Emerging Roles of ADAMTSs in Angiogenesis and Cancer
Abstract
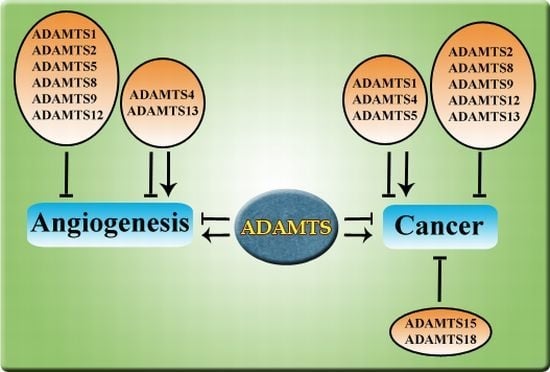
:1. Introduction

2. ADAMTS1
2.1. Structure and Processing
2.2. Regulation and Expression
2.3. Function
| Protein | Alternative Names | Substrates | Knockout phenotype |
|---|---|---|---|
| ADAMTS1 | METH-1, aggrecanase-3 | Aggrecan [22] | a. Growth retardation, changes in kidney structure and impaired female fertility [19,20]. |
| Versican V1 [23] | |||
| Nidogen 1 & 2 | |||
| Tissue Factor Pathway Inhibitor-2 [25] | b. No effect on aggrecan turnover [27]. | ||
| ADAMTS2 | PCINP | Procollagens type I, II, III and V [28,29,30] | a. Fragile skin and male sterility [31]. |
| ADAMTS4 | aggrecanase-1, KIAA0688 | Aggrecan [32,33] | a. Phenotypically normal, no protection against aggrecan degradation [38]. |
| Versican [23] | |||
| Brevican [34] | |||
| Matrilin [35] Hevin [36] Reelin [37] | |||
| ADAMTS5 | aggrecanase-2, ADAMTS11, Implantin | Aggrecan [39], Versican [40], Brevican [41], Neurocan [42]. | a. Normal lifespan, healthy and fertile [43,44]. Syndactyly with a penetrance of 44% [45]. |
| b. Reduced cartilage degradation in a mouse model of osteoarthritis [43]. | |||
| c. Delayed wound healing due to aggrecan deposition [46]. | |||
| d. Double knockout (Adamts4−/−, Adamts5−/−)—Normal, healthy and fertile [47,48]. Reduced body weight in females compared to wild type. Protected against arthritis [47]. | |||
| e. Double knockout (Adamts5−/−, Adamts20−/−)—No gross abnormalities, soft tissue Syndactyly [45]. | |||
| ADAMTS8 | METH-2 | Aggrecan [49]. | - |
| ADAMTS9 | KIAA1312 | Aggrecan [50], | a. Embryonically lethal [51]. |
| Versican [50]. | b. Haploinsufficiency causes increased angiogenesis [52], cardiac and aortic anomalies [53]. | ||
| c. Adamts5−/−, Adamts9+/−: soft tissue Syndactyly [45]. | |||
| ADAMTS12 | - | COMP [54]. | a. Phenotypically normal and fertile [56]. |
| Aggrecan [55] | b. Elevated angiogenesis [56]. | ||
| c. Severe inflammation [57] | |||
| ADAMTS13 | vWFCP | vWF [58]. | a. Viable and fertile [59,60]. |
| ADAMTS15 | - | Aggrecan [61], | Not known |
| Versican [62]. | |||
| ADAMTS18 | - | Not known | Not known |
2.4. Angiogenesis and Cancer
| Protein | Dependence on catalytic activity | Involvement of TSRs | Role in Angiogenesis |
|---|---|---|---|
| ADAMTS1 | Yes (Pro-angiogenic) [64] | Yes (Anti-angiogenic) [66] | Anti-angiogenic |
| Yes (Anti-angiogenic) [63] | - Suppresses EC proliferation in a cell-specific, dose dependent manner [12,63] | ||
| No (Anti-angiogenic) [65] | - Disrupts growth factor induced angiogenesis in vivo in a CAM model and matriplug assay [12]. | ||
| - Suppresses tumor angiogenesis in T47D human breast carcinoma [63] | |||
| - Regulates angiogenesis in ischemic myocardium [67] | |||
| - Suppresses tumor angiogenesis in HT-1080, DU145 and CHO-K1 tumors [65]. | |||
| - Alters blood vessel morphology in prostate tumors [68] | |||
