A Droplet Microfluidics Based Platform for Mining Metagenomic Libraries for Natural Compounds
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmid Construction and Generation of E. Coli Strains
2.2. Cell Culture
2.3. Apoptosis Experiments
2.4. Purified Violacein Experiments
2.5. Microfluidic Chip Fabrication
2.6. Surfactant and Oil System
2.7. Microfluidic Set-Up
2.8. Preparation of Droplet Samples for FACS Analysis
2.9. Preparation of Samples for COPAS Sorting
3. Results
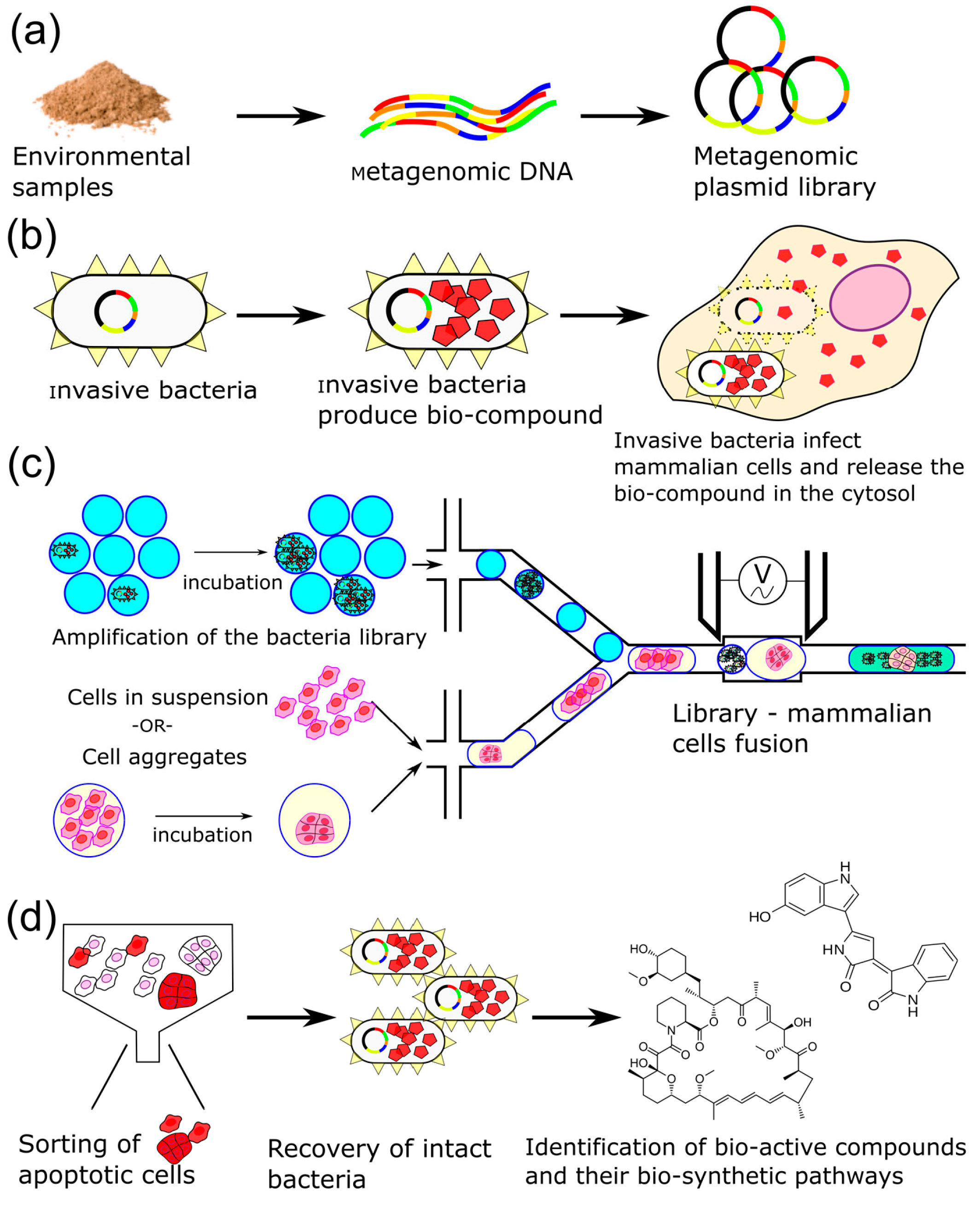
3.1. Strategy
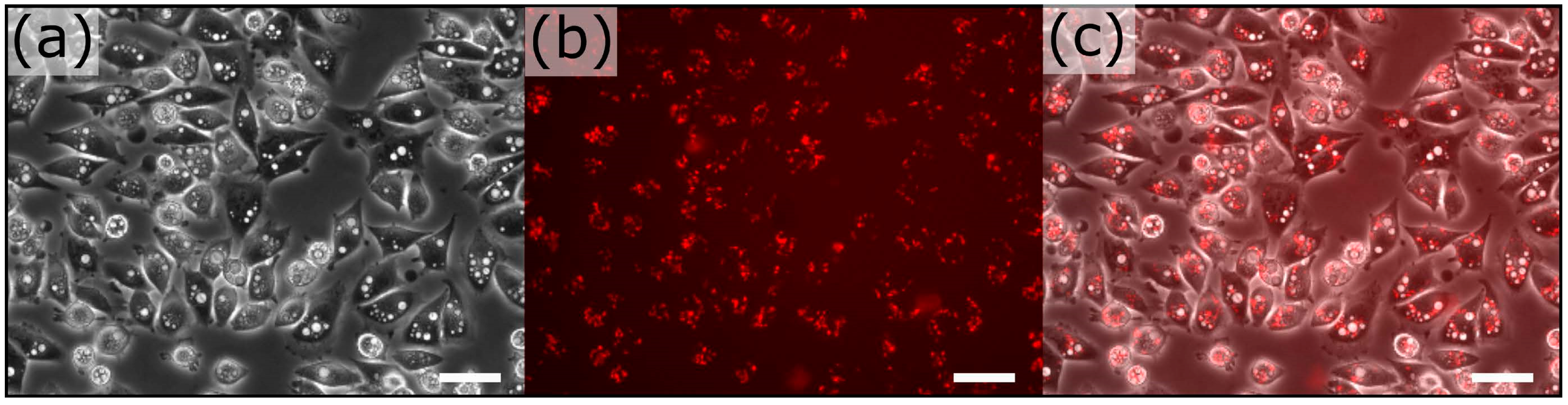
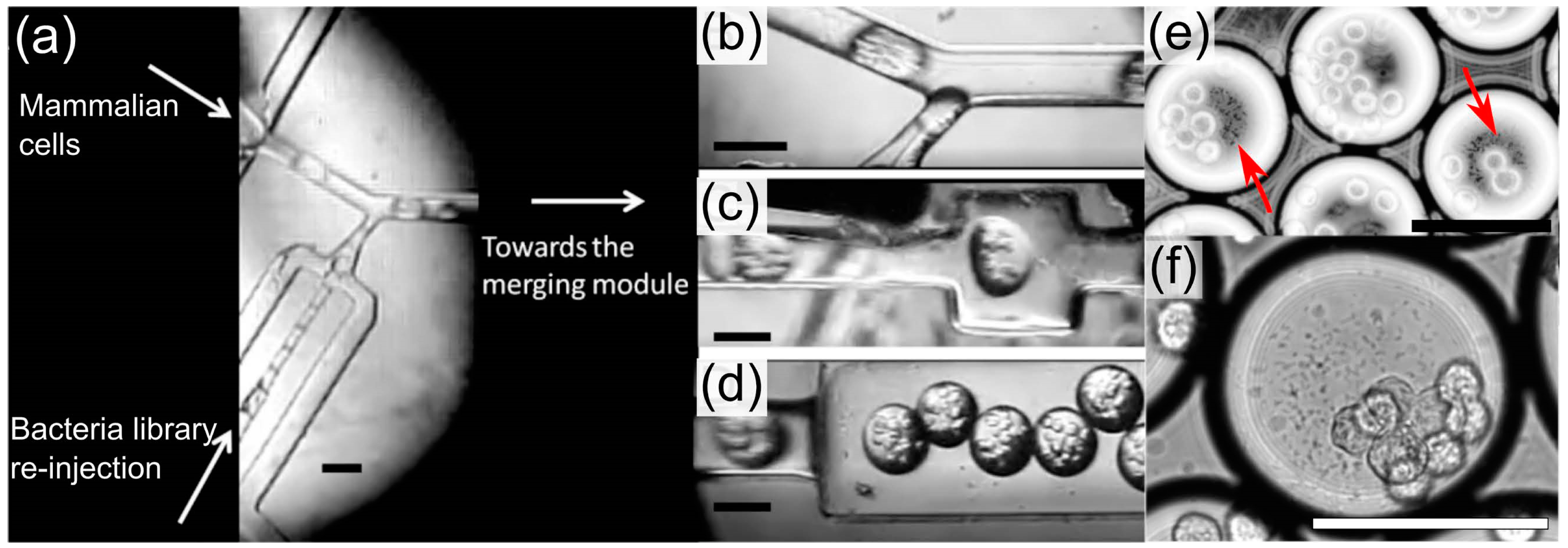
3.2. Extraction-Free Method for Mammalian Cell Delivery of Microbially Produced Natural Products
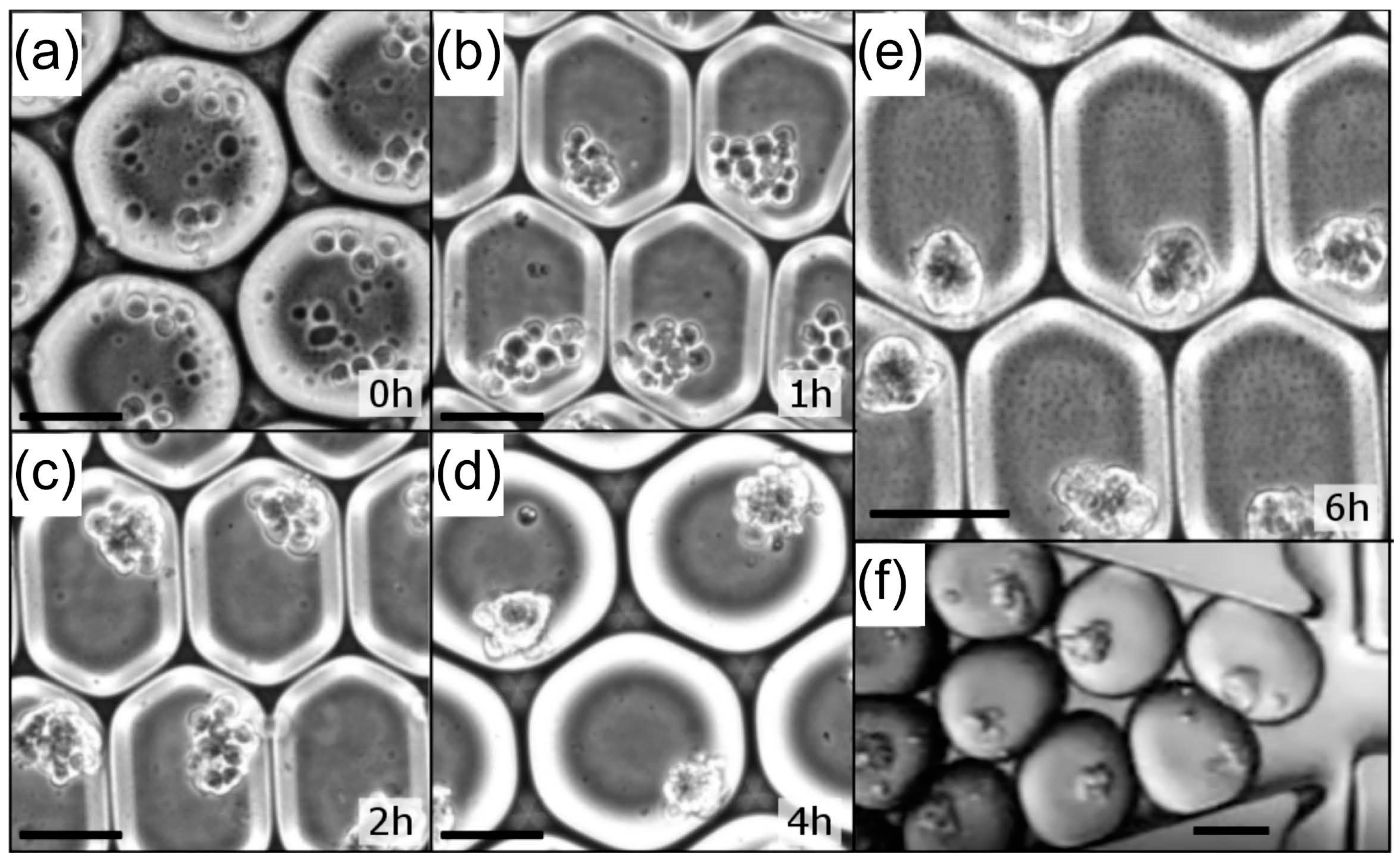
3.3. Droplet Microfluidic Workflow
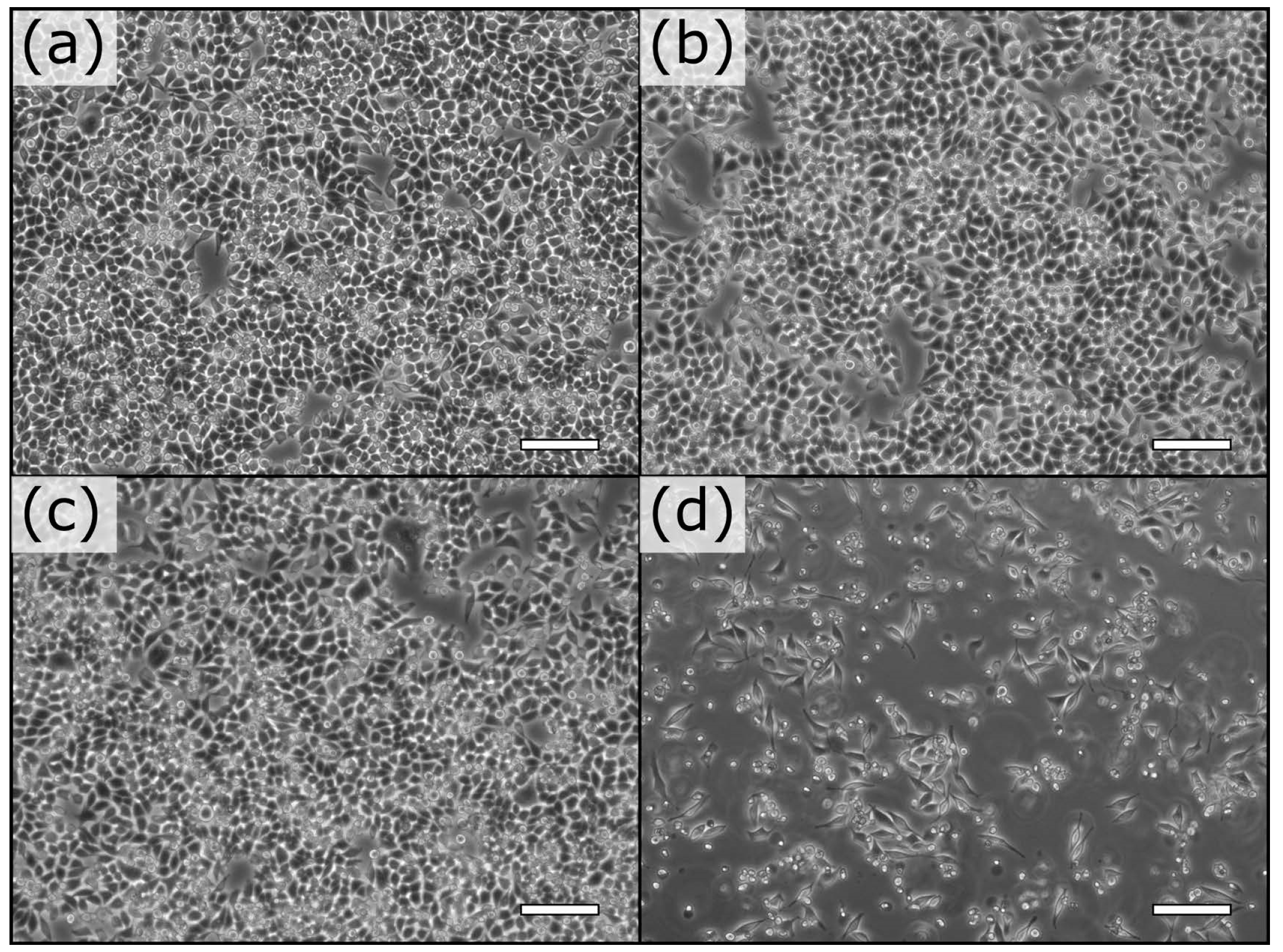
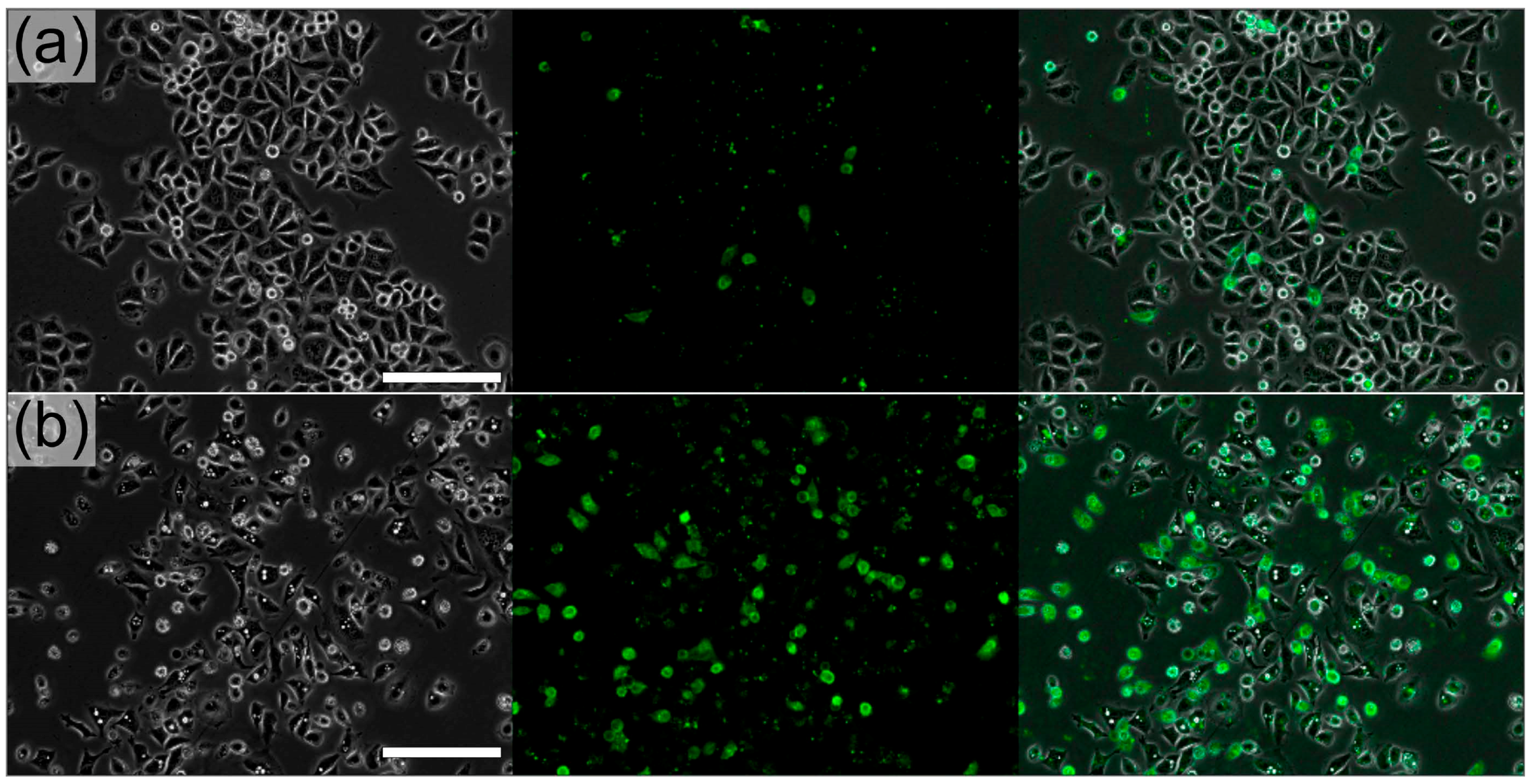
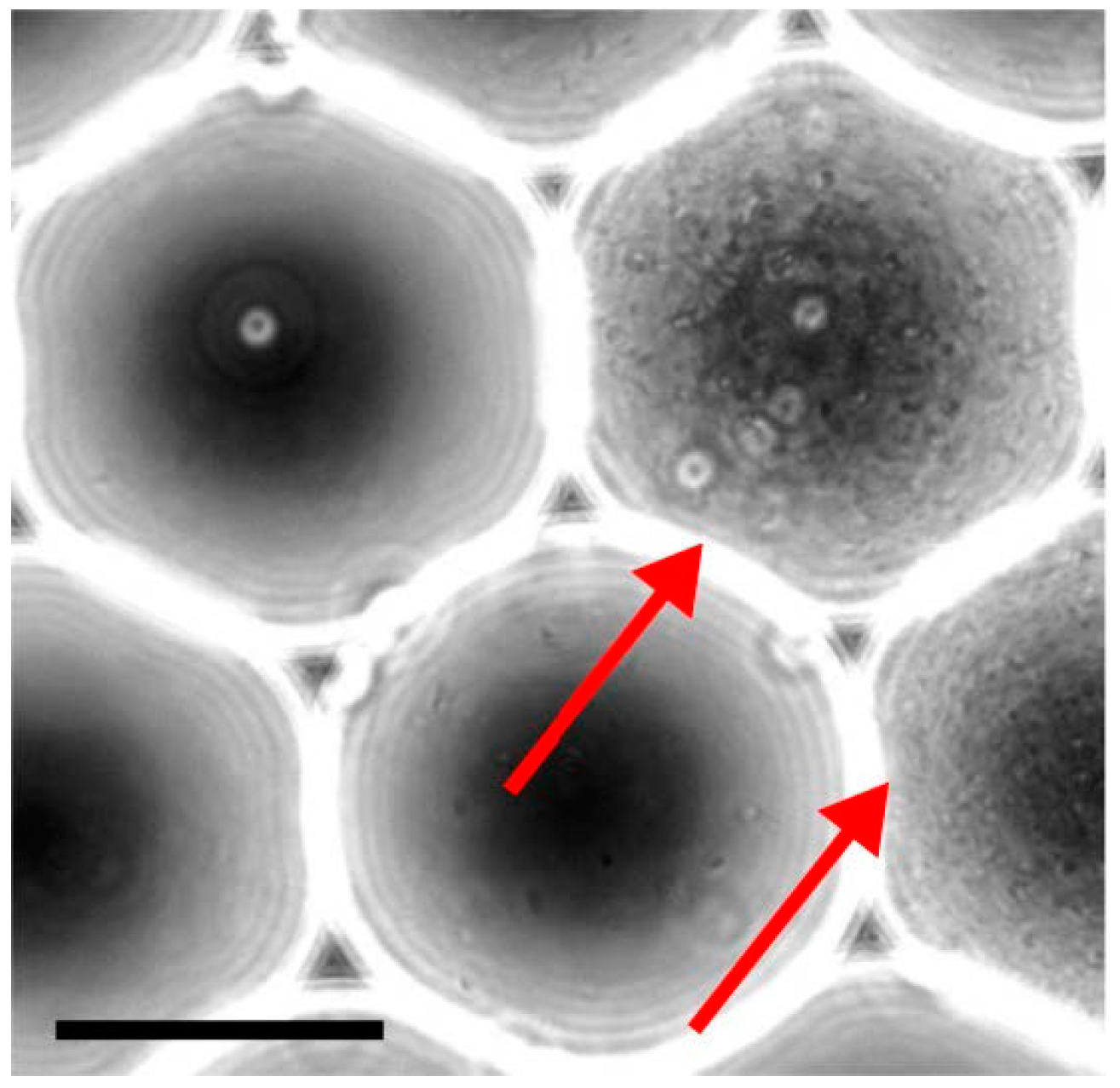
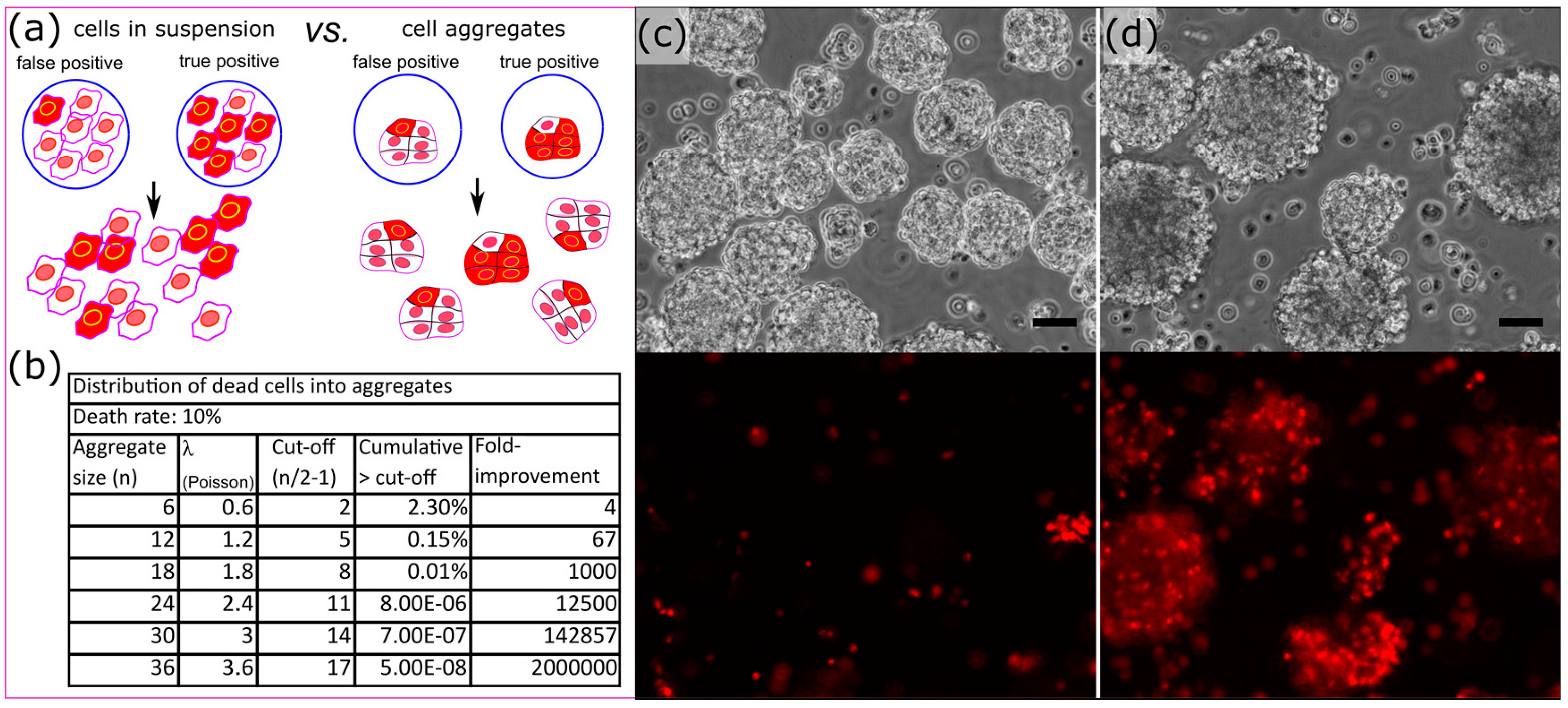
3.4. Cell Aggregates for Efficient Screening of Pro-Apoptotic Compounds
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kino, T.; Hatanaka, H.; Hashimoto, M.; Nishiyama, M.; Goto, T.; Okuhara, M.; Kohsaka, M.; Aoki, H.; Imanaka, H. Fk-506, a novel immunosuppressant isolated from a streptomyces. I. Fermentation, isolation, and physico-chemical and biological characteristics. J. Antibiot. 1987, 40, 1249–1255. [Google Scholar] [CrossRef] [PubMed]
- Paghdal, K.V.; Schwartz, R.A. Sirolimus (rapamycin): From the soil of easter island to a bright future. J. Am. Acad. Dermatol. 2007, 57, 1046–1050. [Google Scholar] [CrossRef] [PubMed]
- Vezina, C.; Kudelski, A.; Sehgal, S.N. Rapamycin (ay-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J. Antibiot. 1975, 28, 721–726. [Google Scholar] [CrossRef] [PubMed]
- Saleh-Lakha, S.; Miller, M.; Campbell, R.G.; Schneider, K.; Elahimanesh, P.; Hart, M.M.; Trevors, J.T. Microbial gene expression in soil: Methods, applications and challenges. J. Microbiol. Methods 2005, 63, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Torsvik, V.; Goksoyr, J.; Daae, F.L. High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 1990, 56, 782–787. [Google Scholar] [PubMed]
- Torsvik, V.; Salte, K.; Sorheim, R.; Goksoyr, J. Comparison of phenotypic diversity and DNA heterogeneity in a population of soil bacteria. Appl. Environ. Microbiol. 1990, 56, 776–781. [Google Scholar] [PubMed]
