Bioapplications of Cell-SELEX-Generated Aptamers in Cancer Diagnostics, Therapeutics, Theranostics and Biomarker Discovery: A Comprehensive Review
Abstract
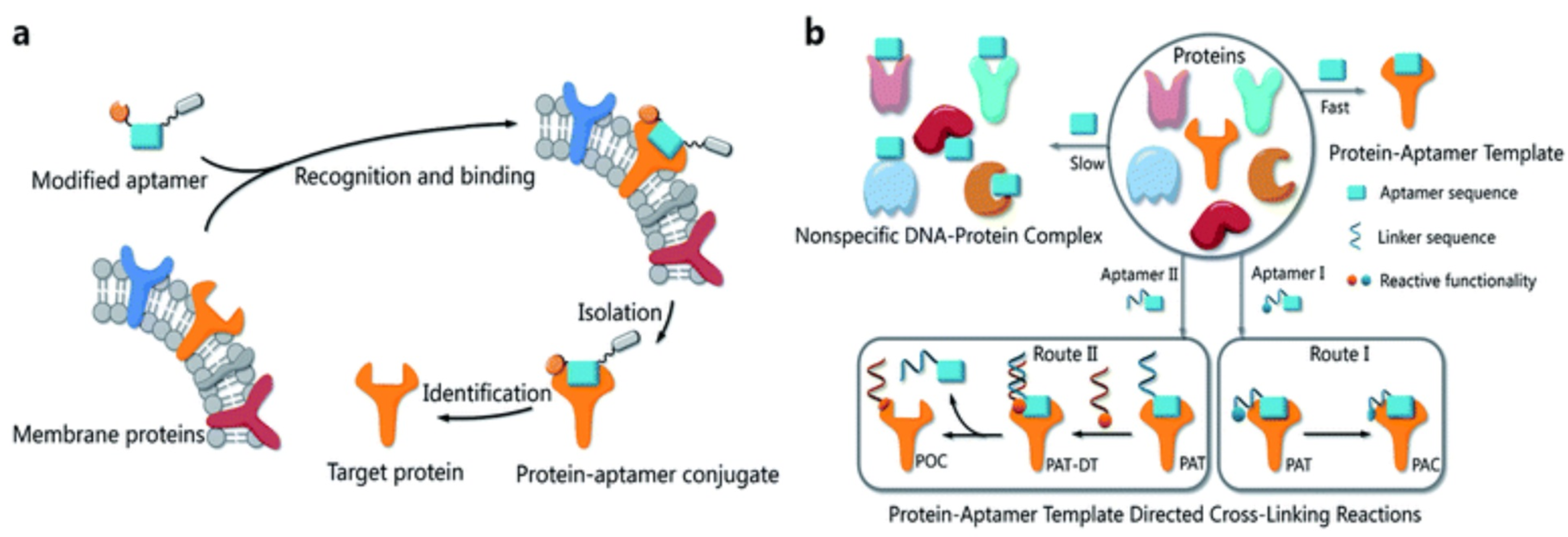
:1. Elucidation
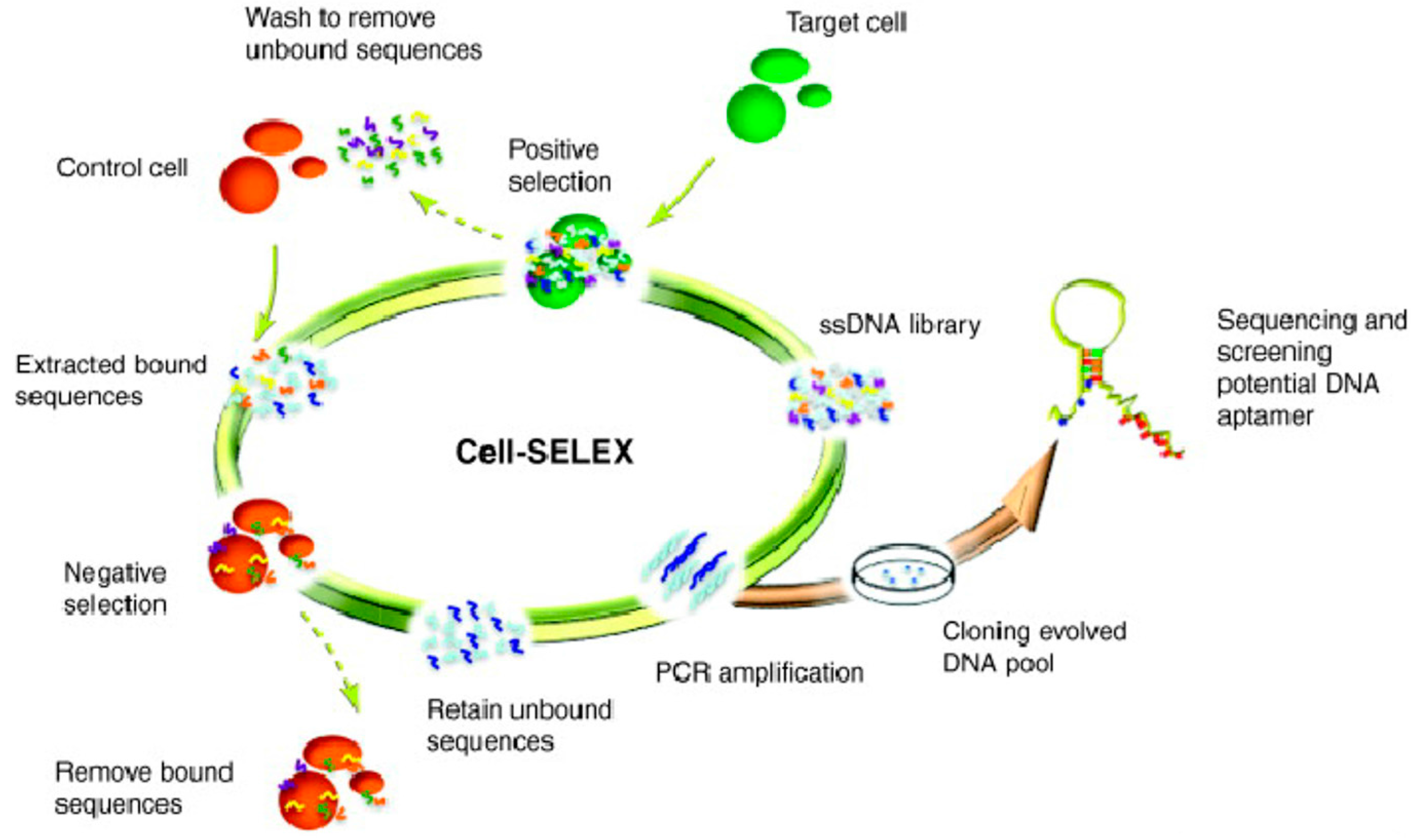
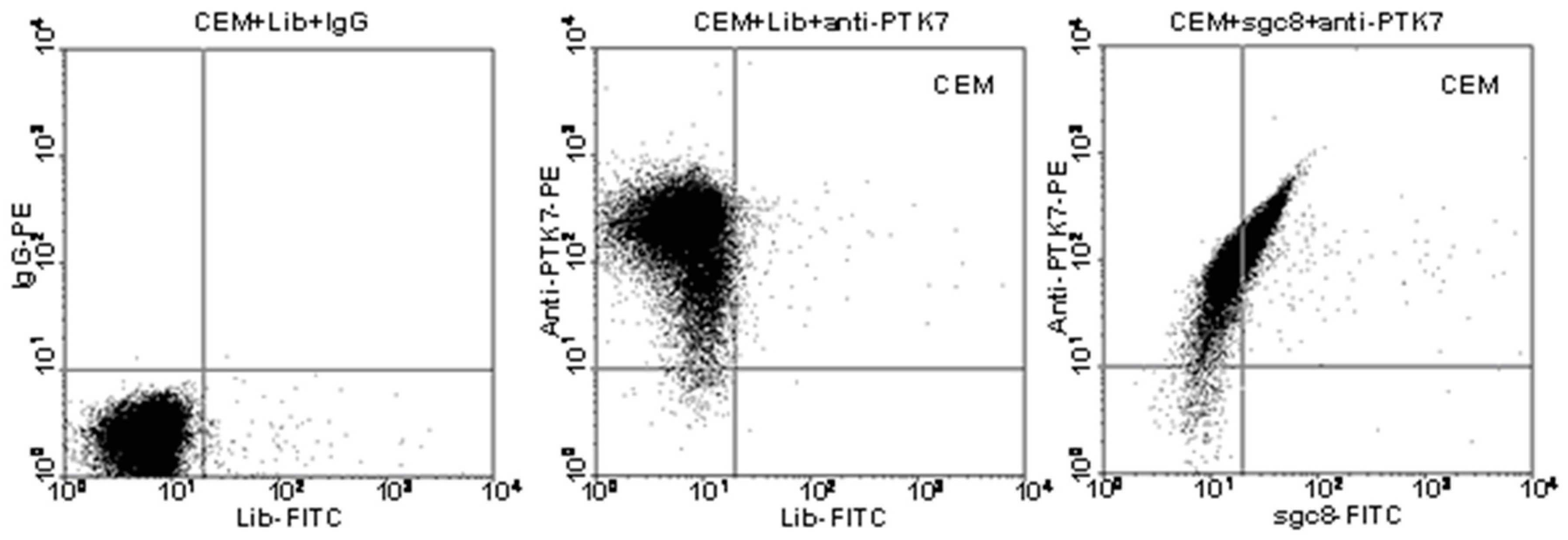
2. Generation of Aptamers by Cell-SELEX
3. Aptamer-Based Applications
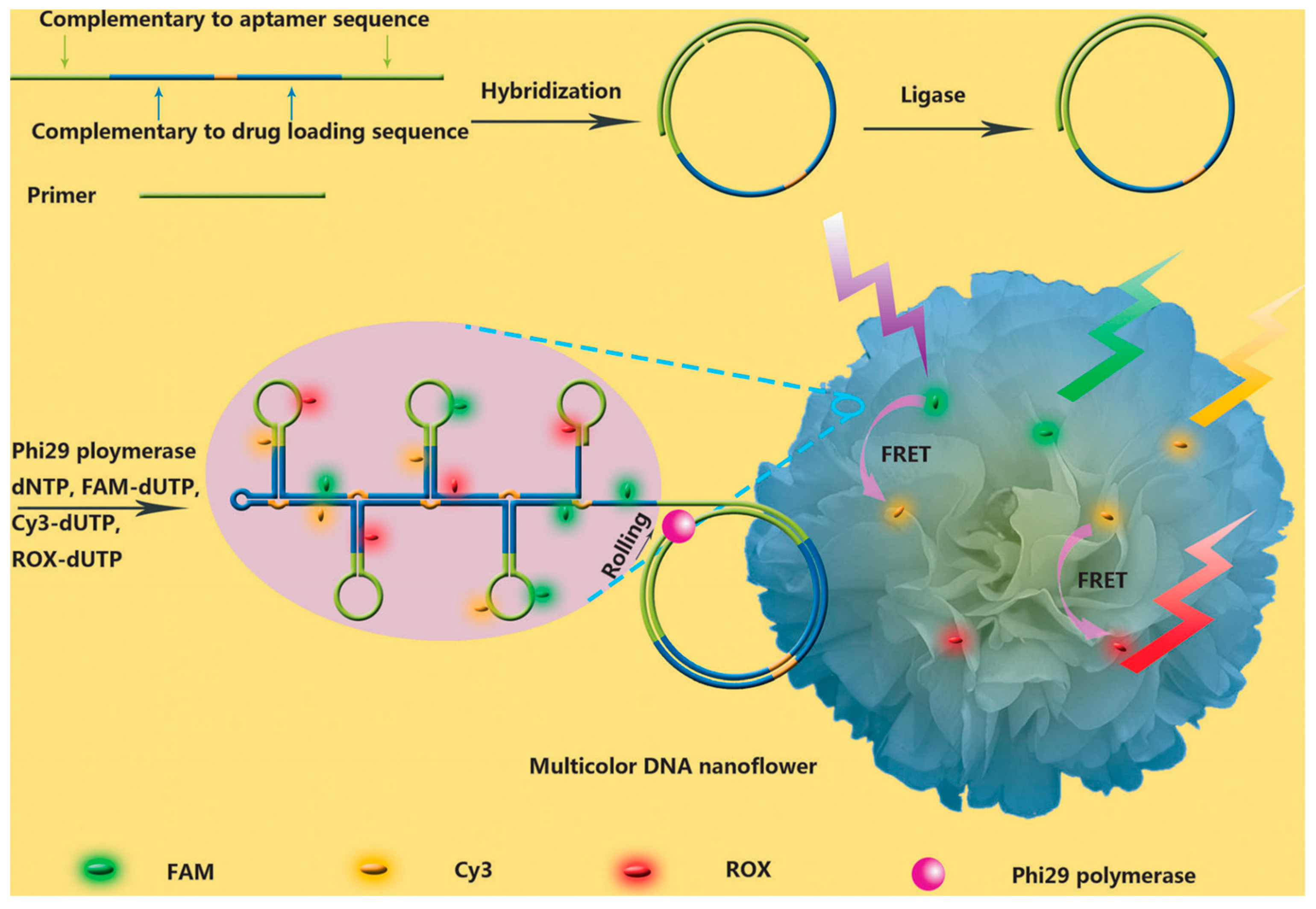
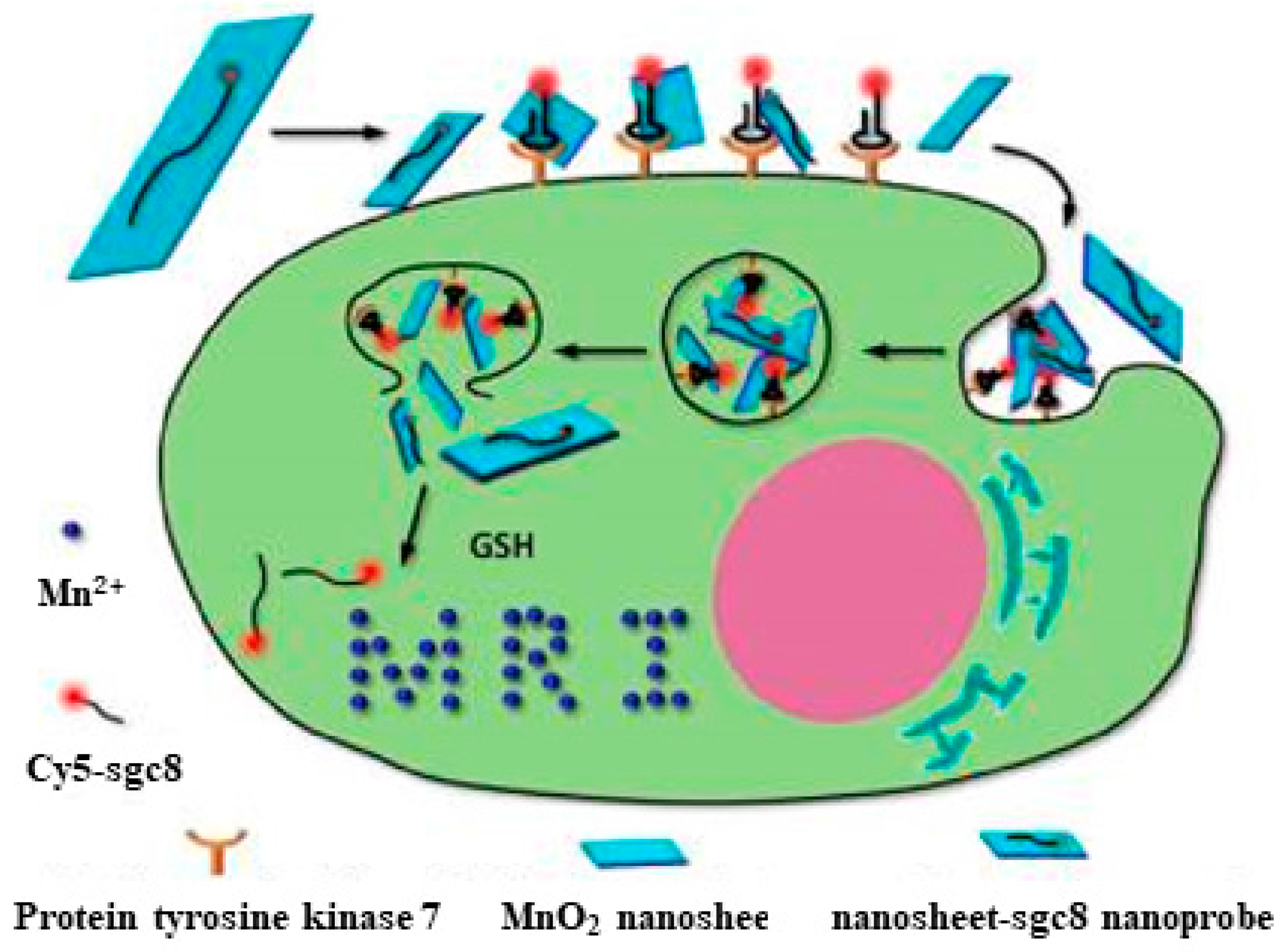
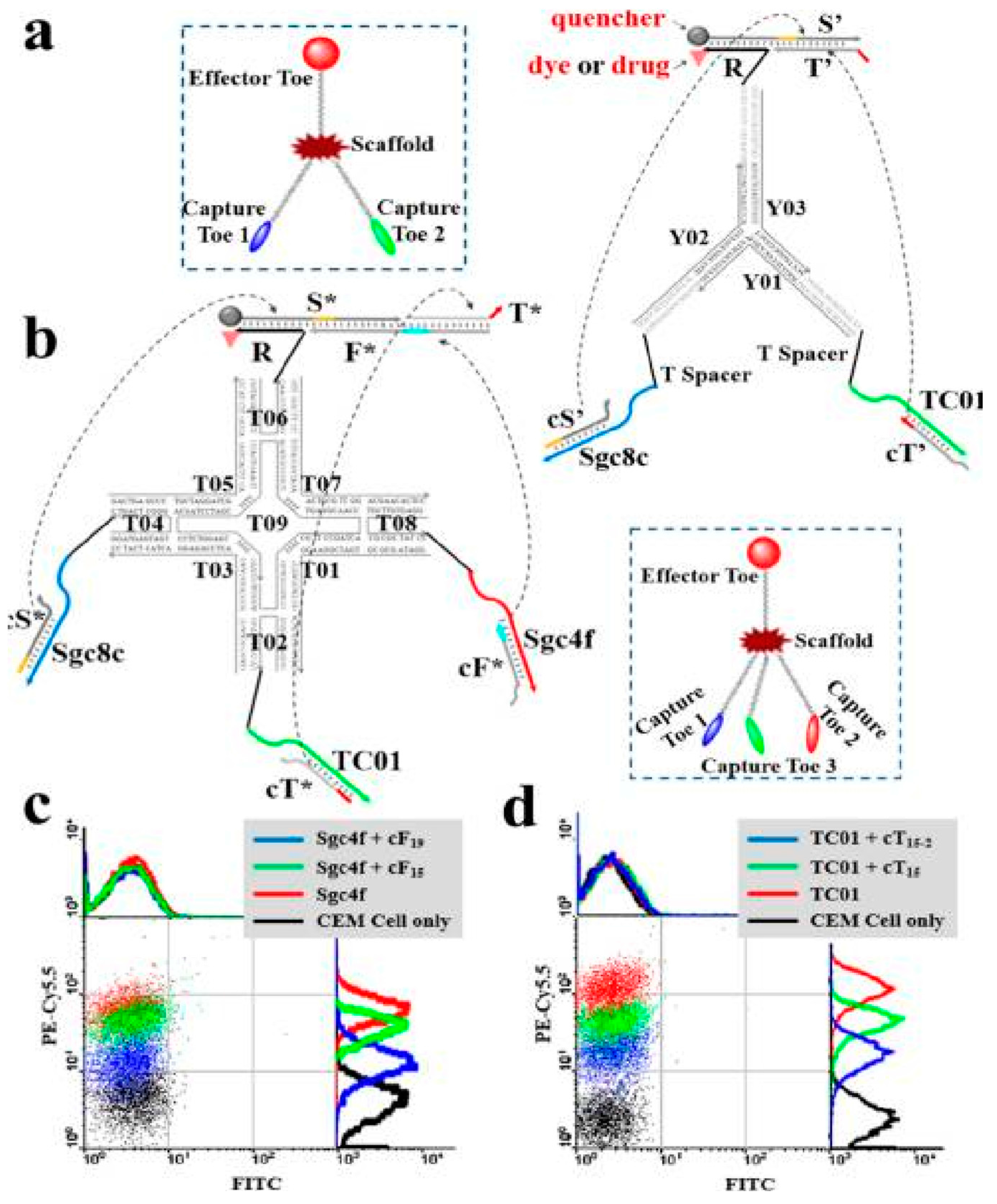
3.1. Imaging
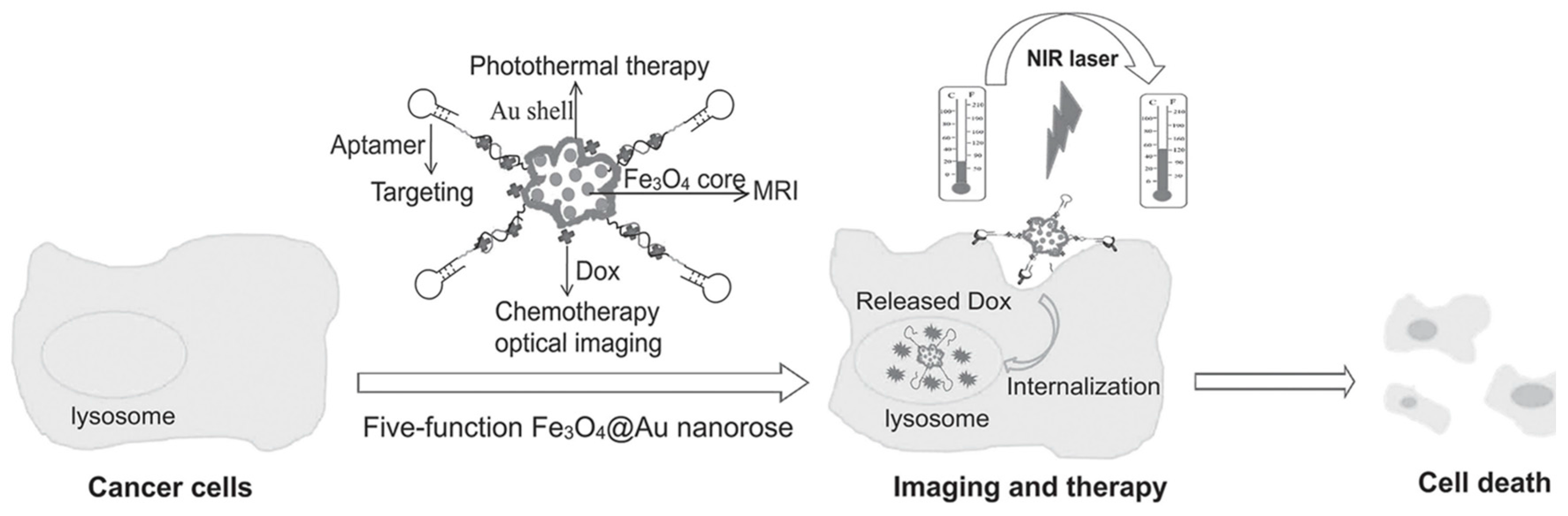
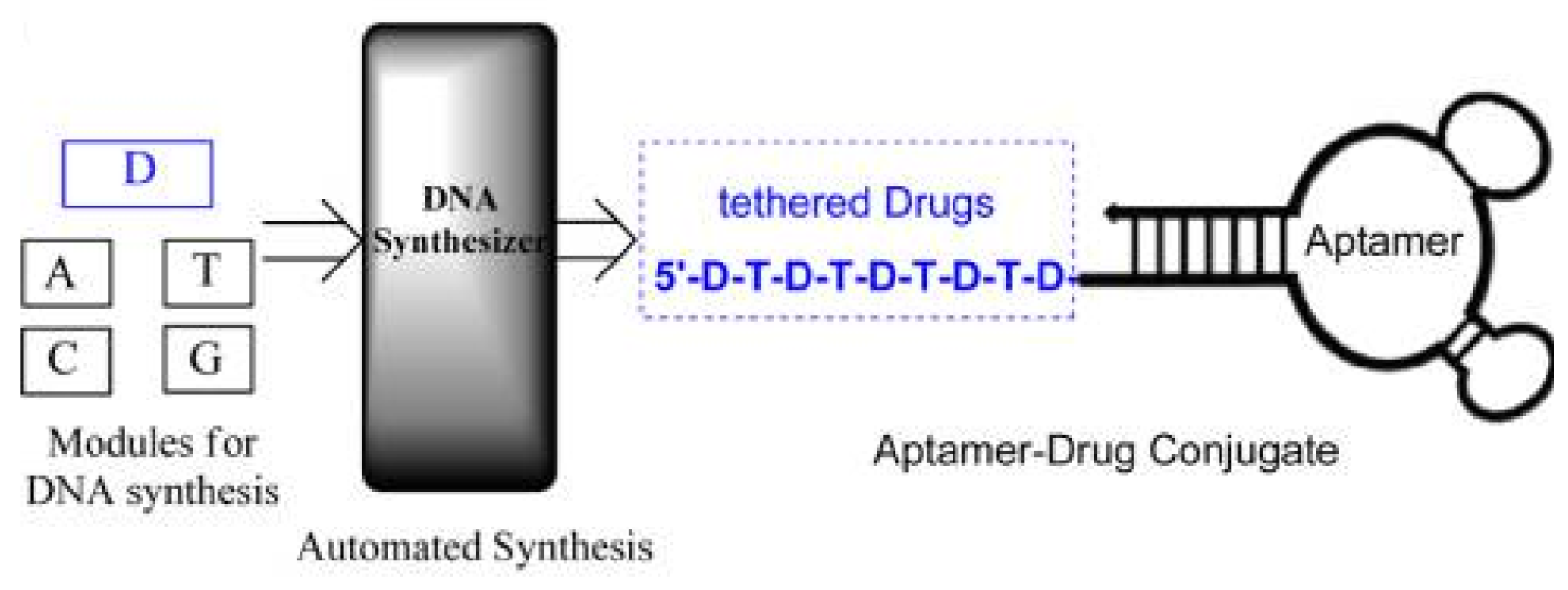
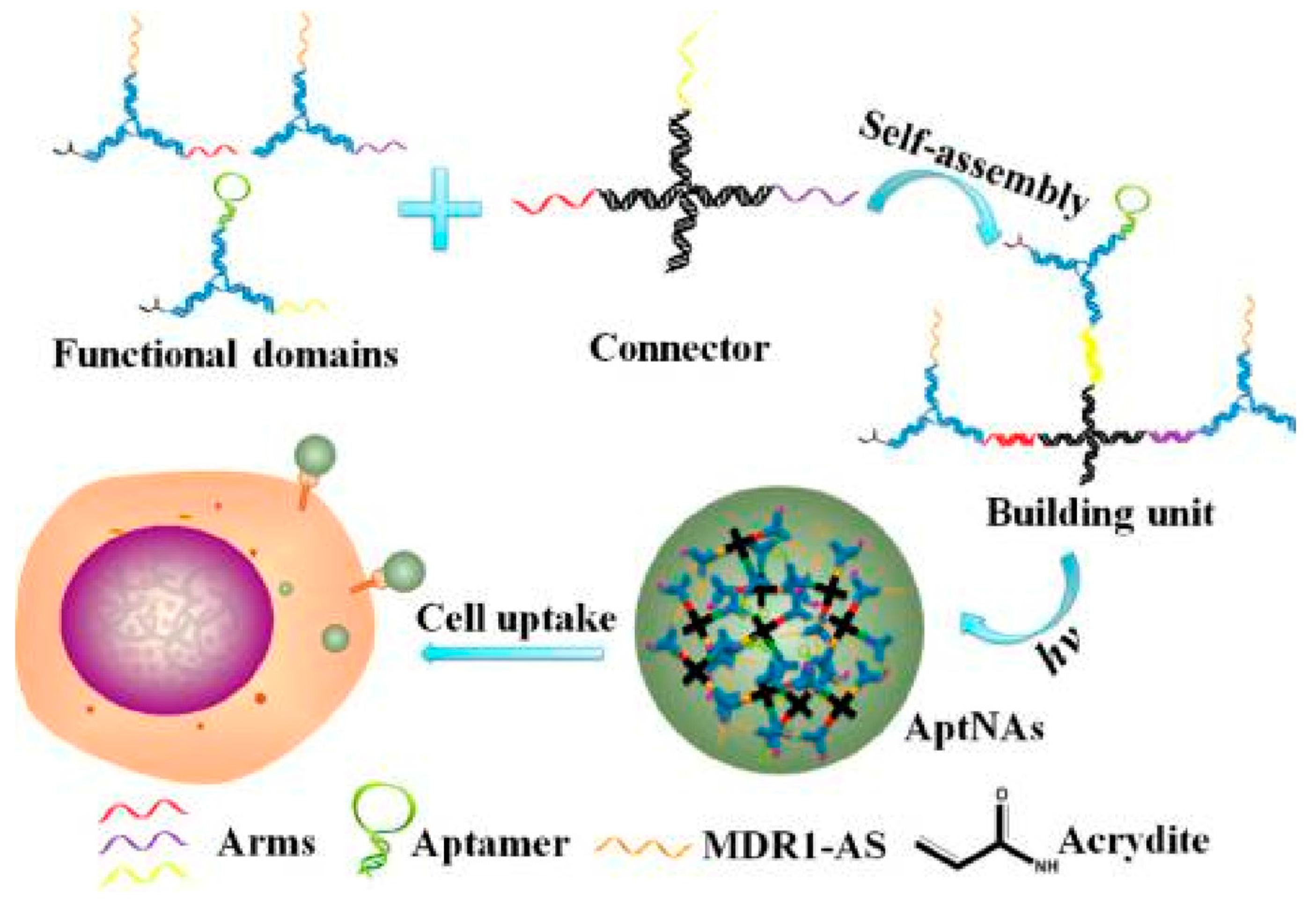
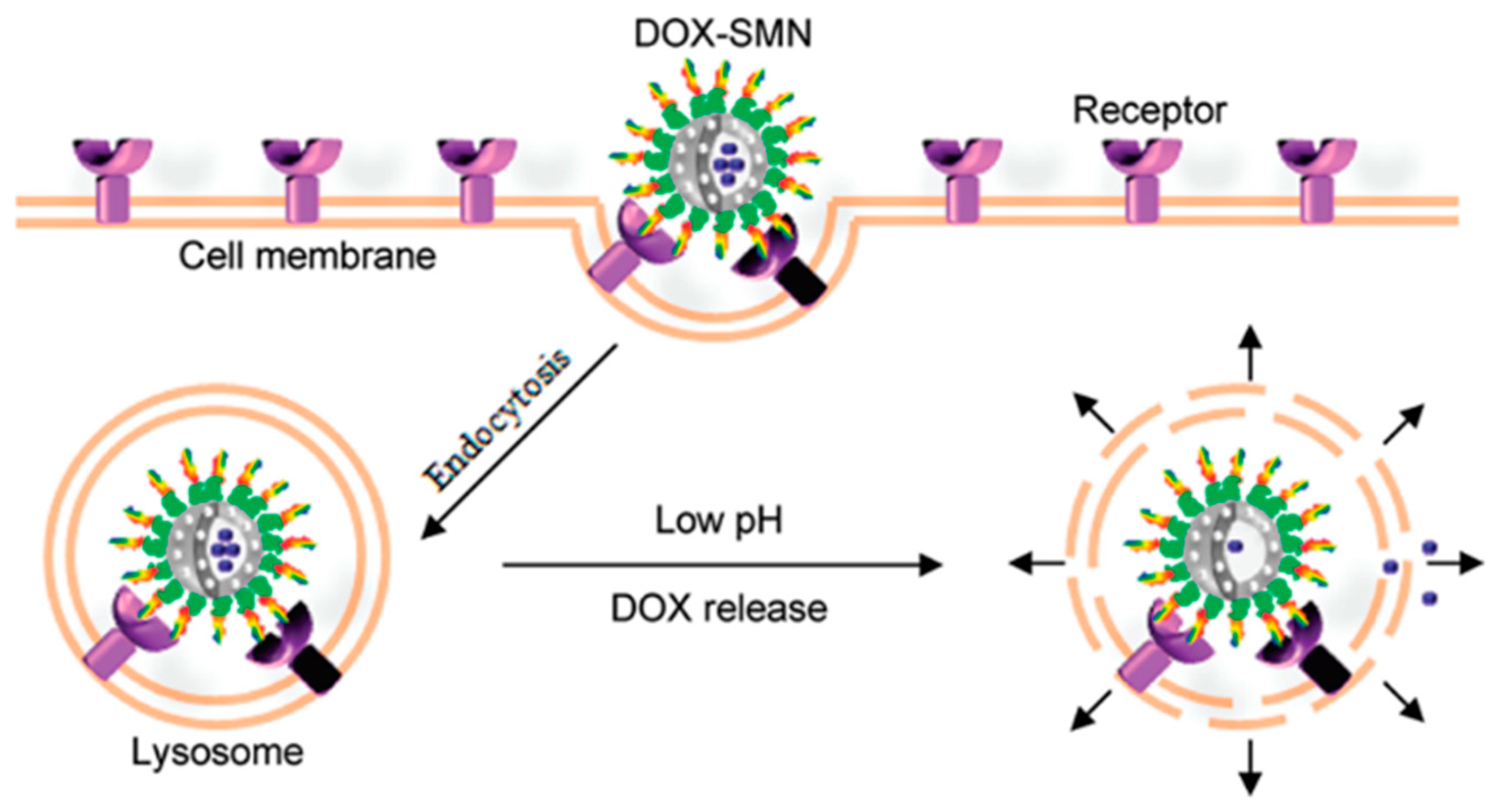
3.2. Targeted Drug Delivery
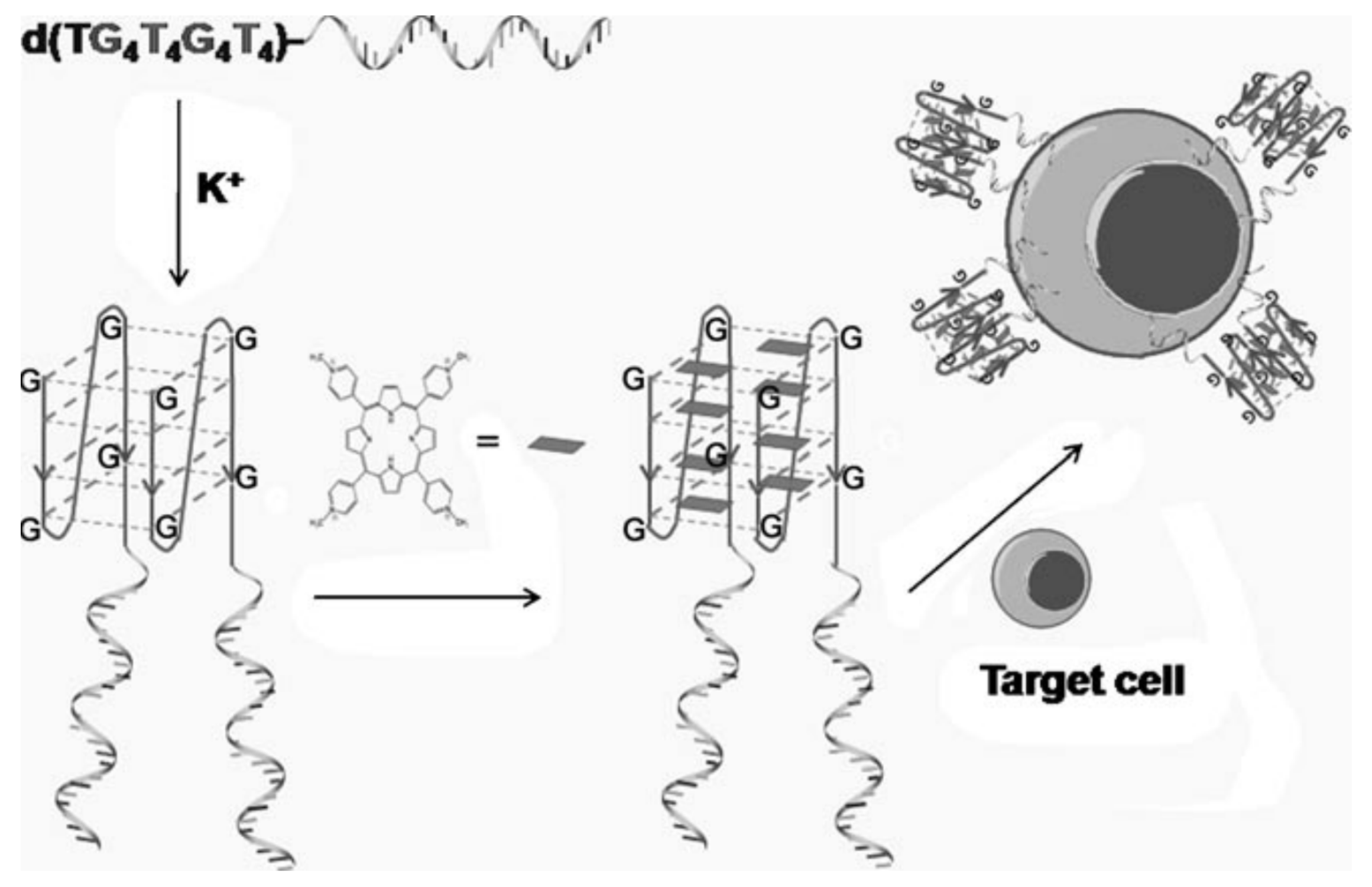
3.3. Therapy
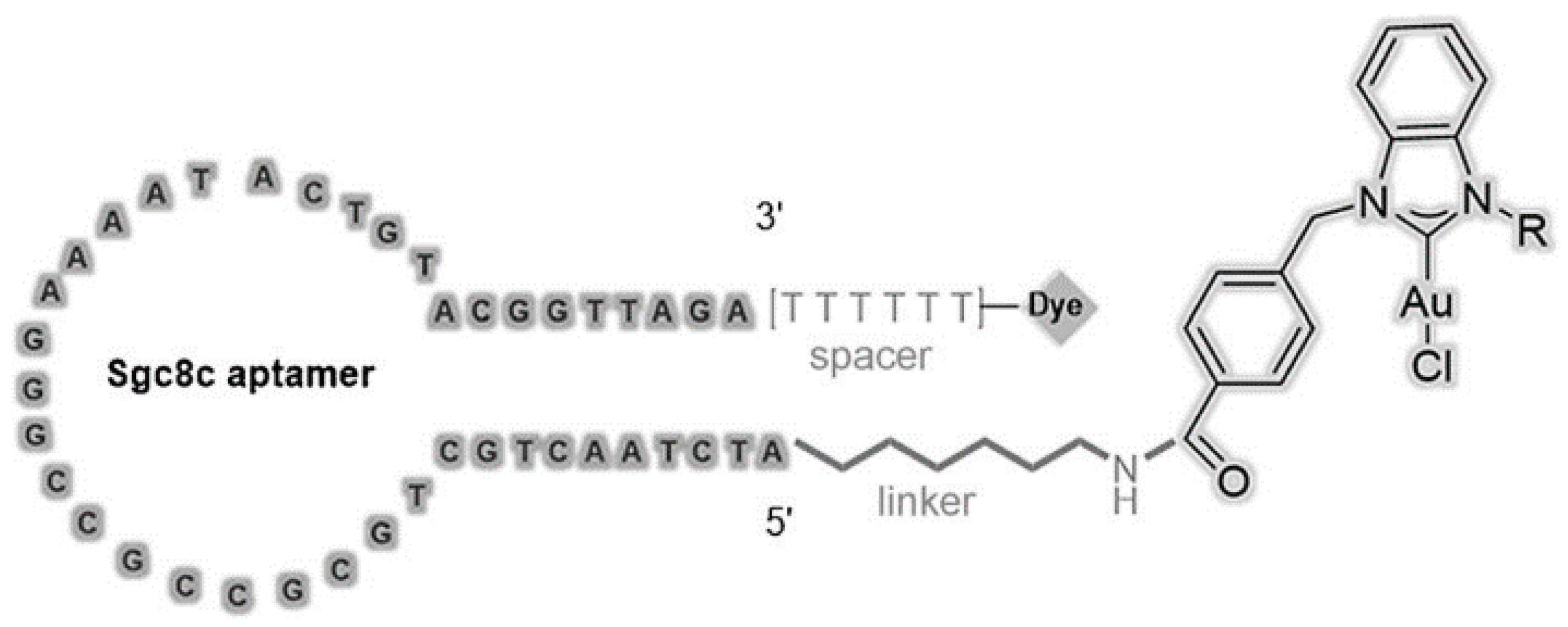
3.3.1. Phototherapy
3.3.2. Gene Therapy
3.3.3. Chemotherapy
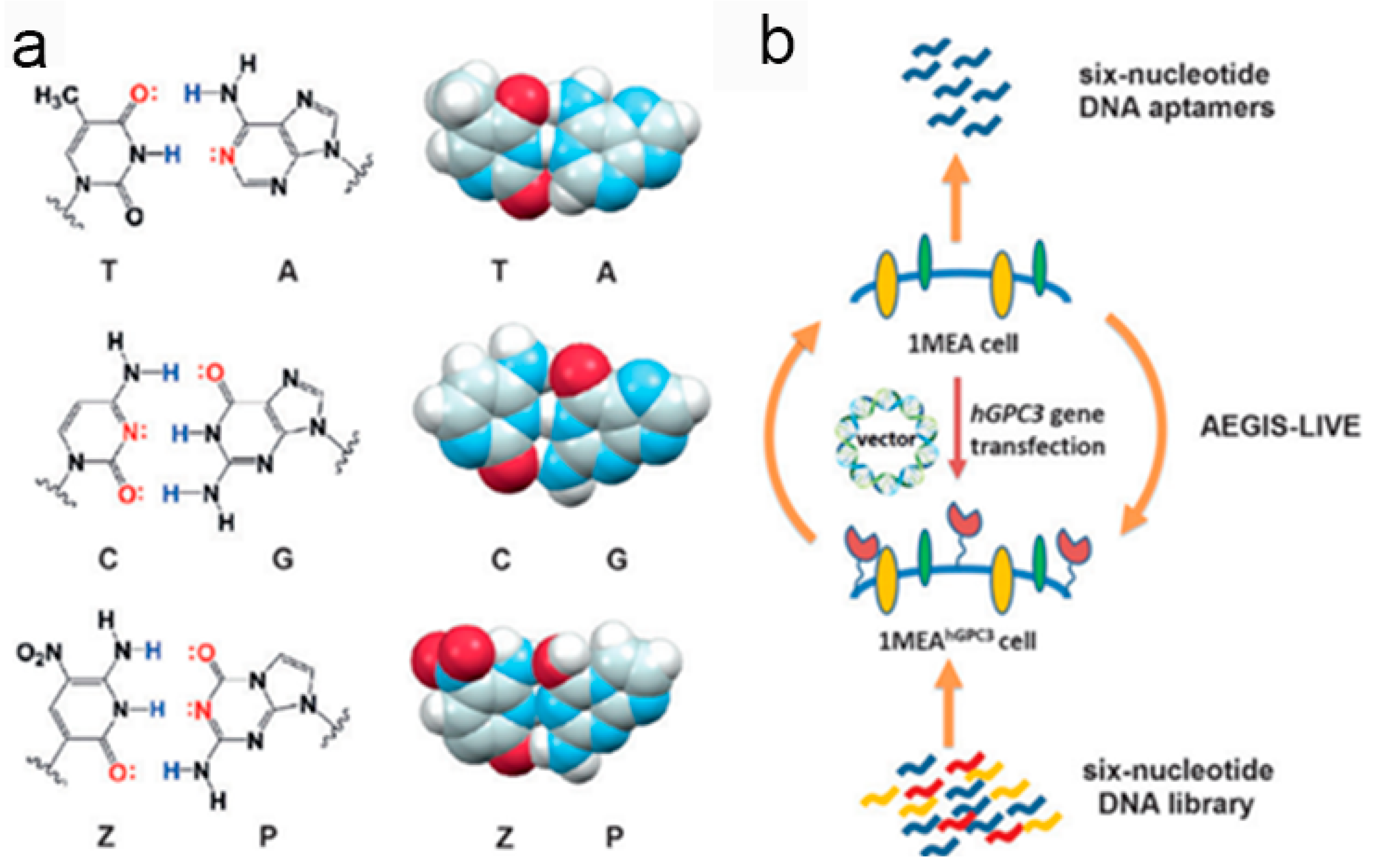
4. Biomarker Discovery for Cancers
5. Conclusions and Perspectives
Acknowledgments
Conflicts of Interest
References
- Jiang, Y.; Zhu, C.; Ling, L.; Wan, L.; Fang, X.; Bai, C. Specific aptamer-protein interaction studied by atomic force microscopy. Anal. Chem. 2003, 75, 2112–2116. [Google Scholar] [CrossRef] [PubMed]
- Famulok, M.; Hartig, J.S.; Mayer, G. Functional aptamers and aptazymes in biotechnology, diagnostics, and therapy. Chem. Rev. 2007, 107, 3715–3743. [Google Scholar] [CrossRef] [PubMed]
- Bunka, D.H.; Stockley, P.G. Aptamers come of age-at last. Nat. Rev. Microbiol. 2006, 4, 588–596. [Google Scholar] [CrossRef] [PubMed]
- Navani, N.K.; Li, Y. Nucleic acid aptamers and enzymes as sensors. Curr. Opin. Chem. Biol. 2006, 10, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Osborne, S.E.; Matsumura, I.; Ellington, A.D. Aptamers as therapeutic and diagnostic reagents: Problems and prospects. Curr. Opin. Chem. Biol. 1997, 1, 5–9. [Google Scholar] [CrossRef]
- Lee, J.H.; Yigit, M.V.; Mazumdar, D.; Lu, Y. Molecular diagnostic and drug delivery agents based on aptamer-nanomaterial conjugates. Adv. Drug Deliv. Rev. 2010, 62, 592–605. [Google Scholar] [CrossRef] [PubMed]
- Shamah, S.M.; Healy, J.M.; Cload, S.T. Complex target SELEX. Acc. Chem. Res. 2008, 41, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Cui, W.; He, X.; Guo, Q.; Wang, K.; Ye, X.; Tang, J. Whole cell-SELEX aptamers for highly specific fluorescence molecular imaging of carcinomas in vivo. PLoS ONE 2013, 8, e70476. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, D.; Meng, L.; Cao, Z.C.; Xiao, Z.; Fang, X.; Li, Y.; Cardona, D.; Witek, R.P.; Liu, C.; Tan, W. Identification of liver cancer-specific aptamers using whole live cells. Anal. Chem. 2008, 80, 721–728. [Google Scholar] [CrossRef] [PubMed]
- Ko, H.Y.; Lee, J.H.; Kang, H.; Ryu, S.H.; Song, I.C.; Lee, D.S.; Kim, S. A nucleolin-targeted multimodal nanoparticle imaging probe for tracking cancer cells using an aptamer. J. Nucl. Med. 2010, 51, 98–105. [Google Scholar]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Hermann, T.; Patel, D.J. Adaptive recognition by nucleic acid aptamers. Science 2000, 287, 820–825. [Google Scholar] [CrossRef] [PubMed]
