miR-9-5p in Nephrectomy Specimens is a Potential Predictor of Primary Resistance to First-Line Treatment with Tyrosine Kinase Inhibitors in Patients with Metastatic Renal Cell Carcinoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients and Samples
2.2. RNA Extraction, MicroRNA Array Cards and miRNA Quantification
2.3. Data Analysis and Statistics
3. Results
3.1. Study Design, Patient and Sample Characteristics
3.2. Array-based Identification of Deregulated miRNAs
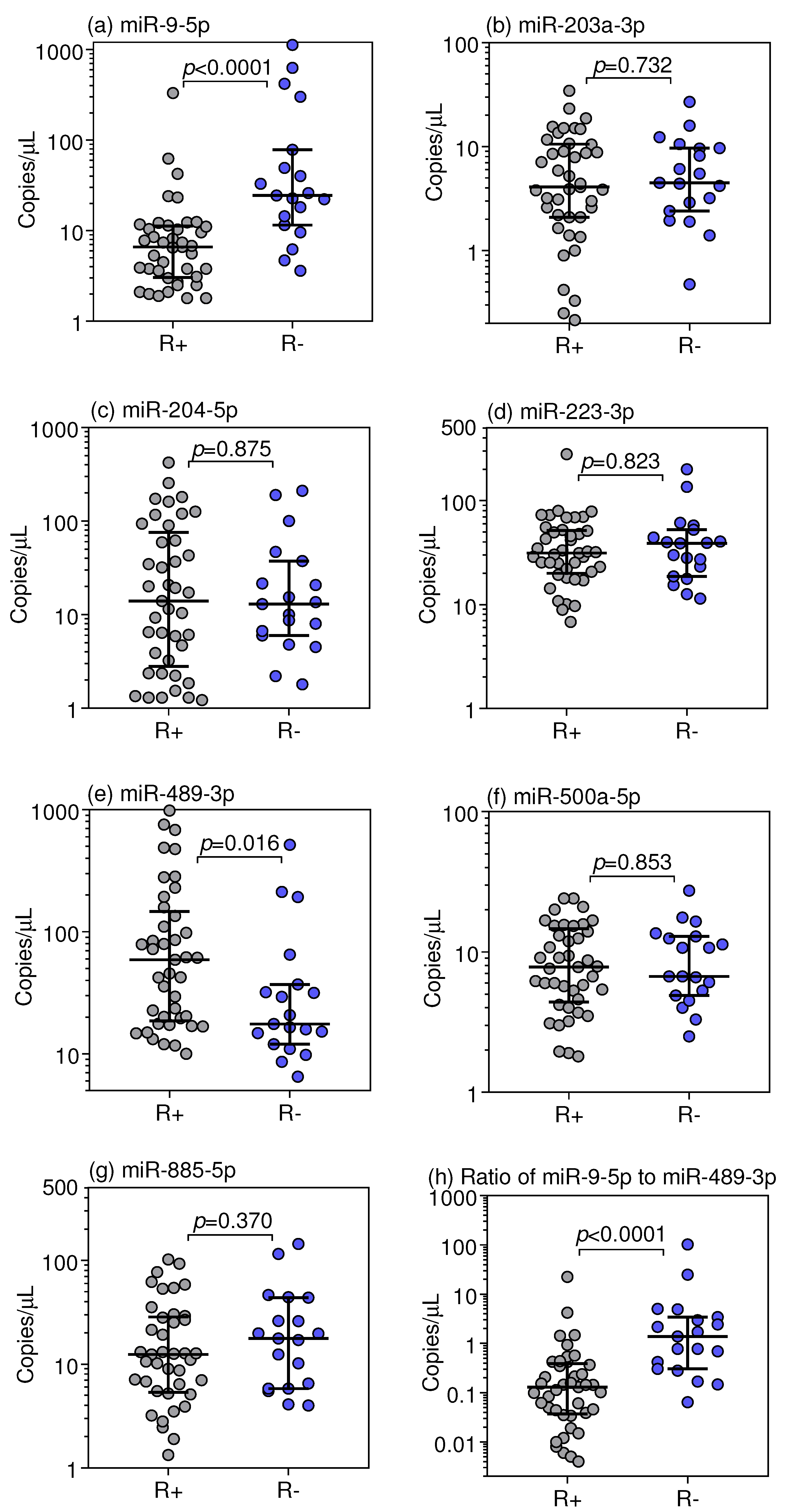
3.3. Droplet Digital RT-qPCR Validation of Selected miRNAs
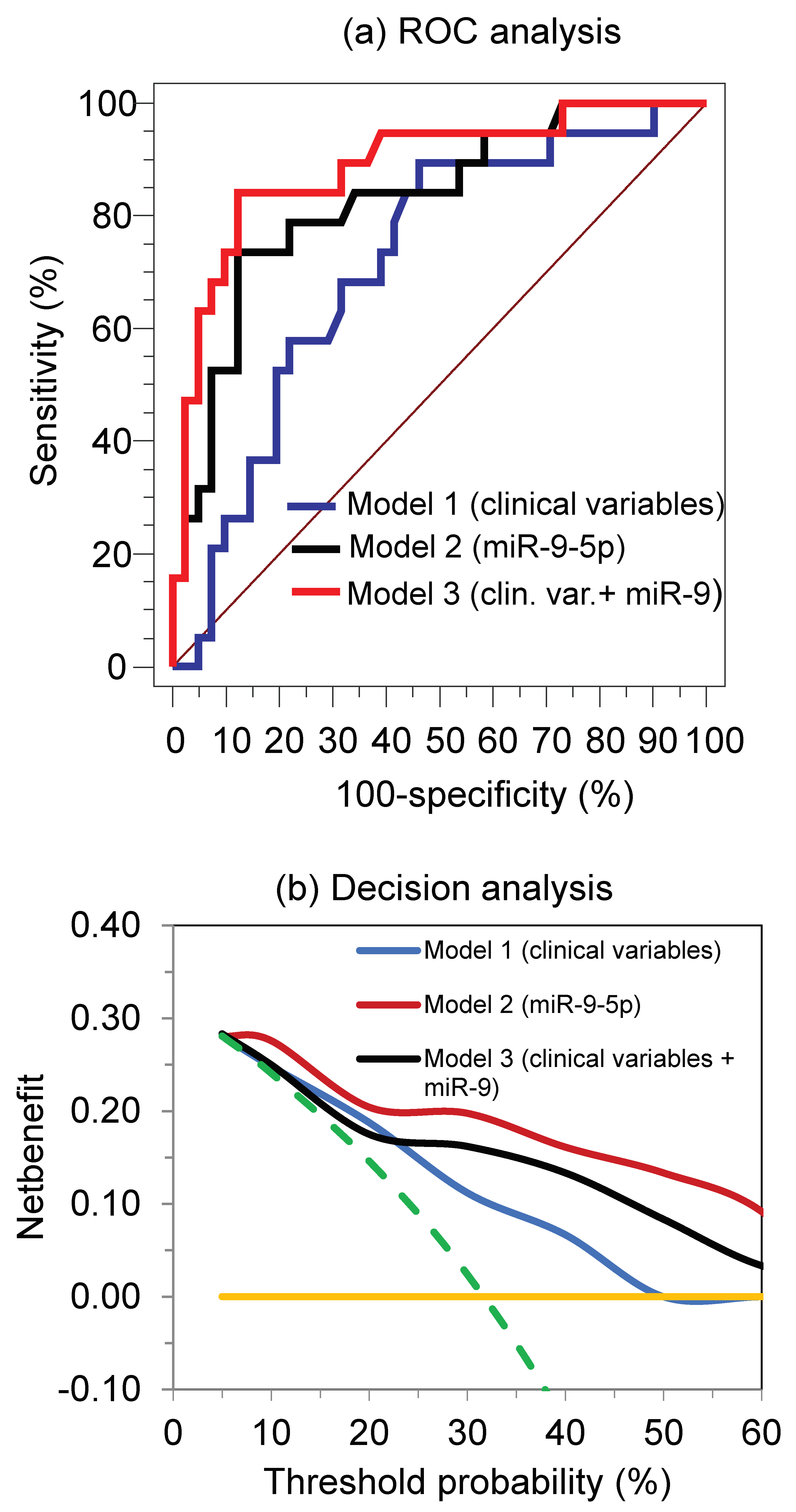
3.4. miRNAs as Predictors of Primary Resistance to the TKI Treatment and Clinical Outcome
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Iacovelli, R.; Sternberg, C.N.; Porta, C.; Verzoni, E.; de Braud, F.; Escudier, B.; Procopio, G. Inhibition of the VEGF/VEGFR pathway improves survival in advanced kidney cancer: A systematic review and meta-analysis. Curr. Drug Targets 2015, 16, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Escudier, B.; Porta, C.; Schmidinger, M.; Algaba, F.; Patard, J.J.; Khoo, V.; Eisen, T.; Horwich, A. Renal cell carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2014, 25 (Suppl. S3), iii49–iii56. [Google Scholar] [CrossRef] [Green Version]
- Powles, T.; Albiges, L.; Staehler, M.; Bensalah, K.; Dabestani, S.; Giles, R.H.; Hofmann, F.; Hora, M.; Kuczyk, M.A.; Lam, T.B.; et al. Updated European Association of Urology Guidelines Recommendations for the Treatment of First-line Metastatic Clear Cell Renal Cancer. Eur. Urol. 2018, 73, 311–315. [Google Scholar] [CrossRef] [PubMed]
- Therasse, P.; Arbuck, S.G.; Eisenhauer, E.A.; Wanders, J.; Kaplan, R.S.; Rubinstein, L.; Verweij, J.; Van, G.M.; van Oosterom, A.T.; Christian, M.C.; et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J. Natl. Cancer Inst. 2000, 92, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef] [PubMed]
- Gore, M.E.; Szczylik, C.; Porta, C.; Bracarda, S.; Bjarnason, G.A.; Oudard, S.; Lee, S.H.; Haanen, J.; Castellano, D.; Vrdoljak, E.; et al. Final results from the large sunitinib global expanded-access trial in metastatic renal cell carcinoma. Br. J. Cancer 2015, 113, 12–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rini, B.I.; Atkins, M.B. Resistance to targeted therapy in renal-cell carcinoma. Lancet Oncol. 2009, 10, 992–1000. [Google Scholar] [CrossRef]