- Davis, K.E.; Joseph, S.J.; Janssen, P.H. Effects of growth medium, inoculum size, and incubation time on culturability and isolation of soil bacteria. Appl. Environ. Microbiol. 2005, 71, 826–834. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, B.S.; Eichorst, S.A.; Wertz, J.T.; Schmidt, T.M.; Breznak, J.A. New strategies for cultivation and detection of previously uncultured microbes. Appl. Environ. Microbiol. 2004, 70, 4748–4755. [Google Scholar] [CrossRef] [PubMed]
- Zengler, K.; Toledo, G.; Rappe, M.; Elkins, J.; Mathur, E.J.; Short, J.M.; Keller, M. Cultivating the uncultured. Proc. Natl. Acad. Sci. USA 2002, 99, 15681–15686. [Google Scholar] [CrossRef] [PubMed]
- Nichols, D.; Cahoon, N.; Trakhtenberg, E.M.; Pham, L.; Mehta, A.; Belanger, A.; Kanigan, T.; Lewis, K.; Epstein, S.S. Use of ichip for high-throughput in situ cultivation of “uncultivable” microbial species. Appl. Environ. Microbiol. 2010, 76, 2445–2450. [Google Scholar] [CrossRef] [PubMed]
- Rondon, M.R.; August, P.R.; Bettermann, A.D.; Brady, S.F.; Grossman, T.H.; Liles, M.R.; Loiacono, K.A.; Lynch, B.A.; MacNeil, I.A.; Minor, C.; et al. Cloning the soil metagenome: A strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 2000, 66, 2541–2547. [Google Scholar] [CrossRef] [PubMed]
- Treusch, A.H.; Kletzin, A.; Raddatz, G.; Ochsenreiter, T.; Quaiser, A.; Meurer, G.; Schuster, S.C.; Schleper, C. Characterization of large-insert DNA libraries from soil for environmental genomic studies of archaea. Environ. Microbiol. 2004, 6, 970–980. [Google Scholar] [CrossRef] [PubMed]
- Van Elsas, J.D.; Speksnijder, A.J.; van Overbeek, L.S. A procedure for the metagenomics exploration of disease-suppressive soils. J. Microbiol. Methods 2008, 75, 515–522. [Google Scholar] [CrossRef] [PubMed]
- Geng, H.; Bruhn, J.B.; Nielsen, K.F.; Gram, L.; Belas, R. Genetic dissection of tropodithietic acid biosynthesis by marine roseobacters. Appl. Environ. Microbiol. 2008, 74, 1535–1545. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, D.E.; Brady, S.F.; Bettermann, A.D.; Cianciotto, N.P.; Liles, M.R.; Rondon, M.R.; Clardy, J.; Goodman, R.M.; Handelsman, J. Isolation of antibiotics turbomycin a and b from a metagenomic library of soil microbial DNA. Appl. Environ. Microbiol. 2002, 68, 4301–4306. [Google Scholar] [CrossRef] [PubMed]
- Ilmberger, N.; Streit, W.R. Screening for cellulase encoding clones in metagenomic libraries. Methods Mol. Biol. 2017, 1539, 205–217. [Google Scholar] [PubMed]