| - Induced by VEGF in ECs and ischemia induced retinal neovascularization [69] | |||
| Pro-angiogenic | |||
| - Induction of ADAMTS1 to degrade basement membrane versican in VEGF-induced pathological angiogenesis [64]. | |||
| - Promotes tumor angiogenesis in TA3 mammary carcinoma and Lewis lung carcinoma [66] | |||
| - Induces endothelial-like phenotype in plastic tumor cells [70] | |||
| ADAMTS2 | No (Anti-angiogenic) [71] | Yes (Anti-angiogenic) [71] | Anti-angiogenic |
| - Suppressed VEGF-stimulated EC proliferation in a cell-specific manner, induces apoptosis and inhibits capillary network formation of HUVEC [71]. | |||
| - Increased blood vessels in vivo in a CAM model in ADAMTS2 knockout mice [71]. | |||
| - Suppressed tumor angiogenesis in ADAMTS2 overexpressing tumors [71]. | |||
| ADAMTS4 | Yes (Pro-angiogenic) [72] | Yes (Anti-angiogenic) [72] | Anti-angiogenic |
| - Anti-angiogenic peptide from ADAMTS4 TSR suppresses EC proliferation and VEGF-induced HUVEC migration [73]. | |||
| - Truncated ADAMTS4 fragment inhibits HuDMEC differentiation and migration in a scratch wound healing assay [74]. | |||
| - ADAMTS4 C-terminal ancillary regions inhibit tumor angiogenesis [72]. | |||
| Pro-angiogenic | |||
| - Full-length ADAMTS4 promotes tumor angiogenesis [72]. | |||
| ADAMTS5 | No (Anti-angiogenic) [75] | Yes (Anti-angiogenic) [75,76] | Anti-angiogenic |
| - ADAMTS5 is anti-angiogenic in vitro and in vivo [75,76]. | |||
| ADAMTS8 | Not known | Yes (Anti-angiogenic) [12] | Anti-angiogenic |
| - Inhibits EC proliferation in a cell-specific reversible manner in vitro [12]. | |||
| - Disrupts growth factor induced angiogenesis in vivo in a CAM model and matriplug assay [12]. | |||
| ADAMTS9 | Yes (Anti-angiogenic) [52] | No (Anti-angiogenic) [52] | Anti-angiogenic |
| - Knockdown of ADAMTS9 in cultured ECs suppresses in vitro capillary network formation and migration [52]. | |||
| - Increased corneal neovascularization and tumor vascularization in in vivo+/− mice compared to the wild type mice [52]. | |||
| - Suppresses oesophageal and nasopharyngeal carcinoma angiogenesis [77] | |||
| ADAMTS12 | No (Anti-angiogenic) [56] | Yes (Anti-angiogenic) [55] | Anti-angiogenic |
| - Inhibits capillary network formation by BAE-1 cells in 3D collagen gels [55]. | |||
| - Adamts12−/− mice showed increased sprout density in both ex vivo and in vivo models of angiogenesis [56]. | |||
| ADAMTS13 | Not known | Yes (Anti-angiogenic) [10] | Anti-angiogenic |
| - ADAMTS13 inhibits VEGF-mediated angiogenesis-mediated HUVEC proliferation, migration and capillary network formation [10]. | |||
| Pro-angiogenic | |||
| - Full-length ADAMTS13 promoted HUVEC tube formation, induces EC proliferation and migration in vitro [10]. | |||
| ADAMTS15 | Not known | Not known | Not known |
| ADAMTS18 | Not known | Not known | Not known |
| Protein | Cancer type | Regulation |
|---|---|---|