- Buczek, M.; Escudier, B.; Bartnik, E.; Szczylik, C.; Czarnecka, A. Resistance to tyrosine kinase inhibitors in clear cell renal cell carcinoma: From the patient’s bed to molecular mechanisms. Biochim. Biophys. Acta 2014, 1845, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Joosten, S.C.; Hamming, L.; Soetekouw, P.M.; Aarts, M.J.; Veeck, J.; van, E.M.; Tjan-Heijnen, V.C. Resistance to sunitinib in renal cell carcinoma: From molecular mechanisms to predictive markers and future perspectives. Biochim. Biophys. Acta 2015, 1855, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Busch, J.; Seidel, C.; Weikert, S.; Wolff, I.; Kempkensteffen, C.; Weinkauf, L.; Hinz, S.; Magheli, A.; Miller, K.; Grunwald, V. Intrinsic resistance to tyrosine kinase inhibitors is associated with poor clinical outcome in metastatic renal cell carcinoma. BMC Cancer 2011, 11, 295. [Google Scholar] [CrossRef] [PubMed]
- Grunwald, V.; McKay, R.R.; Krajewski, K.M.; Kalanovic, D.; Lin, X.; Perkins, J.J.; Simantov, R.; Choueiri, T.K. Depth of remission is a prognostic factor for survival in patients with metastatic renal cell carcinoma. Eur. Urol. 2015, 67, 952–958. [Google Scholar] [CrossRef] [PubMed]
- Heng, D.Y.; Xie, W.; Regan, M.M.; Warren, M.A.; Golshayan, A.R.; Sahi, C.; Eigl, B.J.; Ruether, J.D.; Cheng, T.; North, S.; et al. Prognostic factors for overall survival in patients with metastatic renal cell carcinoma treated with vascular endothelial growth factor-targeted agents: Results from a large, multicenter study. J. Clin. Oncol. 2009, 27, 5794–5799. [Google Scholar] [CrossRef] [PubMed]
- Heng, D.Y.; Xie, W.; Regan, M.M.; Harshman, L.C.; Bjarnason, G.A.; Vaishampayan, U.N.; Mackenzie, M.; Wood, L.; Donskov, F.; Tan, M.H.; et al. External validation and comparison with other models of the International Metastatic Renal-Cell Carcinoma Database Consortium prognostic model: A population-based study. Lancet Oncol. 2013, 14, 141–148. [Google Scholar] [CrossRef]
- Motzer, R.J.; Escudier, B.; Bukowski, R.; Rini, B.I.; Hutson, T.E.; Barrios, C.H.; Lin, X.; Fly, K.; Matczak, E.; Gore, M.E. Prognostic factors for survival in 1059 patients treated with sunitinib for metastatic renal cell carcinoma. Br. J. Cancer 2013, 108, 2470–2477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, M.; Shariat, S.F.; Cheng, C.; Ficarra, V.; Murai, M.; Oudard, S.; Pantuck, A.J.; Zigeuner, R.; Karakiewicz, P.I. Prognostic factors and predictive models in renal cell carcinoma: A contemporary review. Eur. Urol. 2011, 60, 644–661. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, A.; Stephan, C.; Busch, J.; Yousef, G.M.; Jung, K. Diagnostic, prognostic, and therapeutic implications of miRNAs in urologic tumors. Nat. Rev. Urol. 2010, 7, 286–297. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.Y.; Choudhury, Y.; Tan, M.H. Predictive molecular biomarkers to guide clinical decision making in kidney cancer: Current progress and future challenges. Expert Rev. Mol. Diagn. 2015, 15, 631–646. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhu, J.; George, D.J.; Nixon, A.B. Metastatic clear cell renal cell carcinoma: Circulating biomarkers to guide antiangiogenic and immune therapies. Urol. Oncol. 2016, 34, 510–518. [Google Scholar] [CrossRef] [PubMed]