- Knietsch, A.; Waschkowitz, T.; Bowien, S.; Henne, A.; Daniel, R. Construction and screening of metagenomic libraries derived from enrichment cultures: Generation of a gene bank for genes conferring alcohol oxidoreductase activity on Escherichia coli. Appl. Environ. Microbiol. 2003, 69, 1408–1416. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Wang, K.; Liu, Y.H. Molecular cloning and characterization of a novel pyrethroid-hydrolyzing esterase originating from the metagenome. Microb. Cell Fact. 2008, 7, 38. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Choi, J.N.; Kim, P.; Sok, D.E.; Nam, S.W.; Lee, C.H. LC-MS/MS profiling-based secondary metabolite screening of Myxococcus xanthus. J. Microbiol. Biotechnol. 2009, 19, 51–54. [Google Scholar] [PubMed]
- Uchiyama, T.; Abe, T.; Ikemura, T.; Watanabe, K. Substrate-induced gene-expression screening of environmental metagenome libraries for isolation of catabolic genes. Nat. Biotechnol. 2005, 23, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Geng, P.; Bai, F.; Bai, G.; Sun, T.; Li, X.; Shi, L.; Zhong, Q. Draft genome sequence of Streptomyces coelicoflavus ZG0656 reveals the putative biosynthetic gene cluster of acarviostatin family α-amylase inhibitors. Lett. Appl. Microbiol. 2012, 55, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Gao, C.; Tang, N.; Hu, S.; Wu, Q. Identification of genetic variations associated with epsilon-poly-lysine biosynthesis in streptomyces albulus zpm by genome sequencing. Sci. Rep. 2015, 5, 9201. [Google Scholar] [CrossRef] [PubMed]
- Xiang, S.; Fruehauf, J.; Li, C.J. Short hairpin RNA-expressing bacteria elicit RNA interference in mammals. Nat. Biotechnol. 2006, 24, 697–702. [Google Scholar] [CrossRef] [PubMed]
- Dagert, M.; Ehrlich, S.D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene 1979, 6, 23–28. [Google Scholar] [CrossRef]
- Hicks, S.W.; Charron, G.; Hang, H.C.; Galan, J.E. Subcellular targeting of Salmonella virulence proteins by host-mediated S-palmitoylation. Cell Host Microbe 2011, 10, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Beuzon, C.R.; Meresse, S.; Unsworth, K.E.; Ruiz-Albert, J.; Garvis, S.; Waterman, S.R.; Ryder, T.A.; Boucrot, E.; Holden, D.W. Salmonella maintains the integrity of its intracellular vacuole through the action of sifa. EMBO J. 2000, 19, 3235–3249. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, K.; Warburton, P.E. DNA modification and functional delivery into human cells using Escherichia coli DH10B. Nucleic Acids Res. 2003, 31, e51. [Google Scholar] [CrossRef] [PubMed]
- Brouzes, E. Droplet microfluidics for single-cell analysis. Methods Mol. Biol. 2012, 853, 105–139. [Google Scholar] [PubMed]
- McDonald, J.C.; Duffy, D.C.; Anderson, J.R.; Chiu, D.T.; Wu, H.; Schueller, O.J.; Whitesides, G.M. Fabrication of microfluidic systems in poly (dimethylsiloxane). Electrophoresis 2000, 21, 27–40. [Google Scholar] [CrossRef]