| ADAMTS1 | Lung cancer | Down-regulation of ADAMTS1 mRNA in NSCLC cell lines and epigenetic regulation via hypermethylation of its promoter [83] |
| Pancreatic cancer | ADAMTS1 mRNA expression significantly lower in pancreatic cancer compared to noncancerous pancreas [82]. | |
| Enhanced expression of ADAMTS1 mRNA in lymph node metastasis or severe retroperitoneal invasion [82]. | ||
| Prostate cancer | Markedly low protein levels in prostate cancer cells [15]. | |
| Chondrosarcoma | Transcriptional up-regulation in response to TNF-α [16]. | |
| ADAMTS2 | Osteosarcoma | 8-fold increase in ADAMTS2 mRNA levels [28]. |
| ADAMTS4 | Breast cancer | Enhanced mRNA expression in breast cancer compared to normal breast tissue [91]. |
| Head and neck squamous cell carcinoma | Enhanced expression of ADAMTS4 mRNA [92]. | |
| Glioblastoma | Increased expression of ADAMTS4 mRNA [93]. | |
| Ewings sarcoma | Enhanced protein levels serving as a tumor-specific marker [94]. | |
| ADAMTS5 | Breast carcinoma | Down-regulation of ADAMTS5 transcript [91]. |
| Colorectal cancer | Epigenetically silenced by promoter methylation [95]. | |
| Prostate cancer | Down-regulation of ADAMTS5 mRNA in prostate cancer cell lines [42]. | |
| Glioblastoma | Overexpression of ADAMTS5 mRNA and protein [41,93]. | |
| ADAMTS8 | Lung cancer | Down-regulation of ADAMTS8 at the mRNA level in NSCLC [96]. |
| Down-regulation due to epigenetic silencing [97]. | ||
| Brain | Down-regulation due to promoter hypermethylation [98]. | |
| ADAMTS9 | Breast carcinoma | Down-regulation of ADAMTS9 transcript [91]. |
| Esophageal squamous cell carcinoma | Hypermethylation of ADAMTS9 in esophageal tumors [99]. | |
| Nasopharyngeal carcinoma | Promoter hypermethylation and association of lower levels of ADAMTS9 protein with lymph node metastasis in NPC [100]. | |
| Gastric cancer | Epigenetic silencing by promoter hypermethylation [101].Inhibition through Akt/mTOR pathway [102]. | |
| Pancreatic and colorectal cancer | Epigenetic silencing by promoter hypermethylation [101]. | |
| ADAMTS12 | Colorectal cancer | Epigenetic silencing by promoter hypermethylation [103]. |
| ADAMTS13 | Prostate, renal, testicular, head and neck squamous, colorectal, rectal, NSCLC, gastric, melanoma, adenocarcinoma and breast carcinoma. | Correlation of presence or absence of tumor metastasis with lower or higher vWF cleaving respectively [104]. |
| Colon cancer, leukemia, multiple myeloma, breast, stomach cancer, non-Hodgkin’s lymphoma | Decreased activity of ADAMTS13 in plasma of malignant patients [105]. | |
| Brain and prostate cancers | Mild reduction in ADAMTS13 activity but no correlation with malignancy and metastasis [106]. | |
| ADAMTS15 | Colorectal and pancreatic cancer | Inactivation and loss of normal function of the protein due to somatic mutations [107,108]. |
| Colorectal cancer | Loss of heterozygosity in ADAMTS15 locus [109]. | |
| Breast cancer | Grade-specific down-regulation of ADAMTS15 transcript in breast cancer [91]. | |