- Ocana, A.; Ethier, J.L.; Diez-Gonzalez, L.; Corrales-Sanchez, V.; Srikanthan, A.; Gascon-Escribano, M.J.; Templeton, A.J.; Vera-Badillo, F.; Seruga, B.; Niraula, S.; et al. Influence of companion diagnostics on efficacy and safety of targeted anti-cancer drugs: Systematic review and meta-analyses. Oncotarget 2015, 6, 39538–39549. [Google Scholar] [CrossRef] [PubMed]
- Fendler, A.; Stephan, C.; Yousef, G.M.; Jung, K. MiRNAs as regulators of signal transduction in urological tumors. Clin. Chem. 2011, 57, 954–968. [Google Scholar] [CrossRef] [PubMed]
- Berkers, J.; Govaere, O.; Wolter, P.; Beuselinck, B.; Schoffski, P.; van Kempen, L.C.; Albersen, M.; Van den Oord, J.; Roskams, T.; Swinnen, J.; et al. A possible role for microRNA-141 down-regulation in sunitinib resistant metastatic clear cell renal cell carcinoma through induction of epithelial-to-mesenchymal transition and hypoxia resistance. J. Urol. 2013, 189, 1930–1938. [Google Scholar] [CrossRef] [PubMed]
- Prior, C.; Perez-Gracia, J.L.; Garcia-Donas, J.; Rodriguez-Antona, C.; Guruceaga, E.; Esteban, E.; Suarez, C.; Castellano, D.; del Alba, A.G.; Lozano, M.D.; et al. Identification of tissue microRNAs predictive of sunitinib activity in patients with metastatic renal cell carcinoma. PLoS ONE 2014, 9, e86263. [Google Scholar] [CrossRef] [PubMed]
- Khella, H.W.; Butz, H.; Ding, Q.; Rotondo, F.; Evans, K.R.; Kupchak, P.; Dharsee, M.; Latif, A.; Pasic, M.D.; Lianidou, E.; et al. miR-221/222 are involved in response to sunitinib treatment in metastatic renal cell carcinoma. Mol. Ther. 2015, 23, 1748–1758. [Google Scholar] [CrossRef] [PubMed]
- Merhautova, J.; Hezova, R.; Poprach, A.; Kovarikova, A.; Radova, L.; Svoboda, M.; Vyzula, R.; Demlova, R.; Slaby, O. miR-155 and miR-484 are associated with time to progression in metastatic renal cell carcinoma treated with Sunitinib. Biomed. Res. Int. 2015, 2015, 941980. [Google Scholar] [CrossRef] [PubMed]
- Lukamowicz-Rajska, M.; Mittmann, C.; Prummer, M.; Zhong, Q.; Bedke, J.; Hennenlotter, J.; Stenzl, A.; Mischo, A.; Bihr, S.; Schmidinger, M.; et al. MiR-99b-5p expression and response to tyrosine kinase inhibitor treatment in clear cell renal cell carcinoma patients. Oncotarget 2016, 7, 78433–78447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arai, T.; Okato, A.; Kojima, S.; Idichi, T.; Koshizuka, K.; Kurozumi, A.; Kato, M.; Yamazaki, K.; Ishida, Y.; Naya, Y.; et al. Regulation of spindle and kinetochore-associated protein 1 by antitumor miR-10a-5p in renal cell carcinoma. Cancer Sci. 2017, 108, 2088–2101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kovacova, J.; Juracek, J.; Poprach, A.; Buchler, T.; Kopecky, J.; Fiala, O.; Svoboda, M.; Slaby, O. Candidate microRNA biomarkers of therapeutic response to sunitinib in metastatic renal cell carcinoma: A validation study in patients with extremely good and poor response. Anticancer Res. 2018, 38, 2961–2965. [Google Scholar] [PubMed]