- Holtze, C.; Rowat, A.C.; Agresti, J.J.; Hutchison, J.B.; Angile, F.E.; Schmitz, C.H.; Koster, S.; Duan, H.; Humphry, K.J.; Scanga, R.A.; et al. Biocompatible surfactants for water-in-fluorocarbon emulsions. Lab Chip 2008, 8, 1632–1639. [Google Scholar] [CrossRef] [PubMed]
- Pulak, R. Techniques for analysis, sorting, and dispensing of C. elegans on the COPAS™ flow-sorting system. Methods Mol. Biol. 2006, 351, 275–286. [Google Scholar] [PubMed]
- Brouzes, E.; Medkova, M.; Savenelli, N.; Marran, D.; Twardowski, M.; Hutchison, J.B.; Rothberg, J.M.; Link, D.R.; Perrimon, N.; Samuels, M.L. Droplet microfluidic technology for single-cell high-throughput screening. Proc. Natl. Acad. Sci. USA 2009, 106, 14195–14200. [Google Scholar] [CrossRef] [PubMed]
- Chiu, D.T.; Lorenz, R.M. Chemistry and biology in femtoliter and picoliter volume droplets. Acc. Chem. Res. 2009, 42, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Clausell-Tormos, J.; Lieber, D.; Baret, J.C.; El-Harrak, A.; Miller, O.J.; Frenz, L.; Blouwolff, J.; Humphry, K.J.; Koster, S.; Duan, H.; et al. Droplet-based microfluidic platforms for the encapsulation and screening of mammalian cells and multicellular organisms. Chem. Biol. 2008, 15, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Geng, T.; Novak, R.; Mathies, R.A. Single-cell forensic short tandem repeat typing within microfluidic droplets. Anal. Chem. 2014, 86, 703–712. [Google Scholar] [CrossRef] [PubMed]
- Huebner, A.; Sharma, S.; Srisa-Art, M.; Hollfelder, F.; Edel, J.B.; Demello, A.J. Microdroplets: A sea of applications? Lab Chip 2008, 8, 1244–1254. [Google Scholar] [CrossRef] [PubMed]
- Macosko, E.Z.; Basu, A.; Satija, R.; Nemesh, J.; Shekhar, K.; Goldman, M.; Tirosh, I.; Bialas, A.R.; Kamitaki, N.; Martersteck, E.M.; et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 2015, 161, 1202–1214. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Chen, D.L.; Ismagilov, R.F. Reactions in droplets in microfluidic channels. Angew. Chem. 2006, 45, 7336–7356. [Google Scholar] [CrossRef] [PubMed]
- Taly, V.; Kelly, B.T.; Griffiths, A.D. Droplets as microreactors for high-throughput biology. ChemBioChem 2007, 8, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Theberge, A.B.; Courtois, F.; Schaerli, Y.; Fischlechner, M.; Abell, C.; Hollfelder, F.; Huck, W.T.S. Microdroplets in microfluidics: An evolving platform for discoveries in chemistry and biology. Angew. Chem. Int. Ed. 2010, 49, 5846–5868. [Google Scholar] [CrossRef] [PubMed]
