Localized Single-Cell Lysis and Manipulation Using Optothermally-Induced Bubbles
Abstract
:1. Introduction
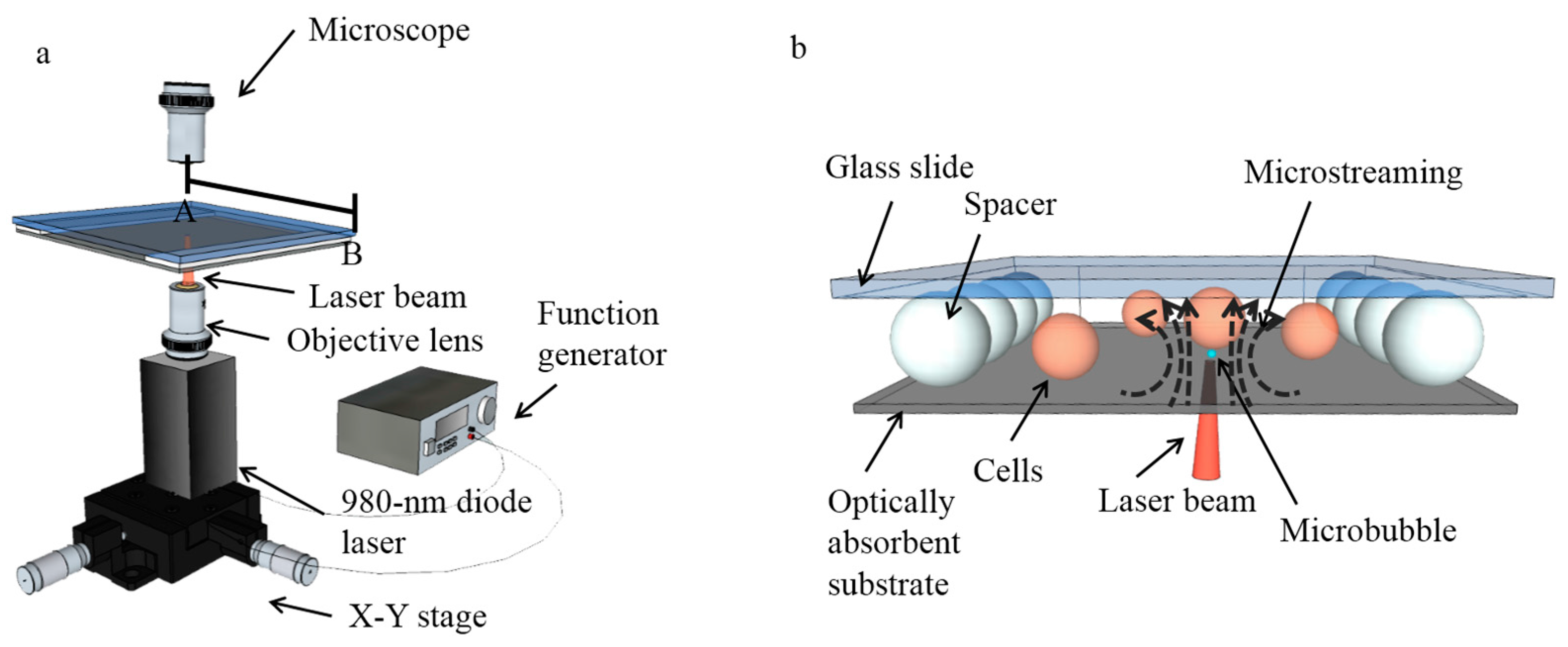
2. Setup and Mechanism
2.1. Setup for Single-Cell Lysis System
2.2. Mechanism
3. Materials and Methods
3.1. Cell Culture
3.2. Cell Lysis
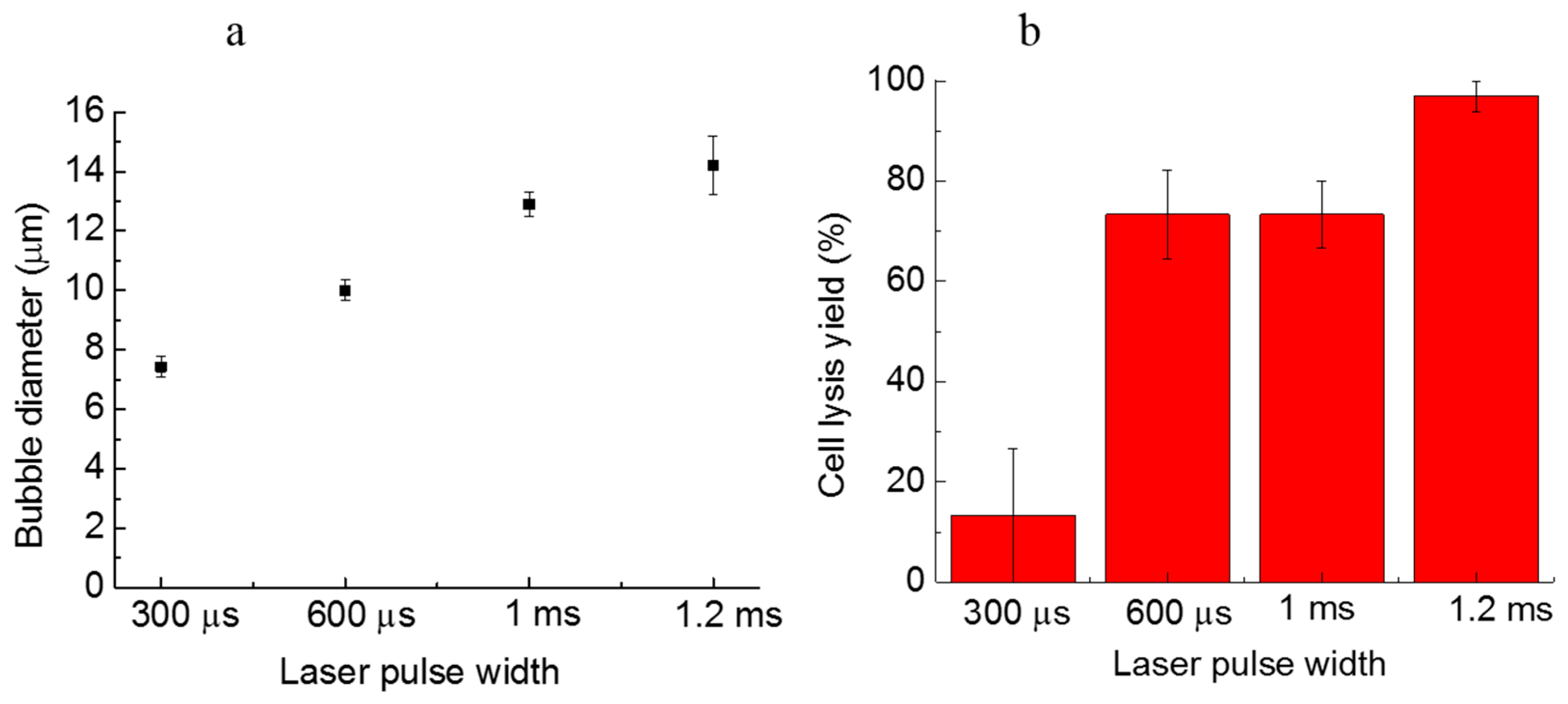
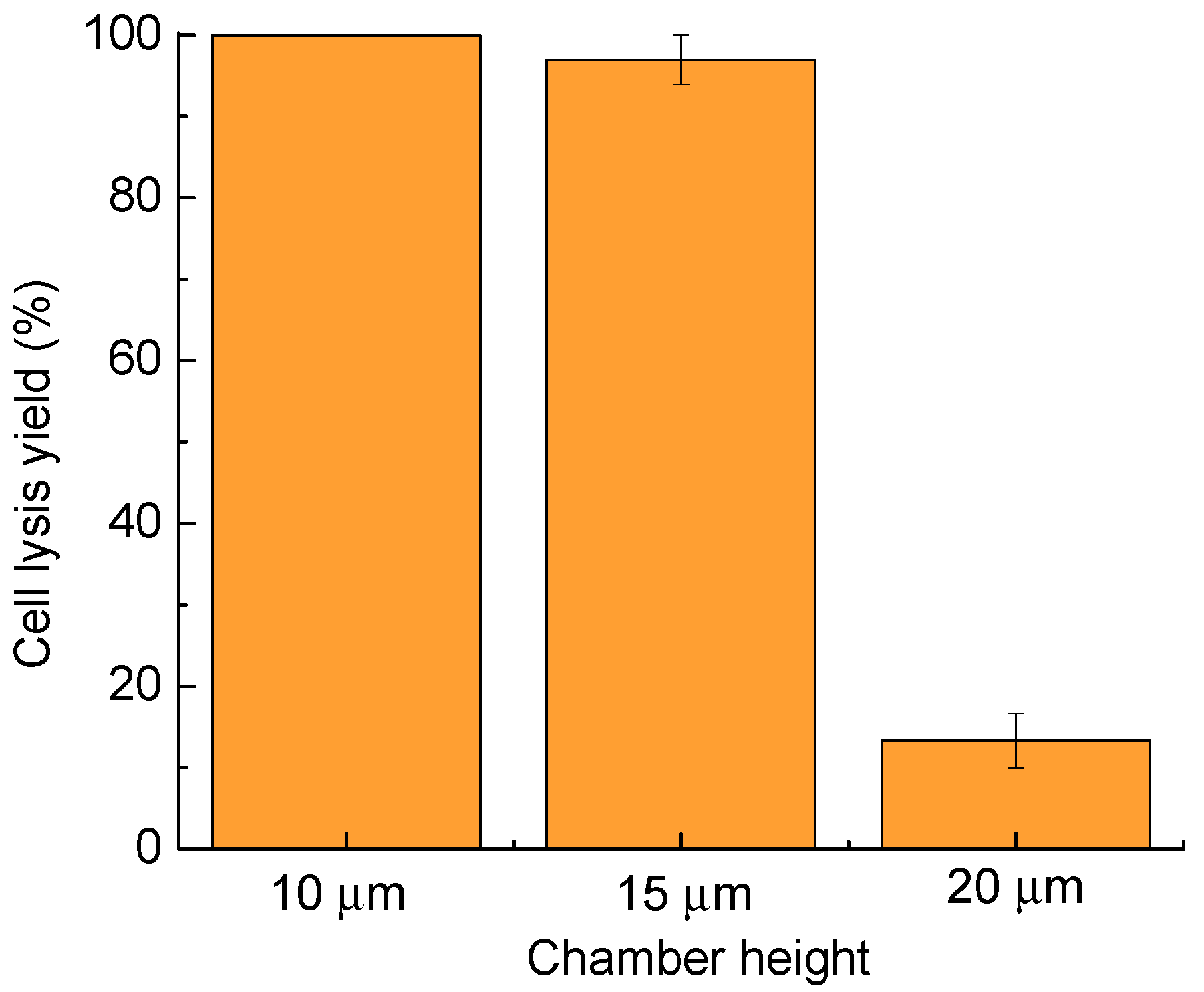
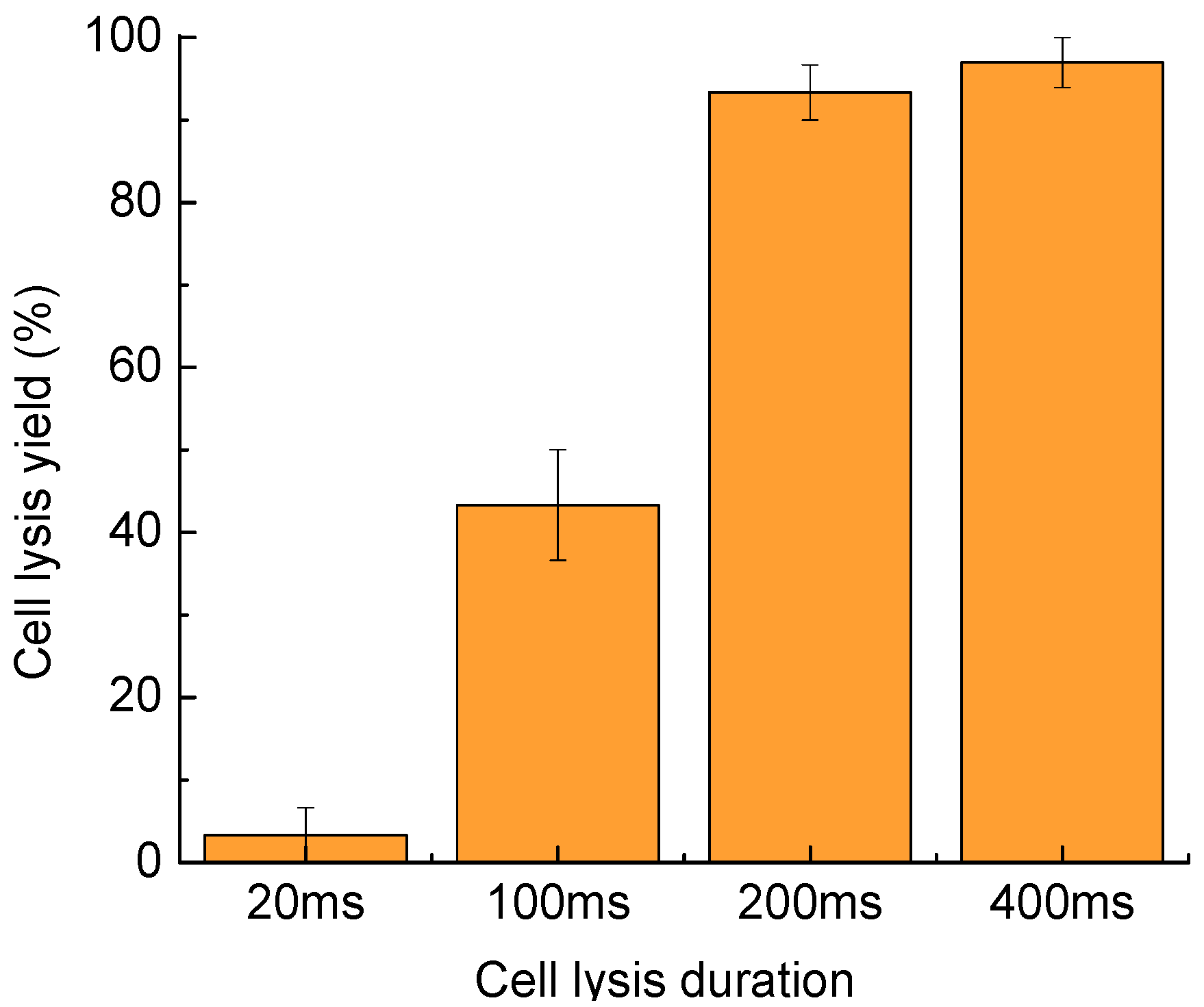
4. Characterization
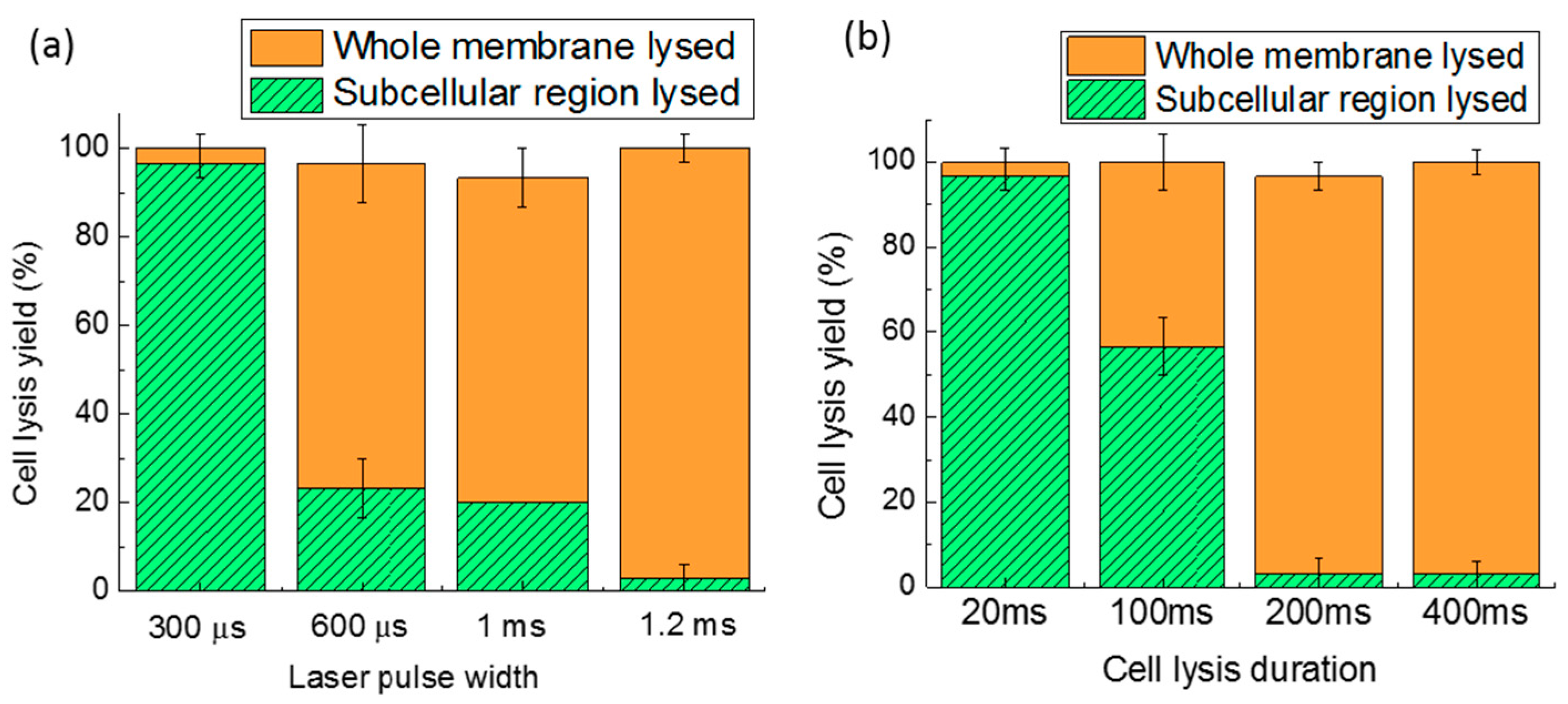
4.1. Laser Pulse Width
4.2. Microfluidic Chamber Height
4.3. Cell Lysis Duration
5. Results and Discussion
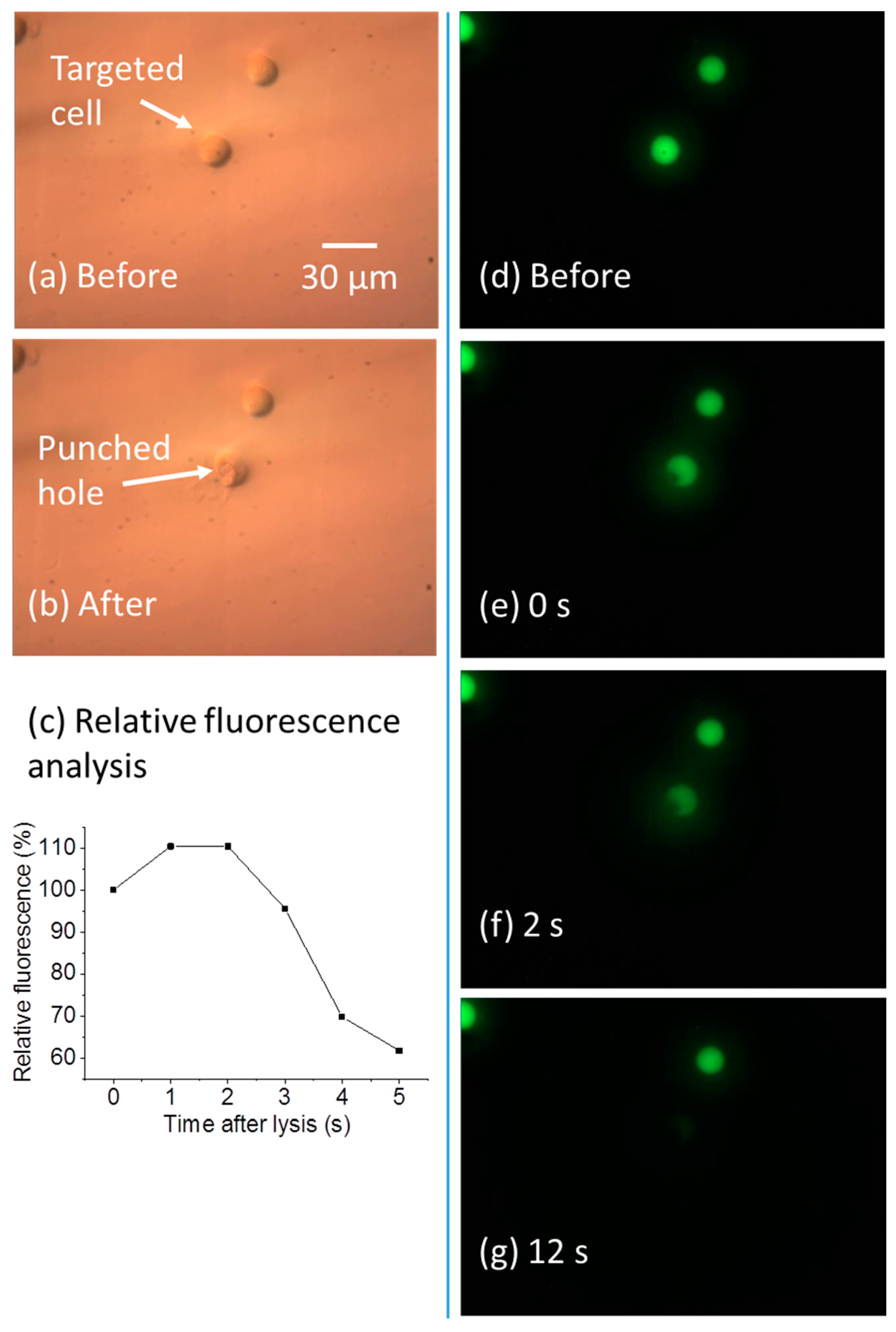
5.1. High-Resolution Single-Cell Lysis
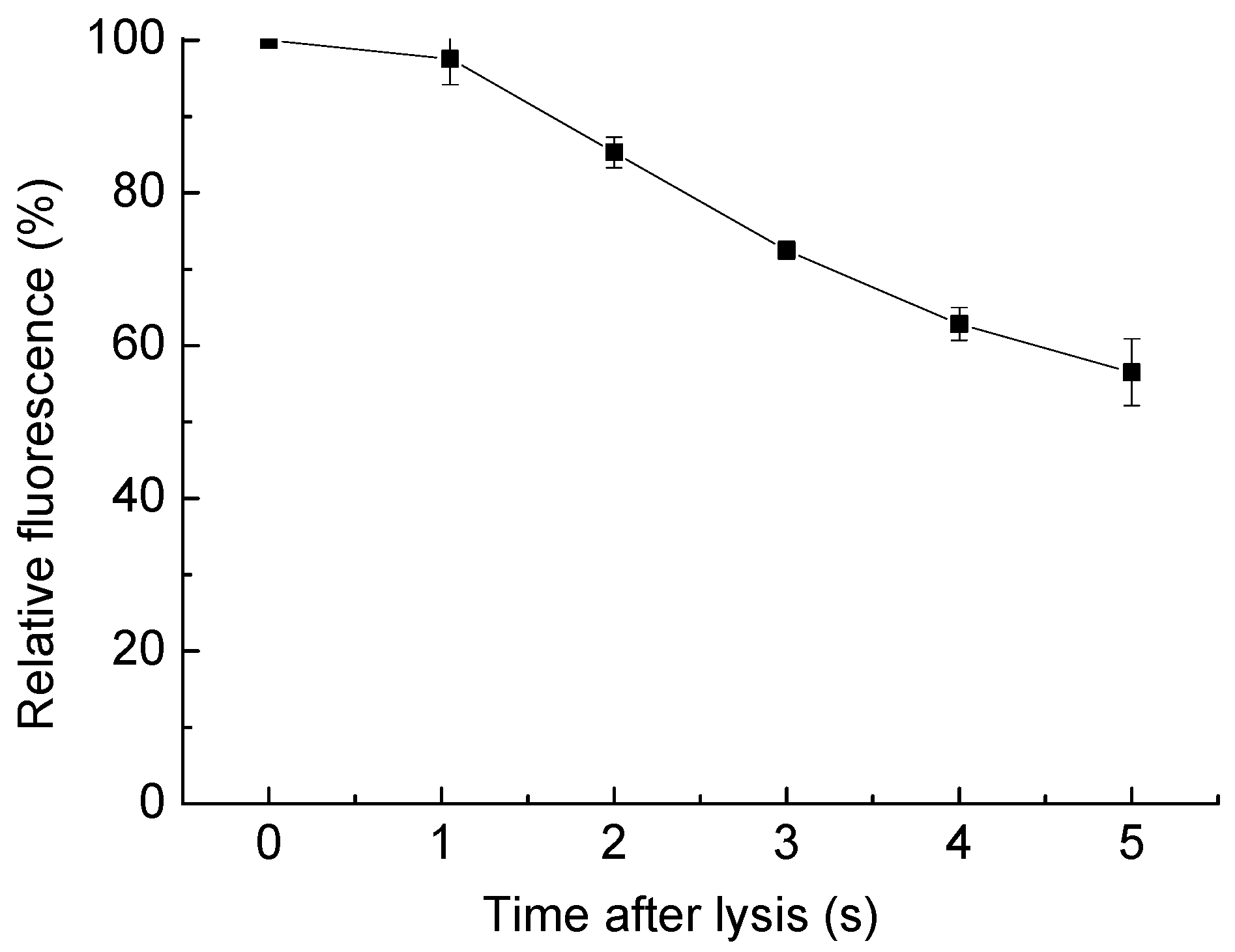
5.2. Dilution of Cell Content
5.3. Lysis of a Subcellular Region
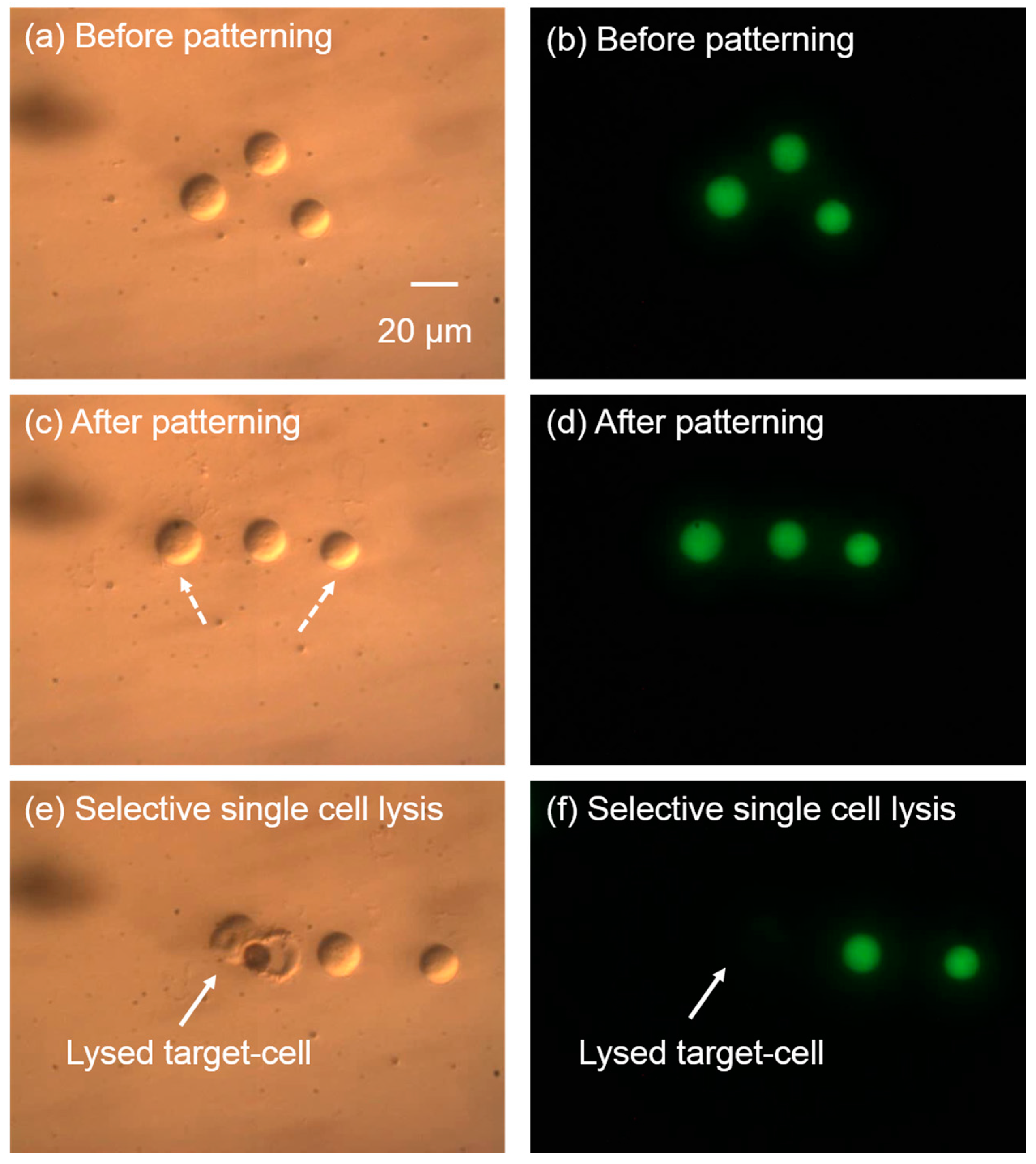
5.4. Integration of Cell Manipulation and Cell Lysis
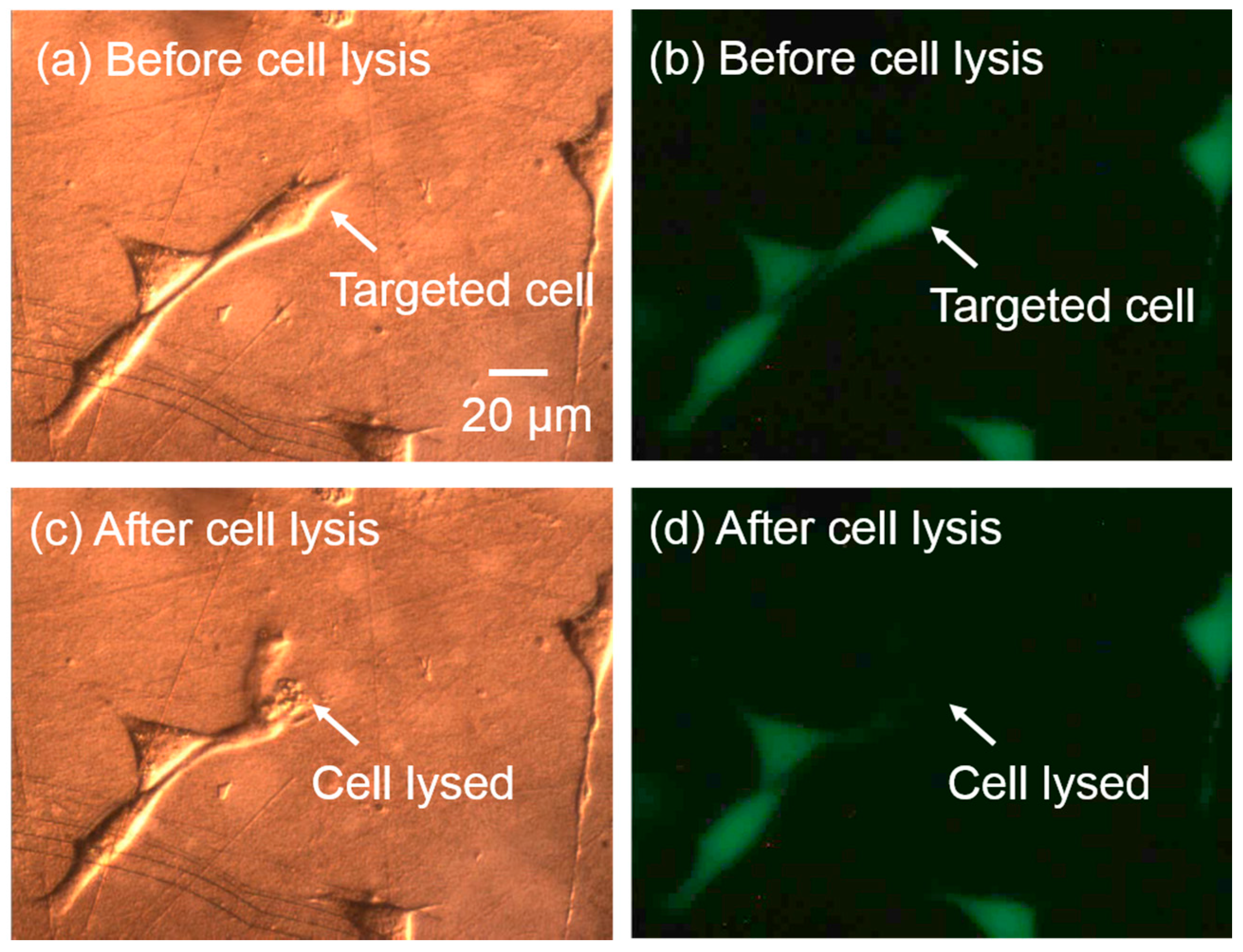
5.5. Adherent Single-Cell Lysis
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Di Carlo, D.; Lee, L.P. Dynamic single-cell analysis for quantitative biology. Anal. Chem. 2006, 78, 7918–7925. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.B.; Audet, J. Current techniques for single-cell lysis. J. R. Soc. Interface R. Soc. 2008, 5 (Suppl. S2), S131–S138. [Google Scholar] [CrossRef] [PubMed]