- Kovacova, J.; Poprach, A.; Buchler, T.; Cho, W.C.; Slaby, O. MicroRNAs as predictive biomarkers of response to tyrosine kinase inhibitor therapy in metastatic renal cell carcinoma. Clin. Chem. Lab. Med. 2018, 56, 1426–1431. [Google Scholar] [CrossRef] [PubMed]
- Altman, D.G.; McShane, L.M.; Sauerbrei, W.; Taube, S.E. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): Explanation and elaboration. PLoS Med. 2012, 9, e1001216. [Google Scholar] [CrossRef] [PubMed]
- Bossuyt, P.M.; Reitsma, J.B.; Bruns, D.E.; Gatsonis, C.A.; Glasziou, P.P.; Irwig, L.M.; Lijmer, J.G.; Moher, D.; Rennie, D.; de Vet, H.C. Towards complete and accurate reporting of studies of diagnostic accuracy: The STARD initiative. Standards for Reporting of Diagnostic Accuracy. Clin. Chem. 2003, 49, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Sobin, L.H.; Wittekind, C. TNM Classification of Malignant Tumours, 6th ed.; Wiley-Liss: New York, NY, USA, 2002; pp. 193–195. [Google Scholar]
- Fuhrman, S.A.; Lasky, L.C.; Limas, C. Prognostic significance of morphologic parameters in renal cell carcinoma. Am. J. Surg. Pathol. 1982, 6, 655–663. [Google Scholar] [CrossRef] [PubMed]
- Busch, J.; Ralla, B.; Jung, M.; Wotschofsky, Z.; Trujillo-Arribas, E.; Schwabe, P.; Kilic, E.; Fendler, A.; Jung, K. Piwi-interacting RNAs as novel prognostic markers in clear cell renal cell carcinomas. J. Exp. Clin. Cancer Res. 2015, 34, 61. [Google Scholar] [CrossRef] [PubMed]
- Chai, H.; Brown, R.E. Field effect in cancer—An update. Ann. Clin. Lab. Sci. 2009, 39, 331–337. [Google Scholar] [PubMed]
- Jung, M.; Mollenkopf, H.J.; Grimm, C.; Wagner, I.; Albrecht, M.; Waller, T.; Pilarsky, C.; Johannsen, M.; Stephan, C.; Lehrach, H.; et al. MicroRNA profiling of clear cell renal cell cancer identifies a robust signature to define renal malignancy. J. Cell. Mol. Med. 2009, 13, 3918–3928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, M.; Schaefer, A.; Steiner, I.; Kempkensteffen, C.; Stephan, C.; Erbersdobler, A.; Jung, K. Robust microRNA stability in degraded RNA preparations from human tissue and cell samples. Clin. Chem. 2010, 56, 998–1006. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, A.; Jung, M.; Mollenkopf, H.J.; Wagner, I.; Stephan, C.; Jentzmik, F.; Miller, K.; Lein, M.; Kristiansen, G.; Jung, K. Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int. J. Cancer 2010, 126, 1166–1176. [Google Scholar] [CrossRef] [PubMed]