- Fajac, I.; Grosse, S.; Collombet, J.M.; Thevenot, G.; Goussard, S.; Danel, C.; Grillot-Courvalin, C. Recombinant Escherichia coli as a gene delivery vector into airway epithelial cells. J. Control. Release 2004, 97, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Grillot-Courvalin, C.; Goussard, S.; Huetz, F.; Ojcius, D.M.; Courvalin, P. Functional gene transfer from intracellular bacteria to mammalian cells. Nat. Biotechnol. 1998, 16, 862–866. [Google Scholar] [CrossRef] [PubMed]
- Isberg, R.R.; Voorhis, D.L.; Falkow, S. Identification of invasin: A protein that allows enteric bacteria to penetrate cultured mammalian cells. Cell 1987, 50, 769–778. [Google Scholar] [CrossRef]
- Hess, D.J.; Henry-Stanley, M.J.; Erickson, E.A.; Wells, C.L. Intracellular survival of staphylococcus aureus within cultured enterocytes. J. Surg. Res. 2003, 114, 42–49. [Google Scholar] [CrossRef]
- Matz, C.; Webb, J.S.; Schupp, P.J.; Phang, S.Y.; Penesyan, A.; Egan, S.; Steinberg, P.; Kjelleberg, S. Marine biofilm bacteria evade eukaryotic predation by targeted chemical defense. PLoS ONE 2008, 3, e2744. [Google Scholar] [CrossRef] [PubMed]
- Masuelli, L.; Pantanella, F.; La Regina, G.; Benvenuto, M.; Fantini, M.; Mattera, R.; Di Stefano, E.; Mattei, M.; Silvestri, R.; Schippa, S.; et al. Violacein, an indole-derived purple-colored natural pigment produced by janthinobacterium lividum, inhibits the growth of head and neck carcinoma cell lines both in vitro and in vivo. Tumour Biol. 2016, 37, 3705–3717. [Google Scholar] [CrossRef] [PubMed]
- Alshatwi, A.A.; Subash-Babu, P.; Antonisamy, P. Violacein induces apoptosis in human breast cancer cells through up regulation of BAX, p53 and down regulation of MDM2. Exp. Toxicol. Pathol. 2016, 68, 89–97. [Google Scholar] [CrossRef] [PubMed]
- De Carvalho, D.D.; Costa, F.T.; Duran, N.; Haun, M. Cytotoxic activity of violacein in human colon cancer cells. Toxicol. In Vitro 2006, 20, 1514–1521. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, C.V.; Bos, C.L.; Versteeg, H.H.; Justo, G.Z.; Duran, N.; Peppelenbosch, M.P. Molecular mechanism of violacein-mediated human leukemia cell death. Blood 2004, 104, 1459–1464. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, P.R.; Rocha-Brito, K.J.; Fernandes, M.R.; Abrantes, J.L.; Duran, N.; Ferreira-Halder, C.V. Violacein induces death of RAS-mutated metastatic melanoma by impairing autophagy process. Tumour Biol. 2016, 37, 14049–14058. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, W.A.; Yusof, N.Z.; Nordin, N.; Zakaria, Z.A.; Rezali, M.F. Production and characterization of violacein by locally isolated chromobacterium violaceum grown in agricultural wastes. Appl. Biochem. Biotechnol. 2012, 167, 1220–1234. [Google Scholar] [CrossRef] [PubMed]