- Jen, C.P.; Hsiao, J.H.; Maslov, N.A. Single-cell chemical lysis on microfluidic chips with arrays of microwells. Sensors 2012, 12, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Wang, Y.; Sims, C.E.; Bachman, M.; Chang, R.; Li, G.P.; Allbritton, N.L. Fast electrical lysis of cells for capillary electrophoresis. Anal. Chem. 2003, 75, 3688–3696. [Google Scholar] [CrossRef] [PubMed]
- Hung, M.-S.; Chang, Y.-T. Single cell lysis and DNA extending using electroporation microfluidic device. Biochip J. 2012, 6, 84–90. [Google Scholar] [CrossRef]
- Jen, C.P.; Amstislavskaya, T.G.; Liu, Y.H.; Hsiao, J.H.; Chen, Y.H. Single-cell electric lysis on an electroosmotic-driven microfluidic chip with arrays of microwells. Sensors 2012, 12, 6967–6977. [Google Scholar] [CrossRef] [PubMed]
- Jokilaakso, N.; Salm, E.; Chen, A.; Millet, L.; Guevara, C.D.; Dorvel, B.; Reddy, B.; Karlstrom, A.E.; Chen, Y.; Ji, H.; et al. Ultra-localized single cell electroporation using silicon nanowires. Lab Chip 2013, 13, 336–339. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-H.; Lee, G.-B. An integrated cell counting and continuous cell lysis device using an optically induced electric field. Sens. Actuators B Chem. 2010, 145, 854–860. [Google Scholar] [CrossRef]
- Kremer, C.; Witte, C.; Neale, S.L.; Reboud, J.; Barrett, M.P.; Cooper, J.M. Shape-dependent optoelectronic cell lysis. Angew. Chem. Int. Ed. Engl. 2014, 53, 842–846. [Google Scholar] [CrossRef] [PubMed]
- Nan, L.; Jiang, Z.; Wei, X. Emerging microfluidic devices for cell lysis: A review. Lab Chip 2014, 14, 1060–1073. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Jin, W. Single-cell analysis by intracellular immuno-reaction and capillary electrophoresis with laser-induced fluorescence detection. J. Chromatogr. A 2006, 1104, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.; Jin, H.; Ma, Y.; Ren, Z.; Xiao, C.; He, Z.; Zhang, F.; Zhu, Q.; Wang, B. A continuous thermal lysis procedure for the large-scale preparation of plasmid DNA. J. Biotechnol. 2005, 118, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Carlo, D.D.; Jeong, K.-H.; Lee, L.P. Reagentless mechanical cell lysis by nanoscale barbs in microchannels for sample preparation. Lab Chip 2003, 3, 287–291. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Hong, J.W.; Kim, D.P.; Shin, J.H.; Park, I. Nanowire-integrated microfluidic devices for facile and reagent-free mechanical cell lysis. Lab Chip 2012, 12, 2914–2921. [Google Scholar] [CrossRef] [PubMed]
- Rau, K.R.; Guerra, A.; Vogel, A.; Venugopalan, V. Investigation of laser-induced cell lysis using time-resolved imaging. Appl. Phys. Lett. 2004, 84, 2940–2942. [Google Scholar] [CrossRef]
- Rau, K.R.; Quinto-Su, P.A.; Hellman, A.N.; Venugopalan, V. Pulsed laser microbeam-induced cell lysis: Time-resolved imaging and analysis of hydrodynamic effects. Biophys. J. 2006, 91, 317–329. [Google Scholar] [CrossRef] [PubMed]
- Hellman, A.N.; Rau, K.R.; Yoon, H.H.; Venugopalan, V. Biophysical response to pulsed laser microbeam-induced cell lysis and molecular delivery. J. Biophotonics 2008, 1, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Lai, H.-H.; Quinto-Su, P.A.; Sims, C.E.; Bachman, M.; Li, G.P.; Venugopalan, V.; Allbritton, N.L. Characterization and use of laser-based lysis for cell analysis on-chip. J. R. Soc. Interface 2008, 5, S113–S121. [Google Scholar] [CrossRef] [PubMed]