- Sefah, K.; Shangguan, D.; Xiong, X.; O’donoghue, M.B.; Tan, W. Development of DNA aptamers using Cell-SELEX. Nat. Protoc. 2010, 5, 1169. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, D.; Li, Y.; Tang, Z.; Cao, Z.C.; Chen, H.W.; Mallikaratchy, P.; Sefah, K.; Yang, C.J.; Tan, W. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc. Natl. Acad. Sci. USA 2006, 103, 11838–11843. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Chen, Y.H.; Lennox, K.A.; Behlke, M.A.; Davidson, B.L. In vivo SELEX for identification of brain-penetrating aptamers. Mol. Ther. Nucleic Acids 2013, 2, e67. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Xu, H.; Ding, H.; Huang, Y.; Cao, X.; Yang, G.; Li, J.; Xie, Z.; Meng, Y.; Li, X. Identification of an aptamer targeting hnRNP A1 by tissue slide-based SELEX. J. Pathol. 2009, 218, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Souza, A.G.; Marangoni, K.; Fujimura, P.T.; Alves, P.T.; Silva, M.J.; Bastos, V.A.F.; Goulart, L.R.; Goulart, V.A. 3D Cell-SELEX: Development of RNA aptamers as molecular probes for PC-3 tumor cell line. Exp. Cell Res. 2016, 341, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Ngubane, N.A.; Gresh, L.; Pym, A.; Rubin, E.J.; Khati, M. Selection of RNA aptamers against the M. tuberculosis EsxG protein using surface plasmon resonance-based SELEX. Biochem. Biophys. Res. Commun. 2014, 449, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Glökler, J.; Schütze, T.; Konthur, Z. Automation in the high-throughput selection of random combinatorial libraries—Different approaches for select applications. Molecules 2010, 15, 2478–2490. [Google Scholar] [CrossRef] [PubMed]
- Mosing, R.K.; Bowser, M.T. Isolating aptamers using capillary electrophoresis–SELEX (CE–SELEX). Nucleic Acid Pept. Aptamers. Methods Protoc. 2009, 535, 33–43. [Google Scholar]
- Xu, Y.; Yang, X.; Wang, E. Aptamers in microfluidic chips. Anal. Chim. Acta 2010, 683, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Mackie, E.J.; Chiquet-Ehrismann, R.; Pearson, C.A.; Inaguma, Y.; Taya, K.; Kawarada, Y.; Sakakura, T. Tenascin is a stromal marker for epithelial malignancy in the mammary gland. Proc. Natl. Acad. Sci. USA 1987, 84, 4621–4625. [Google Scholar] [CrossRef] [PubMed]
- Tolle, F.; Mayer, G. Dressed for success—Applying chemistry to modulate aptamer functionality. Chem. Sci. 2013, 4, 60–67. [Google Scholar] [CrossRef]
- Kim, Y.; Cao, Z.; Tan, W. Molecular assembly for high-performance bivalent nucleic acid inhibitor. Proc. Natl. Acad. Sci. USA 2008, 105, 5664–5669. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, H.; Taira, K.-I.; Sode, K.; Ikebukuro, K. Improvement of aptamer affinity by dimerization. Sensors 2008, 8, 1090–1098. [Google Scholar] [CrossRef] [PubMed]
- Müller, J.; Wulffen, B.; Pötzsch, B.; Mayer, G. Multidomain targeting generates a high-affinity thrombin-inhibiting bivalent aptamer. ChemBioChem 2007, 8, 2223–2226. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yang, Z.; Sefah, K.; Bradley, K.M.; Hoshika, S.; Kim, M.-J.; Kim, H.-J.; Zhu, G.; Jiménez, E.; Cansiz, S. Evolution of functional six-nucleotide DNA. J. Am. Chem. Soc. 2015, 137, 6734–6737. [Google Scholar] [CrossRef] [PubMed]