| Prostate cancer | Down-regulation of ADAMTS15 mRNA linked to poor prognosis in prostate cancer [42]. | |
| ADAMTS18 | Breast cancer | Down-regulation of ADAMTS18 transcript [91]. |
| Pancreatic, gastric and colorectal cancers | Hypermethylation of ADAMTS18 promoter [110] | |
| Kidney and colorectal cancers | Inactivation of ADAMTS18 via somatic mutations [111]. | |
| Melanoma | Somatic mutations in ADAMTS18 linked to higher transformation ability and increased metastases in vivo [112]. |
3. ADAMTS2
3.1. Structure and Processing
3.2. Expression and Regulation
3.3. Function
3.4. Angiogenesis and Cancer
4. ADAMTS4
4.1. Structure and Processing
4.2. Expression and Regulation
4.3. Function
4.4. Angiogenesis and Cancer
5. ADAMTS5
5.1. Structure and Processing
5.2. Expression and Regulation
5.3. Function
5.4. Angiogenesis and Cancer
6. ADAMTS8
6.1. Structure and Processing
6.2. Expression and Regulation
6.3. Function
6.4. Angiogenesis and Cancer
7. ADAMTS9
7.1. Structure and Processing
7.2. Expression and Regulation
7.3. Function
7.4. Angiogenesis and Cancer
8. ADAMTS12
8.1. Structure and Processing
8.2. Expression and Regulation
8.3. Function
8.4. Angiogenesis and Cancer
9. ADAMTS13
9.1. Structure and Processing
9.2. Expression and Regulation
9.3. Function
9.4. Angiogenesis and Cancer
10. ADAMTS15
10.1. Structure and Processing
10.2. Expression and Regulation
10.3. Function
10.4. Angiogenesis and Cancer
11. ADAMTS18
11.1. Structure and Processing
11.2. Expression and Regulation
11.3. Function
11.4. Angiogenesis and Cancer
12. Conclusions

Acknowledgements
References
- Koo, B.-H.; Longpré, J.-M.; Somerville, R.; Alexander, J.; Leduc, R.; Apte, S. Regulation of ADAMTS9 secretion and enzymatic activity by its propeptide. J. Biol. Chem. 2007, 282, 16146–16154. [Google Scholar]
- Majerus, E.; Zheng, X.; Tuley, E.; Sadler, J. Cleavage of the ADAMTS13 propeptide is not required for protease activity. J. Biol. Chem. 2003, 278, 46643–46648. [Google Scholar]
- Porter, S.; Clark, I.M.; Kevorkian, L.; Edwards, D.R. The ADAMTS metalloproteinases. Biochem. J. 2005, 386, 15–27. [Google Scholar] [CrossRef]
- Apte, S. A disintegrin-like and metalloprotease (reprolysin-type) with thrombospondin type 1 motif (ADAMTS) superfamily: Functions and mechanisms. J. Biol. Chem. 2009, 284, 31493–31500. [Google Scholar] [CrossRef]
- Tortorella, M.; Malfait, F.; Barve, R.; Shieh, H.-S.; Malfait, A.-M. A review of the ADAMTS family, pharmaceutical targets of the future. Curr. Pharm. Des. 2009, 15, 2359–2374. [Google Scholar]
- Stanton, H.; Melrose, J.; Little, C.; Fosang, A. Proteoglycan degradation by the ADAMTS family of proteinases. Biochim. Biophys. Acta 2011, 1812, 1616–1629. [Google Scholar] [CrossRef]
- Troeberg, L.; Nagase, H. Proteases involved in cartilage matrix degradation in osteoarthritis. Biochim. Biophys. Acta 2012, 1824, 133–145. [Google Scholar] [CrossRef]