- Quinto-Su, P.A.; Lai, H.-H.; Yoon, H.H.; Sims, C.E.; Allbritton, N.L.; Venugopalan, V. Examination of laser microbeam cell lysis in a PDMS microfluidic channel using time-resolved imaging. Lab Chip 2008, 8, 408–414. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.G.; Liu, A.Q.; Klaseboer, E.; Zhang, J.B.; Ohl, C.D. Single cell membrane poration by bubble-induced microjets in a microfluidic chip. Lab Chip 2013, 13, 1144–1150. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Fan, Q.; Ohta, A.T. An opto-thermocapillary cell micromanipulator. Lab Chip 2013, 13, 2285–2291. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Xie, Y.; Mao, Z.; Zhao, Y.; Rufo, J.; Yang, S.; Guo, F.; Mai, J.D.; Huang, T.J. Theory and experiment on particle trapping and manipulation via optothermally generated bubbles. Lab Chip 2014, 14, 384–391. [Google Scholar] [CrossRef] [PubMed]
- Thery, M. Micropatterning as a tool to decipher cell morphogenesis and functions. J. Cell Sci. 2010, 123, 4201–4213. [Google Scholar] [CrossRef] [PubMed]
- Gautrot, J.E.; Trappmann, B.; Oceguera-Yanez, F.; Connelly, J.; He, X.; Watt, F.M.; Huck, W.T.S. Exploiting the superior protein resistance of polymer brushes to control single cell adhesion and polarisation at the micron scale. Biomaterials 2010, 31, 5030–5041. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Fan, Q.; Ohta, A. Interactive actuation of multiple opto-thermocapillary flow-addressed bubble microrobots. Robot. Biomim. 2014, 1, 14. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Nyborg, W.L. Ultrasound, cavitation bubbles and their interaction with cells. Adv. Drug Deliv. Rev. 2008, 60, 1103–1116. [Google Scholar] [CrossRef] [PubMed]
- Marmottant, P.; Hilgenfeldt, S. Controlled vesicle deformation and lysis by single oscillating bubbles. Nature 2003, 423, 153–156. [Google Scholar] [CrossRef] [PubMed]
- Handa, A.; Emery, A.N.; Spier, R.E. On the evaluation of gas-liquid interfacial effects on hybridoma viability in bubble column bioreactors. Dev. Biol. Stand. 1987, 66, 241–253. [Google Scholar] [PubMed]