- Wotschofsky, Z.; Meyer, H.A.; Jung, M.; Fendler, A.; Wagner, I.; Stephan, C.; Busch, J.; Erbersdobler, A.; Disch, A.C.; Mollenkopf, H.J.; et al. Reference genes for the relative quantification of microRNAs in renal cell carcinomas and their metastases. Anal. Biochem. 2011, 417, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Huggett, J.F.; Foy, C.A.; Benes, V.; Emslie, K.; Garson, J.A.; Haynes, R.; Hellemans, J.; Kubista, M.; Mueller, R.D.; Nolan, T.; et al. The digital MIQE guidelines: Minimum Information for Publication of Quantitative Digital PCR Experiments. Clin. Chem. 2013, 59, 892–902. [Google Scholar] [CrossRef] [PubMed]
- Vickers, A.J.; Elkin, E.B. Decision curve analysis: A novel method for evaluating prediction models. Med. Decis. Mak. 2006, 26, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Stephan, C.; Jung, K.; Semjonow, A.; Schulze-Forster, K.; Cammann, H.; Hu, X.; Meyer, H.-A.; Bögemann, M.; Miller, K.; Friedersdorff, F. Comparative assessment of urinary prostate cancer antigen 3 and TMPRSS2: ERG gene fusion with the serum [-2]proprostate-specific antigen-based prostate health index for detection of prostate cancer. Clin. Chem. 2013, 59, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.H.; Chang, N.W.; Shrestha, S.; Hsu, S.D.; Lin, Y.L.; Lee, W.H.; Yang, C.D.; Hong, H.C.; Wei, T.Y.; Tu, S.J.; et al. miRTarBase 2016: Updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016, 44, D239–D247. [Google Scholar] [CrossRef] [PubMed]
- Dweep, H.; Gretz, N. miRWalk2.0: A comprehensive atlas of microRNA-target interactions. Nat. Methods 2015, 12, 697. [Google Scholar] [CrossRef] [PubMed]
- Vlachos, I.S.; Paraskevopoulou, M.D.; Karagkouni, D.; Georgakilas, G.; Vergoulis, T.; Kanellos, I.; Anastasopoulos, I.L.; Maniou, S.; Karathanou, K.; Kalfakakou, D.; et al. DIANA-TarBase v7.0: Indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res. 2015, 43, D153–D159. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, Y.; Wei, X.; Chu, Y.H.; Ng, L.G.; Tan, H.S.; Koh, V.; Thike, A.A.; Poon, E.; Ng, Q.S.; Toh, C.K.; et al. A multigene assay identifying distinct prognostic subtypes of clear cell renal cell carcinoma with differential response to tyrosine kinase inhibition. Eur. Urol. 2015, 67, 17–20. [Google Scholar] [CrossRef] [PubMed]
- Beuselinck, B.; Vano, Y.A.; Oudard, S.; Wolter, P.; De, S.R.; Depoorter, L.; Teghom, C.; Karadimou, A.; Zucman-Rossi, J.; Debruyne, P.R.; et al. Prognostic impact of baseline serum C-reactive protein in patients with metastatic renal cell carcinoma (RCC) treated with sunitinib. BJU Int. 2014, 114, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Gamez-Pozo, A.; Anton-Aparicio, L.M.; Bayona, C.; Borrega, P.; Gallegos Sancho, M.I.; Garcia-Dominguez, R.; de, P.T.; Ramos-Vazquez, M.; Perez-Carrion, R.; Bolos, M.V.; et al. MicroRNA expression profiling of peripheral blood samples predicts resistance to first-line sunitinib in advanced renal cell carcinoma patients. Neoplasia 2012, 14, 1144–1152. [Google Scholar] [CrossRef] [PubMed]