- Wagstaff, L.; Kelwick, R.; Decock, J.; Edwards, D.R. The roles of ADAMTS metalloproteinases in tumorigenesis and metastasis. Front. Biosci. 2011, 16, 1861–1872. [Google Scholar]
- Salter, R.; Ashlin, T.; Kwan, A.; Ramji, D. ADAMTS proteases: Key roles in atherosclerosis? J. Mol. Med. 2010, 88, 1203–1211. [Google Scholar] [CrossRef]
- Lee, M.; Rodansky, E.; Smith, J.; Rodgers, G. ADAMTS13 promotes angiogenesis and modulates VEGF-induced angiogenesis. Microvasc. Res. 2012, 84, 109–115. [Google Scholar]
- Kuno, K.; Kanada, N.; Nakashima, E.; Fujiki, F.; Ichimura, F.; Matsushima, K. Molecular cloning of a gene encoding a new type of metalloproteinase-disintegrin family protein with thrombospondin motifs as an inflammation associated gene. J. Biol. Chem. 1997, 272, 556–618. [Google Scholar]
- Vazquez, F.; Hastings, G.; Ortega, M.A.; Lane, T.F.; Oikemus, S.; Lombardo, M.; Iruela-Arispe, M.L. METH-1, a human ortholog of ADAMTS-1, and METH-2 are members of a new family of proteins with angio-inhibitory activity. J. Biol. Chem. 1999, 274, 23349–23357. [Google Scholar]
- Kuno, K.; Matsushima, K. ADAMTS-1 protein anchors at the extracellular matrix through the thrombospondin type I motifs and its spacing region. J. Biol. Chem. 1998, 273, 13912–13917. [Google Scholar]
- Thai, S.N.; Iruela-Arispe, M.L. Expression of ADAMTS1 during murine development. Mech. Dev. 2002, 115, 181–185. [Google Scholar] [CrossRef]
- Gustavsson, H.; Wang, W.; Jennbacken, K.; Welén, K.; Damber, J.-E. ADAMTS1, a putative anti-angiogenic factor, is decreased in human prostate cancer. BJU Int. 2009, 104, 1786–1790. [Google Scholar] [CrossRef]
- Hatipoglu, O.F.; Hirohata, S.; Yaykasli, K.O.; Cilek, M.Z.; Demircan, K.; Shinohata, R.; Yonezawa, T.; Oohashi, T.; Kusachi, S.; Ninomiya, Y. The 3'-untranslated region of ADAMTS1 regulates its mRNA stability. Acta Med. Okayama 2009, 63, 79–85. [Google Scholar]
- Kalinski, T.; Krueger, S.; Sel, S.; Werner, K.; Röpke, M.; Roessner, A. ADAMTS1 is regulated by interleukin-1beta, not by hypoxia, in chondrosarcoma. Hum. Pathol. 2007, 38, 86–94. [Google Scholar] [CrossRef]
- Lee, N.; Rodriguez-Manzaneque, J.; Thai, S.; Twal, W.; Luque, A.; Lyons, K.; Argraves, W.; Iruela-Arispe, M. Fibulin-1 acts as a cofactor for the matrix metalloprotease ADAMTS-1. J. Biol. Chem. 2005, 280, 34796–34804. [Google Scholar]
- Shindo, T.; Kurihara, H.; Kuno, K.; Yokoyama, H.; Wada, T.; Kurihara, Y.; Imai, T.; Wang, Y.; Ogata, M.; Nishimatsu, H.; et al. DAMTS-1: A metalloproteinase-disintegrin essential for normal growth, fertility, and organ morphology and function. J. Clin. Invest. 2000, 105, 1345–1352. [Google Scholar]
- Mittaz, L.; Russell, D.; Wilson, T.; Brasted, M.; Tkalcevic, J.; Salamonsen, L.; Hertzog, P.; Pritchard, M. Adamts-1 is essential for the development and function of the urogenital system. Biol. Reprod. 2004, 70, 1096–1105. [Google Scholar]