- Sefah, K.; Yang, Z.; Bradley, K.M.; Hoshika, S.; Jiménez, E.; Zhang, L.; Zhu, G.; Shanker, S.; Yu, F.; Turek, D. In vitro selection with artificial expanded genetic information systems. Proc. Natl. Acad. Sci. USA 2014, 111, 1449–1454. [Google Scholar] [CrossRef] [PubMed]
- Kimoto, M.; Yamashige, R.; Matsunaga, K.-I.; Yokoyama, S.; Hirao, I. Generation of high-affinity DNA aptamers using an expanded genetic alphabet. Nat. Biotechnol. 2013, 31, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Gedi, V.; Kim, Y.-P. Detection and characterization of cancer cells and pathogenic bacteria using aptamer-based nano-conjugates. Sensors 2014, 14, 18302–18327. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.S.; Huh, Y.M.; Kim, S.; Lee, D.-K. Isolation of RNA aptamers targeting HER-2-overexpressing breast cancer cells using cell-SELEX. Bull. Korean Chem. Soc. 2009, 30, 1827–1831. [Google Scholar]
- Chen, H.W.; Medley, C.D.; Sefah, K.; Shangguan, D.; Tang, Z.; Meng, L.; Smith, J.E.; Tan, W. Molecular recognition of small-cell lung cancer cells using aptamers. ChemMedChem 2008, 3, 991–1001. [Google Scholar] [CrossRef] [PubMed]
- Sefah, K.; Tang, Z.; Shangguan, D.; Chen, H.; Lopez-Colon, D.; Li, Y.; Parekh, P.; Martin, J.; Meng, L.; Tan, W. Molecular recognition of acute myeloid leukemia using aptamers. Leukemia 2009, 23, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Charlton, J.; Sennello, J.; Smith, D. In vivo imaging of inflammation using an aptamer inhibitor of human neutrophil elastase. Chem. Biol. 1997, 4, 809–816. [Google Scholar] [CrossRef]
- Babendure, J.R.; Adams, S.R.; Tsien, R.Y. Aptamers switch on fluorescence of triphenylmethane dyes. J. Am. Chem. Soc. 2003, 125, 14716–14717. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Ebright, J.N.; Stovall, G.M.; Chen, X.; Nguyen, H.H.; Singh, A.; Syrett, A.; Ellington, A.D. Technical and biological issues relevant to cell typing with aptamers. J. Proteome Res. 2009, 8, 2438–2448. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Zhang, X.; Zhao, Z.; Zhu, G.; Chen, T.; Fu, T.; Tan, W. DNA nanoflowers for multiplexed cellular imaging and traceable targeted drug delivery. Angew. Chem. Int. Ed. 2014, 126, 5821–5826. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.Z.; Bagalkot, V.; Vasilliou, C.C.; Gu, F.; Alexis, F.; Zhang, L.; Shaikh, M.; Yuet, K.; Cima, M.J.; Langer, R. Superparamagnetic iron oxide nanoparticle—Aptamer bioconjugates for combined prostate cancer imaging and therapy. ChemMedChem 2008, 3, 1311–1315. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Chen, T.; Ocsoy, I.; Zhu, G.; Yasun, E.; You, M.; Wu, C.; Zheng, J.; Song, E.; Huang, C.Z. Gold-coated Fe3O4 nanoroses with five unique functions for cancer cell targeting, imaging, and therapy. Adv. Funct. Mater. 2014, 24, 1772–1780. [Google Scholar] [CrossRef] [PubMed]
- Kuai, H.; Zhao, Z.; Mo, L.; Liu, H.; Hu, X.; Fu, T.; Zhang, X.; Tan, W. Circular bivalent aptamers enable in vivo stability and recognition. J. Am. Chem. Soc. 2017, 139, 9128–9131. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Fan, H.; Zhou, G.; Bai, H.; Liang, H.; Wang, R.; Zhang, X.; Tan, W. Activatable fluorescence/MRI bimodal platform for tumor cell imaging via MnO2 nanosheet—Aptamer nanoprobe. J. Am. Chem. Soc. 2014, 136, 11220–11223. [Google Scholar] [CrossRef] [PubMed]