- Monzon, J.G.; Heng, D.Y. Management of metastatic kidney cancer in the era of personalized medicine. Crit. Rev. Clin. Lab. Sci. 2014, 51, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Busch, J.; Jung, M.; Rabenhorst, S.; Ralla, B.; Kilic, E.; Mergemeier, S.; Budach, N.; Fendler, A.; Jung, K. Diagnostic and prognostic potential of circulating cell-free genomic and mitochondrial DNA fragments in clear cell renal cell carcinoma patients. Clin. Chim. Acta 2016, 452, 109–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerszten, R.E.; Wang, T.J. The search for new cardiovascular biomarkers. Nature 2008, 451, 949–952. [Google Scholar] [CrossRef] [PubMed]
- Drakaki, A.; Hatziapostolou, M.; Polytarchou, C.; Vorvis, C.; Poultsides, G.A.; Souglakos, J.; Georgoulias, V.; Iliopoulos, D. Functional microRNA high throughput screening reveals miR-9 as a central regulator of liver oncogenesis by affecting the PPARA-CDH1 pathway. BMC Cancer 2015, 15, 542. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Cai, X. Up-regulation of miR-9 expression predicate advanced clinicopathological features and poor prognosis in patients with hepatocellular carcinoma. Diagn. Pathol. 2014, 9, 1000. [Google Scholar] [CrossRef] [PubMed]
- Gwak, J.M.; Kim, H.J.; Kim, E.J.; Chung, Y.R.; Yun, S.; Seo, A.N.; Lee, H.J.; Park, S.Y. MicroRNA-9 is associated with epithelial-mesenchymal transition, breast cancer stem cell phenotype, and tumor progression in breast cancer. Breast Cancer Res. Treat. 2014, 147, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Li, J.; Zhu, Y.; Dai, Y.; Zeng, T.; Liu, L.; Li, J.; Wang, H.; Qin, Y.; Zeng, M.; et al. MicroRNA-9 promotes tumor metastasis via repressing E-cadherin in esophageal squamous cell carcinoma. Oncotarget 2014, 5, 11669–11680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bandres, E.; Agirre, X.; Bitarte, N.; Ramirez, N.; Zarate, R.; Roman-Gomez, J.; Prosper, F.; Garcia-Foncillas, J. Epigenetic regulation of microRNA expression in colorectal cancer. Int. J. Cancer 2009, 125, 2737–2743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, L.M.; Pu, Y.; Han, Z.; Liu, T.; Li, Y.X.; Liu, M.; Li, X.; Tang, H. MicroRNA-9 inhibits ovarian cancer cell growth through regulation of NF-kappaB1. FEBS J. 2009, 276, 5537–5546. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.Y.; Guo, L.M.; Liu, T.; Liu, M.; Li, X.; Tang, H. Regulation of the transcription factor NF-kappaB1 by microRNA-9 in human gastric adenocarcinoma. Mol. Cancer 2010, 9, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schiffgen, M.; Schmidt, D.H.; von, R.A.; Muller, S.C.; Ellinger, J. Epigenetic regulation of microRNA expression in renal cell carcinoma. Biochem. Biophys. Res. Commun. 