- Balibar, C.J.; Walsh, C.T. In vitro biosynthesis of violacein from L-tryptophan by the enzymes VioA-E from Chromobacterium violaceum. Biochemistry 2006, 45, 15444–15457. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, Y.; Zhao, S.; Zhang, J.; Wu, Y.; Zeng, S. Imidazole inhibits autophagy flux by blocking autophagic degradation and triggers apoptosis via increasing FoxO3a-Bim expression. Int. J. Oncol. 2015, 46, 721–731. [Google Scholar] [CrossRef] [PubMed]
- Huebner, A.; Srisa-Art, M.; Holt, D.; Abell, C.; Hollfelder, F.; deMello, A.J.; Edel, J.B. Quantitative detection of protein expression in single cells using droplet microfluidics. Chem. Commun. 2007, 1218–1220. [Google Scholar] [CrossRef] [PubMed]
- Anna, S.L.; Bontoux, N.; Stone, H.A. Formation of dispersions using “flow focusing” in microchannels. Appl. Phys. Lett. 2003, 82, 364–366. [Google Scholar] [CrossRef]
- Thorsen, T.; Roberts, R.W.; Arnold, F.H.; Quake, S.R. Dynamic pattern formation in a vesicle-generating microfluidic device. Phys. Rev. Lett. 2001, 86, 4163–4166. [Google Scholar] [CrossRef] [PubMed]
- Ahn, K.; Agresti, J.; Chong, H.; Marquez, M.; Weitz, D.A. Electrocoalescence of drops synchronized by size-dependent flow in microfluidic channels. Appl. Phys. Lett. 2006, 88, 264105. [Google Scholar] [CrossRef]
- McBurney, M.W.; Jones-Villeneuve, E.M.; Edwards, M.K.; Anderson, P.J. Control of muscle and neuronal differentiation in a cultured embryonal carcinoma cell line. Nature 1982, 299, 165–167. [Google Scholar] [CrossRef] [PubMed]
- McFall, R.C.; Sery, T.W.; Makadon, M. Characterization of a new continuous cell line derived from a human retinoblastoma. Cancer Res. 1977, 37, 1003–1010. [Google Scholar] [PubMed]
- Pfragner, R.; Hofler, H.; Behmel, A.; Ingolic, E.; Walser, V. Establishment and characterization of continuous cell line MTC-SK derived from a human medullary thyroid carcinoma. Cancer Res. 1990, 50, 4160–4166. [Google Scholar] [PubMed]
- Martins, A.H.; Resende, R.R.; Majumder, P.; Faria, M.; Casarini, D.E.; Tarnok, A.; Colli, W.; Pesquero, J.B.; Ulrich, H. Neuronal differentiation of P19 embryonal carcinoma cells modulates kinin B2 receptor gene expression and function. J. Biol. Chem. 2005, 280, 19576–19586. [Google Scholar] [CrossRef] [PubMed]
- Marikawa, Y.; Tamashiro, D.A.; Fujita, T.C.; Alarcon, V.B. Aggregated P19 mouse embryonal carcinoma cells as a simple in vitro model to study the molecular regulations of mesoderm formation and axial elongation morphogenesis. Genesis 2009, 47, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J. Natural products as leads to potential drugs: An old process or the new hope for drug discovery? J. Med. Chem. 2008, 51, 2589–2599. [Google Scholar] [CrossRef] [PubMed]