- Farokhzad, O.C.; Jon, S.; Khademhosseini, A.; Tran, T.-N.T.; LaVan, D.A.; Langer, R. Nanoparticle-aptamer bioconjugates. Cancer Res. 2004, 64, 7668–7672. [Google Scholar] [CrossRef] [PubMed]
- Chaloin, L.; Lehmann, M.J.; Sczakiel, G.; Restle, T. Endogenous expression of a high-affinity pseudoknot RNA aptamer suppresses replication of HIV-1. Nucleic Acids Res. 2002, 30, 4001–4008. [Google Scholar] [CrossRef] [PubMed]
- Gu, F.; Zhang, L.; Teply, B.A.; Mann, N.; Wang, A.; Radovic-Moreno, A.F.; Langer, R.; Farokhzad, O.C. Precise engineering of targeted nanoparticles by using self-assembled biointegrated block copolymers. Proc. Natl. Acad. Sci. USA 2008, 105, 2586–2591. [Google Scholar] [CrossRef] [PubMed]
- Chu, T.C.; Twu, K.Y.; Ellington, A.D.; Levy, M. Aptamer mediated siRNA delivery. Nucleic Acids Res. 2006, 34, e73. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Yang, J.; Park, J.; Kim, S.; Kim, N.H.; Yook, J.I.; Suh, J.-S.; Haam, S.; Huh, Y.-M. Consecutive targetable smart nanoprobe for molecular recognition of cytoplasmic microRNA in metastatic breast cancer. ACS Nano 2012, 6, 8525–8535. [Google Scholar] [CrossRef] [PubMed]
- Niu, W.; Chen, X.; Tan, W.; Veige, A.S. N-heterocyclic carbene-gold (I) complexes conjugated to a leukemia-specific DNA aptamer for targeted drug delivery. Angew. Chem. Int. Ed. 2016, 55, 8889–8893. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhu, G.; Mei, L.; Xie, Y.; Ma, H.; Ye, M.; Qing, F.-L.; Tan, W. Automated modular synthesis of aptamer-drug conjugates for targeted drug delivery. J. Am. Chem. Soc. 2014, 136, 2731–2734. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Tang, Z.; Phillips, J.A.; Yang, R.; Wang, H.; Tan, W. Regulation of singlet oxygen generation using single-walled carbon nanotubes. J. Am. Chem. Soc. 2008, 130, 10856–10857. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; You, M.; Chen, Y.; Han, D.; Zhu, Z.; Huang, J.; Williams, K.; Yang, C.J.; Tan, W. Self-assembly of a biofunctional DNA carrier for drug delivery. Angew. Chem. Int. Ed. 2011, 50, 6098–6101. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Zheng, J.; Song, E.; Donovan, M.; Zhang, K.; Liu, C.; Tan, W. Self-assembled, aptamer-tethered DNA nanotrains for targeted transport of molecular drugs in cancer theranostics. Proc. Natl. Acad. Sci. USA 2013, 110, 7998–8003. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Zhu, G.; Li, Y.; Li, C.; You, M.; Chen, T.; Song, E.; Yang, R.; Tan, W. A spherical nucleic acid platform based on self-assembled DNA biopolymer for high-performance cancer therapy. ACS Nano 2013, 7, 6545–6554. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhu, G.; You, M.; Song, E.; Shukoor, M.I.; Zhang, K.; Altman, M.B.; Chen, Y.; Zhu, Z.; Huang, C.Z. Assembly of aptamer switch probes and photosensitizer on gold nanorods for targeted photothermal and photodynamic cancer therapy. ACS Nano 2012, 6, 5070–5077. [Google Scholar] [CrossRef] [PubMed]