2013, 436, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Peters, I.; Dubrowinskaja, N.; Abbas, M.; Seidel, C.; Kogosov, M.; Scherer, R.; Gebauer, K.; Merseburger, A.S.; Kuczyk, M.A.; Grunwald, V.; et al. DNA methylation biomarkers predict progression-free and overall survival of metastatic renal cell cancer (mRCC) treated with antiangiogenic therapies. PLoS ONE 2014, 9, e91440. [Google Scholar] [CrossRef] [PubMed]
- Hildebrandt, M.A.; Gu, J.; Lin, J.; Ye, Y.; Tan, W.; Tamboli, P.; Wood, C.G.; Wu, X. Hsa-miR-9 methylation status is associated with cancer development and metastatic recurrence in patients with clear cell renal cell carcinoma. Oncogene 2010, 29, 5724–5728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, L.; Young, J.; Prabhala, H.; Pan, E.; Mestdagh, P.; Muth, D.; Teruya-Feldstein, J.; Reinhardt, F.; Onder, T.T.; Valastyan, S.; et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat. Cell. Biol. 2010, 12, 247–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thiele, S.; Wittmann, J.; Jack, H.M.; Pahl, A. miR-9 enhances IL-2 production in activated human CD4(+) T cells by repressing Blimp-1. Eur. J. Immunol. 2012, 42, 2100–2108. [Google Scholar] [CrossRef] [PubMed]
- Kroesen, B.J.; Teteloshvili, N.; Smigielska-Czepiel, K.; Brouwer, E.; Boots, A.M.; van den Berg, A.; Kluiver, J. Immuno-miRs: Critical regulators of T-cell development, function and ageing. Immunology 2015, 144, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Prahallad, A.; Heynen, G.J.; Germano, G.; Willems, S.M.; Evers, B.; Vecchione, L.; Gambino, V.; Lieftink, C.; Beijersbergen, R.L.; Di, N.F.; et al. PTPN11 is a central node in intrinsic and acquired resistance to targeted cancer drugs. Cell. Rep. 2015, 12, 1978–1985. [Google Scholar] [CrossRef] [PubMed]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; Tarpey, P.; et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Ljungberg, B.; Mehle, C.; Stenling, R.; Roos, G. Heterogeneity in renal cell carcinoma and its impact no prognosis—A flow cytometric study. Br. J. Cancer 1996, 74, 123–127. [Google Scholar] [CrossRef] [PubMed]
- Burke, H.B. Predicting clinical outcomes using molecular biomarkers. Biomark. Cancer 2016, 8, 89–99. [Google Scholar] [CrossRef] [PubMed]



| Patient Characteristics | TKI Therapy Groups | p Value * | |
|---|---|---|---|
| Responders | Non-Responders | ||
| Number | 41 | 19 | |
| Sex | 0.081 | ||
| female | 13 | 2 | |
| male | 28 | 17 | |
| Age, median years (95% CI) | 62.0 (59.0–67.2) | 59.0 (53.6–67.0) | 0.427 |
| Tumor characteristics | 0.678 | ||
| clear cell carcinoma | 38 | 17 | |
| papillary carcinoma | 3 | 2 | |
| Tumor stage | 0.226 | ||
| pT1 | 8 | 4 | |
