed]
- Ward, K.A.; Li, W.I.; Zimmer, S.; Davis, T. Viscoelastic properties of transformed cells: Role in tumor cell progression and metastasis formation. Biorheology 1990, 28, 301–313. [Google Scholar]
- Guck, J.; Schinkinger, S.; Lincoln, B.; Wottawah, F.; Ebert, S.; Romeyke, M.; Lenz, D.; Erickson, H.M.; Ananthakrishnan, R.; Mitchell, D. Optical Deformability as an inherent cell marker for testing malignant transformation and metastatic competence. Biophys. J. 2005, 88, 3689–3698. [Google Scholar] [CrossRef] [PubMed]
- Goldmann, W.H.; Galneder, R.; Ludwig, M.; Xu, W.; Adamson, E.D.; Wang, N.; Ezzell, R.M. Differences in elasticity of vinculin-deficient F9 cells measured by magnetometry and atomic force microscopy. Exp. Cell Res. 1998, 239, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsova, T.G.; Starodubtseva, M.N.; Yegorenkov, N.I.; Chizhik, S.A.; Zhdanov, R.I. Atomic force microscopy probing of cell elasticity. Micron 2007, 38, 824–833. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Chen, P.; Li, J. Spatial distribution of cytoskeletal mechanical properties in vascular smooth muscle cells. J. Biomed. Sci. Eng. 2015, 8, 350. [Google Scholar] [CrossRef]
- Rebelo, L.M.; Desousa, J.S.; Mendes, F.J.; Radmacher, M. Comparison of the viscoelastic properties of cells from different kidney cancer phenotypes measured with atomic force microscopy. Nanotechnology 2013, 24, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Cross, S.E.; Jin, Y.S.; Tondre, J.; Wong, R.; Rao, J.; Gimzewski, J.K. AFM-based analysis of human metastatic cancer cells. Nanotechnology 2008, 19, 384003. [Google Scholar] [CrossRef] [PubMed]
- Costa, K.D.; Sim, A.J.; Yin, F.C. Non-Hertzian approach to analyzing mechanical properties of endothelial cells probed by atomic force microscopy. J. Biomech. Eng. 2006, 128, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Sosa, M.S.; Bragado, P.; Aguirreghiso, J.A. Mechanisms of disseminated cancer cell dormancy: An awakening field. Nat. Rev. Cancer 2014, 14, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Mavrakis, M.; Azougros, Y.; Tsai, F.C.; Bertin, A.; Alvarado, J.; lv, F.; Kress, A.; Brasselet, S.; Lecuit, T. Septins promote F-actin ring formation by crosslinking actin filaments into curved bundles. Nat. Cell Biol. 2014, 16, 322–334. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, O.M.; Baum, B. Shaping up to divide: Coordinating actin and microtubule cytoskeletal remodelling during mitosis. Semin. Cell Dev. Biol. 2014, 34, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Heng, Y.W.; Koh, C.G. Actin cytoskeleton dynamics and the cell division cycle. Int. J. Biochem. Cell Biol. 2010, 42, 1622–1633. [Google Scholar] [CrossRef] [PubMed]
- Matzke, R.; Jacobson, K.; Radmacher, M. Direct, high-resolution measurement of furrow stiffening during division of adherent cells. Nat. Cell Biol. 2001, 3, 607–610. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, P.S.; Loboa, E.G. Cytoskeletal and focal adhesion influences on mesenchymal stem cell shape, mechanical properties, and differentiation down osteogenic, adipogenic, and chondrogenic pathways. Tissue Eng. Part B. Rev. 2012, 18, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Stewart, M.P.; Toyoda, Y.; Hyman, A.A. Tracking mechanics and volume of globular cells with atomic force microscopy using a constant-height clamp. Nat. Protoc. 2012, 7, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Hutter, J.L.; Bechhoefer, J. Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 1993, 64, 1868–1873. [Google Scholar] [CrossRef]
- Sneddon, I.N. The relation between load and penetration in the axisymmetric boussinesq problem for a punch of arbitrary profile. Int. J. Eng. Sci. 1965, 3, 47–57. [Google Scholar] [CrossRef]
- Alcaraz, J.; Buscemi, L.; Grabulosa, M.; Trepat, X.; Fabry, B.; Farre, R.; Navajas, D. Microrheology of human lung epithelial cells measured by atomic force microscopy. Biophys. J. 2003, 84, 2071–2079. [Google Scholar] [CrossRef] [Green Version]
- Lekka, M. Discrimination between normal and cancerous cells using AFM. Bionanoscience 2016, 6, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Suresh, S. Biomechanics and biophysics of cancer cells. Acta Biomater. 2007, 3, 413–438. [Google Scholar] [CrossRef] [PubMed]
- Clark, A.G.; Paluch, E. Mechanics and regulation of cell shape during the cell cycle. Results. Probl. Cell Differ. 2011, 53, 31–73. [Google Scholar] [PubMed]
- Pogoda, K.; Jaczewska, J.; Wiltowska-Zuber, J.; Klymenko, O.; Zuber, K.; Fornal, M.; Lekka, M. Depth-sensing analysis of cytoskeleton organization based on AFM data. Biophys. Struct. Mech. 2012, 41, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Haupt, B.J.; Pelling, A.E.; Horton, M.A. Integrated confocal and scanning probe microscopy for biomedical research. Sci. World J. 2006, 6, 1609–1618. [Google Scholar] [CrossRef] [PubMed]
- Stewart, M.P.; Helenius, J.; Toyoda, Y.; Ramanathan, S.P.; Muller, D.J.; Hyman, A.A. Hydrostatic pressure and the actomyosin cortex drive mitotic cell rounding. Nature 2011, 469, 226–230. [Google Scholar] [CrossRef] [PubMed]
- Pollard, T.D.; Borisy, G.G. Cellular motility driven by assembly and disassembly of actin filaments. Cell 2003, 112, 453–465. [Google Scholar] [CrossRef]
- Sanger, J.M.; Sanger, J.W. Assembly of cytoskeletal proteins into cleavage furrows of tissue culture cells. Microsc. Res. Tech. 2000, 49, 190–201. [Google Scholar] [CrossRef]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, G.; Fan, N.; Lv, X.; Liu, Y.; Guo, J.; Yang, L.; Peng, B.; Jiang, H. Investigation of the Mechanical Properties of the Human Osteosarcoma Cell at Different Cell Cycle Stages †. Micromachines 2017, 8, 89. https://doi.org/10.3390/mi8030089
Zhang G, Fan N, Lv X, Liu Y, Guo J, Yang L, Peng B, Jiang H. Investigation of the Mechanical Properties of the Human Osteosarcoma Cell at Different Cell Cycle Stages †. Micromachines. 2017; 8(3):89. https://doi.org/10.3390/mi8030089
Chicago/Turabian StyleZhang, Guocheng, Na Fan, Xiaoying Lv, Yiyao Liu, Jian Guo, Longxiang Yang, Bei Peng, and Hai Jiang. 2017. "Investigation of the Mechanical Properties of the Human Osteosarcoma Cell at Different Cell Cycle Stages †" Micromachines 8, no. 3: 89. https://doi.org/10.3390/mi8030089