- Arcamone, F.; Cassinelli, G.; Fantini, G.; Grein, A.; Orezzi, P.; Pol, C.; Spalla, C. Adriamycin, 14-hydroxydaunomycin, a new antitumor antibiotic from S. peucetius var. caesius. Biotechnol. Bioeng. 1969, 11, 1101–1110. [Google Scholar] [CrossRef] [PubMed]
- Arcamone, F.; Cassinelli, G.; Fantini, G.; Grein, A.; Orezzi, P.; Pol, C.; Spalla, C. Adriamycin, 14-hydroxydaunomycin, a new antitumor antibiotic from S. peucetius var. caesius. Reprinted from biotechnology and bioengineering, vol. Xi, issue 6, pages 1101–1110 (1969). Biotechnol. Bioeng. 2000, 67, 704–713. [Google Scholar] [CrossRef]
- Ling, L.L.; Schneider, T.; Peoples, A.J.; Spoering, A.L.; Engels, I.; Conlon, B.P.; Mueller, A.; Schaberle, T.F.; Hughes, D.E.; Epstein, S.; et al. A new antibiotic kills pathogens without detectable resistance. Nature 2015, 517, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Chen, R.; Si, S.; Sun, C.; Xu, H. A new nucleosidyl-peptide antibiotic, sansanmycin. J. Antibiot. 2007, 60, 158–161. [Google Scholar] [CrossRef] [PubMed]
- Kevany, B.M.; Rasko, D.A.; Thomas, M.G. Characterization of the complete zwittermicin a biosynthesis gene cluster from bacillus cereus. Appl. Environ. Microbiol. 2009, 75, 1144–1155. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Shah, S.; Chung, L.; Carney, J.; Katz, L.; Khosla, C.; Julien, B. Cloning and heterologous expression of the epothilone gene cluster. Science 2000, 287, 640–642. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Allen, H.K.; Klimowicz, A.K.; Mlot, C.; Gross, J.A.; Savengsuksa, S.; McEllin, J.; Clardy, J.; Ruess, R.W.; Handelsman, J. Psychrotrophic strain of janthinobacterium lividum from a cold alaskan soil produces prodigiosin. DNA Cell Biol. 2010, 29, 533–541. [Google Scholar] [CrossRef] [PubMed]
- Rosenberger, C.M.; Finlay, B.B. Phagocyte sabotage: Disruption of macrophage signalling by bacterial pathogens. Nat. Rev. Mol. Cell Biol. 2003, 4, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Benoit, D.S.; Tripodi, M.C.; Blanchette, J.O.; Langer, S.J.; Leinwand, L.A.; Anseth, K.S. Integrin-linked kinase production prevents anoikis in human mesenchymal stem cells. J. Biomed. Mater. Res. Part A 2007, 81, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Marchionini, D.M.; Collier, T.J.; Camargo, M.; McGuire, S.; Pitzer, M.; Sortwell, C.E. Interference with anoikis-induced cell death of dopamine neurons: Implications for augmenting embryonic graft survival in a rat model of parkinson’s disease. J. Comp. Neurol. 2003, 464, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Weber, L.M.; Hayda, K.N.; Anseth, K.S. Cell-matrix interactions improve beta-cell survival and insulin secretion in three-dimensional culture. Tissue Eng. Part A 2008, 14, 1959–1968. [Google Scholar] [CrossRef] [PubMed]