- Özalp, V.C.; Schäfer, T. Aptamer-Based Switchable Nanovalves for Stimuli-Responsive Drug Delivery. Chem. Asian J. 2011, 17, 9893–9896. [Google Scholar] [CrossRef] [PubMed]
- Qiao, G.; Zhuo, L.; Gao, Y.; Yu, L.; Li, N.; Tang, B. A tumor mRNA-dependent gold nanoparticle—Molecular beacon carrier for controlled drug release and intracellular imaging. Chem. Commun. 2011, 47, 7458–7460. [Google Scholar] [CrossRef] [PubMed]
- Fan, P.; Suri, A.K.; Fiala, R.; Live, D.; Patel, D.J. Molecular recognition in the FMN-RNA aptamer complex. J. Mol. Biol. 1996, 258, 480–500. [Google Scholar] [CrossRef] [PubMed]
- Morishita, R.; Gibbons, G.H.; Horiuchi, M.; Ellison, K.E.; Nakama, M.; Zhang, L.; Kaneda, Y.; Ogihara, T.; Dzau, V.J. A gene therapy strategy using a transcription factor decoy of the E2F binding site inhibits smooth muscle proliferation in vivo. Proc. Natl. Acad. Sci. USA 1995, 92, 5855–5859. [Google Scholar] [CrossRef] [PubMed]
- Oseroff, A.R.; Ohuoha, D.; Hasan, T.; Bommer, J.C.; Yarmush, M.L. Antibody-targeted photolysis: Selective photodestruction of human T-cell leukemia cells using monoclonal antibody-chlorin e6 conjugates. Proc. Natl. Acad. Sci. USA 1986, 83, 8744–8748. [Google Scholar] [CrossRef] [PubMed]
- Dolmans, D.E.; Fukumura, D.; Jain, R.K. Photodynamic therapy for cancer. Nat. Rev. Cancer 2003, 3, 380–387. [Google Scholar] [CrossRef] [PubMed]
- You, M.; Zhu, G.; Chen, T.; Donovan, M.J.; Tan, W. DNA “Nano-Claw”: Logic-Based Autonomous Cancer Targeting and Therapy. J. Am. Chem. Soc. 2014, 136, 1256–1259. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Han, D.; Chen, T.; Peng, L.; Zhu, G.; You, M.; Qiu, L.; Sefah, K.; Zhang, X.; Tan, W. Building a multifunctional aptamer-based DNA nanoassembly for targeted cancer therapy. J. Am. Chem. Soc. 2013, 135, 18644–18650. [Google Scholar] [CrossRef] [PubMed]
- Mallikaratchy, P.; Tang, Z.; Tan, W. Cell specific aptamer-photosensitizer conjugates as a molecular tool in photodynamic therapy. ChemMedChem 2008, 3, 425–428. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-F.; Sefah, K.; Bamrungsap, S.; Chang, H.-T.; Tan, W. Selective photothermal therapy for mixed cancer cells using aptamer-conjugated nanorods. Langmuir 2008, 24, 11860–11865. [Google Scholar] [CrossRef] [PubMed]
- Hao, L.; Patel, P.C.; Alhasan, A.H.; Giljohann, D.A.; Mirkin, C.A. Nucleic acid-gold nanoparticle conjugates as mimics of microRNA. Small 2011, 7, 3158–3162. [Google Scholar] [CrossRef] [PubMed]
- Seferos, D.S.; Giljohann, D.A.; Hill, H.D.; Prigodich, A.E.; Mirkin, C.A. Nano-flares: Probes for transfection and mRNA detection in living cells. J. Am. Chem. Soc. 2007, 129, 15477–15479. [Google Scholar] [CrossRef] [PubMed]
- Giljohann, D.A.; Seferos, D.S.; Prigodich, A.E.; Patel, P.C.; Mirkin, C.A. Gene regulation with polyvalent siRNA−nanoparticle conjugates. J. Am. Chem. Soc. 2009, 131, 2072–2073. [Google Scholar] [CrossRef] [PubMed]
- Horton, H.M.; Anderson, D.; Hernandez, P.; Barnhart, K.M.; Norman, J.A.; Parker, S.E. A gene therapy for cancer using intramuscular injection of plasmid DNA encoding interferon α. Proc. Natl. Acad. Sci. USA 1999, 96, 1553–1558. [Google Scholar] [CrossRef] [PubMed]