| pT2 | 5 | - | |
| pT3 | 25 | 15 | |
| pT4 | 3 | - | |
| Regional/distant metastasis † | 0.033 | ||
| pN0/M0 | 23 | 5 | |
| pN1/M1 | 18 | 14 | |
| Residual tumor | 0.528 | ||
| R0/Rx | 32 | 16 | |
| R1/2 | 9 | 3 | |
| Fuhrman grade | 0.229 | ||
| FG1/2 | 25 | 9 | |
| FG3/4 | 14 | 10 | |
| FGx ‡ | 2 | - | |
| First-line TKI therapy | 0.313 | ||
| Sunitinib | 35 | 16 | |
| Pazopanib | 3 | 3 | |
| Sorafenib | 3 | - | |
| Response to targeted therapy | |||
| Partial response | 11 | - | |
| Stable disease | 30 | - | |
| Progressive disease | - | 19 | |
| Median months survival (95% CI) § | |||
| Progression-free | 16.2 (11.4–21.0) | 2.6 (2.3–3.5) | <0.0001 |
| Overall | 67.6 (40.1–96.2) | 20.9 (13.3–37.4) | <0.0001 |
| miRNA | AUC (95% CI) | p Value Different to AUC = 0.5 | Differentiating Ability at the Youden Index * | Overall Correct Classification (%) | |
|---|---|---|---|---|---|
| Sensitivity (%) | Specificity (%) | ||||
| Single miRNAs | |||||
| miR-9-5p | 0.83 (0.71–0.91) | <0.0001 | 74 | 88 | 73 |
| miR-203a-3p | 0.53 (0.40–0.66) | 0.718 | 63 | 51 | 68 |
| miR-204-5p | 0.51 (0.38–0.65) | 0.861 | 89 | 29 | 68 |
| miR-223-3p | 0.52 (0.03–0.65) | 0.871 | 53 | 61 | 68 |
| miR-489-3p | 0.69 (0.58–0.80) | 0.014 | 79 | 59 | 68 |
| miR-500a-5p | 0.52 (0.38–0.65) | 0.846 | 16 | 73 | 68 |
| miR-885-5p | 0.57 (0.44–0.70) | 0.353 | 58 | 63 | 72 |
| Combined miRNAs | |||||
| All miRNAs † | 0.79 (0.66–0.88) | <0.0001 | 95 | 59 | 77 |
| miR-9/miR-489 | 0.83 (0.73–0.93) | <0.0001 | 69 | 87 | 73 |
| miR-9 + miR-489 † | 0.82 (0.71–1.00) | <0.0001 | 97 | 21 | 72 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ralla, B.; Busch, J.; Flörcken, A.; Westermann, J.; Zhao, Z.; Kilic, E.; Weickmann, S.; Jung, M.; Fendler, A.; Jung, K. miR-9-5p in Nephrectomy Specimens is a Potential Predictor of Primary Resistance to First-Line Treatment with Tyrosine Kinase Inhibitors in Patients with Metastatic Renal Cell Carcinoma. Cancers 2018, 10, 321. https://doi.org/10.3390/cancers10090321
Ralla B, Busch J, Flörcken A, Westermann J, Zhao Z, Kilic E, Weickmann S, Jung M, Fendler A, Jung K. miR-9-5p in Nephrectomy Specimens is a Potential Predictor of Primary Resistance to First-Line Treatment with Tyrosine Kinase Inhibitors in Patients with Metastatic Renal Cell Carcinoma. Cancers. 2018; 10(9):321. https://doi.org/10.3390/cancers10090321
Chicago/Turabian StyleRalla, Bernhard, Jonas Busch, Anne Flörcken, Jörg Westermann, Zhongwei Zhao, Ergin Kilic, Sabine Weickmann, Monika Jung, Annika Fendler, and Klaus Jung. 2018. "miR-9-5p in Nephrectomy Specimens is a Potential Predictor of Primary Resistance to First-Line Treatment with Tyrosine Kinase Inhibitors in Patients with Metastatic Renal Cell Carcinoma" Cancers 10, no. 9: 321. https://doi.org/10.3390/cancers10090321