- Frisch, S.M.; Francis, H. Disruption of epithelial cell-matrix interactions induces apoptosis. J. Cell Biol. 1994, 124, 619–626. [Google Scholar] [CrossRef] [PubMed]
- Van Wezel, A.L. Growth of cell-strains and primary cells on micro-carriers in homogeneous culture. Nature 1967, 216, 64–65. [Google Scholar] [CrossRef] [PubMed]
- 7Bardouille, C.; Lehmann, J.; Heimann, P.; Jockusch, H. Growth and differentiation of permanent and secondary mouse myogenic cell lines on microcarriers. Appl. Microbiol. Biotechnol. 2001, 55, 556–562. [Google Scholar] [CrossRef]
- Wang, Q.; Slegers, H.; Clauwaert, J. Isolation of plasma membranes from rat C6 glioma cells cultivated on microcarriers. Acta Histochem. 1999, 101, 327–339. [Google Scholar] [CrossRef]
- Leon, F.; Roy, G. Isolation of human small bowel intraepithelial lymphocytes by annexin V-coated magnetic beads. Lab. Investig. 2004, 84, 804–809. [Google Scholar] [CrossRef] [PubMed]
- Sciambi, A.; Abate, A.R. Accurate microfluidic sorting of droplets at 30 kHz. Lab Chip 2015, 15, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Agresti, J.J.; Antipov, E.; Abate, A.R.; Ahn, K.; Rowat, A.C.; Baret, J.C.; Marquez, M.; Klibanov, A.M.; Griffiths, A.D.; Weitz, D.A. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc. Natl. Acad. Sci. USA 2010, 107, 4004–4009. [Google Scholar] [CrossRef] [PubMed]
- Sjostrom, S.L.; Bai, Y.; Huang, M.; Liu, Z.; Nielsen, J.; Joensson, H.N.; Andersson Svahn, H. High-throughput screening for industrial enzyme production hosts by droplet microfluidics. Lab Chip 2014, 14, 806–813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Theodorou, E.; Scanga, R.; Twardowski, M.; Snyder, M.P.; Brouzes, E. A Droplet Microfluidics Based Platform for Mining Metagenomic Libraries for Natural Compounds. Micromachines 2017, 8, 230. https://doi.org/10.3390/mi8080230
Theodorou E, Scanga R, Twardowski M, Snyder MP, Brouzes E. A Droplet Microfluidics Based Platform for Mining Metagenomic Libraries for Natural Compounds. Micromachines. 2017; 8(8):230. https://doi.org/10.3390/mi8080230
Chicago/Turabian StyleTheodorou, Elias, Randall Scanga, Mariusz Twardowski, Michael P. Snyder, and Eric Brouzes. 2017. "A Droplet Microfluidics Based Platform for Mining Metagenomic Libraries for Natural Compounds" Micromachines 8, no. 8: 230. https://doi.org/10.3390/mi8080230