- Paddison, P.J.; Caudy, A.A.; Bernstein, E.; Hannon, G.J.; Conklin, D.S. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002, 16, 948–958. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, A.; Tanabe, K.; Inasaki, T.; Saito, I. Phototriggered drug release from functionalized oligonucleotides by a molecular beacon strategy. Angew. Chem. Int. Ed. 2003, 115, 2606–2608. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, P.; Mao, H.; Leong, K. Enhanced gene expression in mouse muscle by sustained release of plasmid DNA using PPE-EA as a carrier. Gene Ther. 2002, 9, 1254–1261. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zheng, C.; Cansiz, S.; Wu, C.; Xu, J.; Cui, C.; Liu, Y.; Hou, W.; Wang, Y.; Zhang, L. Self-assembly of DNA nanohydrogels with controllable size and stimuli-responsive property for targeted gene regulation therapy. J. Am. Chem. Soc. 2015, 137, 1412–1415. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Wu, C.S.; Jimenez, E.; Zhu, Z.; Dajac, J.G.; You, M.X.; Han, D.; Zhang, X.B.; Tan, W.H. DNA micelle flares for intracellular mRNA imaging and gene therapy. Angew. Chem. Int. Ed. 2013, 52, 2012–2016. [Google Scholar] [CrossRef] [PubMed]
- Farokhzad, O.C.; Cheng, J.; Teply, B.A.; Sherifi, I.; Jon, S.; Kantoff, P.W.; Richie, J.P.; Langer, R. Targeted nanoparticle-aptamer bioconjugates for cancer chemotherapy in vivo. Proc. Natl. Acad. Sci. USA 2006, 103, 6315–6320. [Google Scholar] [CrossRef] [PubMed]
- Chu, T.C.; Marks, J.W.; Lavery, L.A.; Faulkner, S.; Rosenblum, M.G.; Ellington, A.D.; Levy, M. Aptamer: Toxin conjugates that specifically target prostate tumor cells. Cancer Res. 2006, 66, 5989–5992. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Shukoor, M.I.; Wang, R.; Zhao, Z.; Yuan, Q.; Bamrungsap, S.; Xiong, X.; Tan, W. Smart multifunctional nanostructure for targeted cancer chemotherapy and magnetic resonance imaging. ACS Nano 2011, 5, 7866–7873. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.F.; Shangguan, D.; Liu, H.; Phillips, J.A.; Zhang, X.; Chen, Y.; Tan, W. Molecular assembly of an aptamer-drug conjugate for targeted drug delivery to tumor cells. ChemBioChem 2009, 10, 862–868. [Google Scholar] [CrossRef] [PubMed]
- Strimbu, K.; Tavel, J.A. What are biomarkers? Curr. Opin. HIV AIDS 2010, 5, 463–466. [Google Scholar] [CrossRef] [PubMed]
- Henry, N.L.; Hayes, D.F. Cancer biomarkers. Mol. Oncol. 2012, 6, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Frangogiannis, N.G. Biomarkers: Hopes and challenges in the path from discovery to clinical practice. Transl. Res. J. Lab. Clin. Med. 2012, 159, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Berezovski, M.V.; Lechmann, M.; Musheev, M.U.; Mak, T.W.; Krylov, S.N. Aptamer-facilitated biomarker discovery (AptaBiD). J. Am. Chem. Soc. 2008, 130, 9137–9143. [Google Scholar] [CrossRef] [PubMed]
- Negm, R.S.; Verma, M.; Srivastava, S. The promise of biomarkers in cancer screening and detection. Trends Mol. Med. 2002, 8, 288–293. [Google Scholar] [CrossRef]