- Michaels, J.D.; Nowak, J.E.; Mallik, A.K.; Koczo, K.; Wasan, D.T.; Papoutsakis, E.T. Analysis of cell-to-bubble attachment in sparged bioreactors in the presence of cell-protecting additives. Biotechnol. Bioeng. 1995, 47, 407–419. [Google Scholar] [CrossRef] [PubMed]
- Meredith, G.D.; Sims, C.E.; Soughayer, J.S.; Allbritton, N.L. Measurement of kinase activation in single mammalian cells. Nat. Biotechnol. 2000, 18, 309–312. [Google Scholar] [PubMed]
- Li, H.; Wu, H.Y.; Wang, Y.; Sims, C.E.; Allbritton, N.L. Improved capillary electrophoresis conditions for the separation of kinase substrates by the laser micropipet system. J. Chromatogr. B Biomed. Sci. Appl. 2001, 757, 79–88. [Google Scholar] [CrossRef]
- Li, H.; Sims, C.E.; Kaluzova, M.; Stanbridge, E.J.; Allbritton, N.L. A quantitative single-cell assay for protein kinase B reveals important insights into the biochemical behavior of an intracellular substrate peptide. Biochemistry 2004, 43, 1599–1608. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Sims, C.E.; Wu, H.Y.; Allbritton, N.L. Spatial control of cellular measurements with the laser micropipet. Anal. Chem. 2001, 73, 4625–4631. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.; Hu, W.; Ohta, A.T. Efficient single-cell poration by microsecond laser pulses. Lab Chip 2015, 15, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Pan, Q.; Lee, L.P. Single-cell level co-culture platform for intercellular communication. Integr. Biol. 2012, 4, 374–380. [Google Scholar] [CrossRef] [PubMed]
- Mellors, J.S.; Jorabchi, K.; Smith, L.M.; Ramsey, J.M. Integrated microfluidic device for automated single cell analysis using electrophoretic separation and electrospray ionization mass spectrometry. Anal. Chem. 2010, 82, 967–973. [Google Scholar] [CrossRef] [PubMed]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, Q.; Hu, W.; Ohta, A.T. Localized Single-Cell Lysis and Manipulation Using Optothermally-Induced Bubbles. Micromachines 2017, 8, 121. https://doi.org/10.3390/mi8040121
Fan Q, Hu W, Ohta AT. Localized Single-Cell Lysis and Manipulation Using Optothermally-Induced Bubbles. Micromachines. 2017; 8(4):121. https://doi.org/10.3390/mi8040121
Chicago/Turabian StyleFan, Qihui, Wenqi Hu, and Aaron T. Ohta. 2017. "Localized Single-Cell Lysis and Manipulation Using Optothermally-Induced Bubbles" Micromachines 8, no. 4: 121. https://doi.org/10.3390/mi8040121