- Wu, C.C.; Yates, J.R. The application of mass spectrometry to membrane proteomics. Nat. Biotechnol. 2003, 21, 262–267. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, D.; Cao, Z.; Meng, L.; Mallikaratchy, P.; Sefah, K.; Wang, H.; Li, Y.; Tan, W. Cell-specific aptamer probes for membrane protein elucidation in cancer cells. J.Proteome Res. 2008, 7, 2133–2139. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; He, X.; Wang, K.; Wu, X.; Ye, X.; Guo, Q.; Tan, W.; Qing, Z.; Yang, X.; Zhou, B. Activatable aptamer probe for contrast-enhanced in vivo cancer imaging based on cell membrane protein-triggered conformation alteration. Proc. Natl. Acad. Sci. USA 2011, 108, 3900–3905. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Jacobson, O.; Avdic, D.; Rotstein, B.H.; Weiss, I.D.; Collier, L.; Chen, X.; Vasdev, N.; Liang, S.H. Ortho-stabilized 18F-azido click agents and their application in PET imaging with single-stranded DNA aptamers. Angew. Chem. Int. Ed. 2015, 54, 12777–12781. [Google Scholar] [CrossRef] [PubMed]
- Pu, Y.; Liu, Z.; Lu, Y.; Yuan, P.; Liu, J.; Yu, B.; Wang, G.; Yang, C.J.; Liu, H.; Tan, W. Using DNA aptamer probe for immunostaining of cancer frozen tissues. Anal. Chem. 2015, 87, 1919–1924. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Liu, H.; Zhao, Z.; Altman, M.B.; Lopez-Colon, D.; Yang, C.J.; Chang, L.J.; Liu, C.; Tan, W. DNA aptamer-mediated cell targeting. Angew. Chem. Int. Ed. 2013, 52, 1472–1476. [Google Scholar] [CrossRef] [PubMed]
- Douglas, S.M.; Bachelet, I.; Church, G.M. A logic-gated nanorobot for targeted transport of molecular payloads. Science 2012, 335, 831–834. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Hu, R.; Zhao, Z.; Chen, Z.; Zhang, X.; Tan, W. Noncanonical self-assembly of multifunctional DNA nanoflowers for biomedical applications. J. Am. Chem. Soc. 2013, 135, 16438–16445. [Google Scholar] [CrossRef] [PubMed]
- You, M.; Peng, L.; Shao, N.; Zhang, L.; Qiu, L.; Cui, C.; Tan, W. DNA “nano-claw”: Logic-based autonomous cancer targeting and therapy. J. Am. Chem. Soc. 2014, 136, 1256–1259. [Google Scholar] [CrossRef] [PubMed]
- Mallikaratchy, P.; Tang, Z.; Kwame, S.; Meng, L.; Shangguan, D.; Tan, W. Aptamer directly evolved from live cells recognizes membrane bound immunoglobin heavy mu chain in Burkitt’s lymphoma cells. Mol. Cell. Proteom. 2007, 6, 2230–2238. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.D.; Srivastava, B.; Allman, D. Regulation of peripheral B cell maturation. Cell. Immunol. 2006, 239, 92–102. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Tan, W. Aptamers generated from cell-SELEX for molecular medicine: A chemical biology approach. Acc. Chem. Res. 2009, 43, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wan, S.; Jiang, Y.; Wang, Y.; Fu, T.; Liu, Q.; Cao, Z.; Qiu, L.; Tan, W. Molecular elucidation of disease biomarkers at the interface of chemistry and biology. J. Am. Chem. Soc. 2017, 139, 2532–2540. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yang, Z.; Le Trinh, T.; Teng, I.; Wang, S.; Bradley, K.M.; Hoshika, S.; Wu, Q.; Cansiz, S.; Rowold, D.J. Aptamers against cells overexpressing Glypican3 from expanded genetic systems combined with cell engineering and laboratory evolution. Angew. Chem. Int. Ed. 2016, 55, 12372–12375. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Lu, D.; Bai, H.; Jin, C.; Yan, G.; Ye, M.; Qiu, L.; Chang, R.; Cui, C.; Liang, H. Using modified aptamers for site specific protein-aptamer conjugations. Chem. Sci. 2016, 7, 2157–2161. [Google Scholar] [CrossRef] [PubMed]
- Bing, T.; Shangguan, D.; Wang, Y. Facile discovery of cell-surface protein targets of cancer cell aptamers. Mol. Cell. Proteom. 2015, 14, 2692–2700. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Zhu, Z.; An, Y.; Zhang, W.; Zhang, H.; Liu, D.; Yu, C.; Duan, W.; Yang, C.J. Selection of DNA aptamers against epithelial cell adhesion molecule for cancer cell imaging and circulating tumor cell capture. Anal. Chem. 2013, 85, 4141–4149. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, N.; Sreemanthula, J.B.; Balaji, B.; Kanwar, J.R.; Biswas, J.; Krishnakumar, S. A strain-promoted alkyne—Azide cycloaddition (SPAAC) reaction of a novel EpCAM aptamer—Fluorescent conjugate for imaging of cancer cells. Chem. Commun. 2014, 50, 11810–11813. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Lu, Y.; Pu, Y.; Liu, J.; Liu, B.; Yu, B.; Chen, K.; Fu, T.; Yang, C.J.; Liu, H. Using aptamers to elucidate esophageal cancer clinical samples. Sci. Rep. 2015, 5, 18516. [Google Scholar] [CrossRef] [PubMed]
- Mahlknecht, G.; Maron, R.; Mancini, M.; Schechter, B.; Sela, M.; Yarden, Y. Aptamer to ErbB-2/HER2 enhances degradation of the target and inhibits tumorigenic growth. Proc. Natl. Acad. Sci. USA 2013, 110, 8170–8175. [Google Scholar] [CrossRef] [PubMed]
- Tolle, F.; Brändle, G.M.; Matzner, D.; Mayer, G. A Versatile approach towards nucleobase-modified aptamers. Angew. Chem. Int. Ed. 2015, 54, 10971–10974. [Google Scholar] [CrossRef] [PubMed]
- Robinson, P.V.; de Almeida-Escobedo, G.; de Groot, A.E.; McKechnie, J.L.; Bertozzi, C.R. Live-cell labeling of specific protein glycoforms by proximity-enhanced bioorthogonal ligation. J. Am. Chem. Soc. 2015, 137, 10452–10455. [Google Scholar] [CrossRef] [PubMed]
- Vinkenborg, J.L.; Mayer, G.; Famulok, M. Aptamer-based affinity labeling of proteins. Angew. Chem. Int. Ed. 2012, 51, 9176–9180. [Google Scholar] [CrossRef] [PubMed]



© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pang, X.; Cui, C.; Wan, S.; Jiang, Y.; Zhang, L.; Xia, L.; Li, L.; Li, X.; Tan, W. Bioapplications of Cell-SELEX-Generated Aptamers in Cancer Diagnostics, Therapeutics, Theranostics and Biomarker Discovery: A Comprehensive Review. Cancers 2018, 10, 47. https://doi.org/10.3390/cancers10020047
Pang X, Cui C, Wan S, Jiang Y, Zhang L, Xia L, Li L, Li X, Tan W. Bioapplications of Cell-SELEX-Generated Aptamers in Cancer Diagnostics, Therapeutics, Theranostics and Biomarker Discovery: A Comprehensive Review. Cancers. 2018; 10(2):47. https://doi.org/10.3390/cancers10020047
Chicago/Turabian StylePang, Xuehui, Cheng Cui, Shuo Wan, Ying Jiang, Liangliang Zhang, Lian Xia, Long Li, Xiaowei Li, and Weihong Tan. 2018. "Bioapplications of Cell-SELEX-Generated Aptamers in Cancer Diagnostics, Therapeutics, Theranostics and Biomarker Discovery: A Comprehensive Review" Cancers 10, no. 2: 47. https://doi.org/10.3390/cancers10020047