- Yokoyama, H.; Wada, T.; Kobayashi, K.-I.; Kuno, K.; Kurihara, H.; Shindo, T.; Matsushima, K. A disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS)-1 null mutant mice develop renal lesions mimicking obstructive nephropathy. Nephrol. Dial. Transplant. 2002, 17, 39–41. [Google Scholar]
- Rodríguez-Manzaneque, J.; Westling, J.; Thai, S.; Luque, A.; Knauper, V.; Murphy, G.; Sandy, J.; Iruela-Arispe, M. ADAMTS1 cleaves aggrecan at multiple sites and is differentially inhibited by metalloproteinase inhibitors. Biochem. Biophys. Res. Commun. 2002, 293, 501–508. [Google Scholar]
- Sandy, J.D.; Westling, J.; Kenagy, R.D.; Iruela-Arispe, M.L.; Verscharen, C.; Rodriguez-Mazaneque, J.C.; Zimmermann, D.R.; Lemire, J.M.; Fischer, J.W.; Wight, T.N.; et al. Versican V1 proteolysis in human aorta in vivo occurs at the Glu(441)-Ala(442) bond, a site that is cleaved by recombinant ADAMTS-1 and ADAMTS-4. J. Biol. Chem. 2001, 276, 13372–13378. [Google Scholar]
- Canals, F.; Colomé, N.; Ferrer, C.; Plaza-Calonge, M.; Rodríguez-Manzaneque, J. Identification of substrates of the extracellular protease ADAMTS1 by DIGE proteomic analysis. Proteomics 2006, 6, 35. [Google Scholar]
- Torres-Collado, A.; Kisiel, W.; Iruela-Arispe, M.; Rodríguez-Manzaneque, J. ADAMTS1 interacts with, cleaves, and modifies the extracellular location of the matrix inhibitor tissue factor pathway inhibitor-2. J. Biol. Chem. 2006, 281, 17827–17837. [Google Scholar]
- Krampert, M.; Kuenzle, S.; Thai, S.; Lee, N.; Iruela-Arispe, M.; Werner, S. ADAMTS1 proteinase is up-regulated in wounded skin and regulates migration of fibroblasts and endothelial cells. J. Biol. Chem. 2005, 280, 23844–23852. [Google Scholar]
- Little, C.; Mittaz, L.; Belluoccio, D.; Rogerson, F.; Campbell, I.; Meeker, C.; Bateman, J.; Pritchard, M.; Fosang, A. ADAMTS-1-knockout mice do not exhibit abnormalities in aggrecan turnover in vitro or in vivo. Arthritis Rheum. 2005, 52, 1461–1472. [Google Scholar]
- Wang, W.-M.; Lee, S.; Steiglitz, B.; Scott, I.; Lebares, C.; Allen, M.; Brenner, M.; Takahara, K.; Greenspan, D. Transforming growth factor-beta induces secretion of activated ADAMTS-2. A procollagen III N-proteinase. J. Biol. Chem. 2003, 278, 19549–19557. [Google Scholar]
- Colige, A.; Sieron, A.; Li, S.; Schwarze, U.; Petty, E.; Wertelecki, W.; Wilcox, W.; Krakow, D.; Cohn, D.; Reardon, W.; et al. Human Ehlers-Danlos syndrome type VII C and bovine dermatosparaxis are caused by mutations in the procollagen I N-proteinase gene. Am. J. Hum. Genet. 1999, 65, 308–317. [Google Scholar] [CrossRef]
- Colige, A.; Ruggiero, F.; Vandenberghe, I.; Dubail, J.; Kesteloot, F.; van Beeumen, J.; Beschin, A.; Brys, L.; Lapière, C.; Nusgens, B. Domains and maturation processes that regulate the activity of ADAMTS-2, a metalloproteinase cleaving the aminopropeptide of fibrillar procollagens types I–III and V. J. Biol. Chem. 2005, 280, 34397–34408. [Google Scholar]
- Li, S.; Arita, M.; Fertala, A.; Bao, Y.; Kopen, G.; Långsjö, T.; Hyttinen, M.; Helminen, H.; Prockop, D. Transgenic mice with inactive alleles for procollagen N-proteinase (ADAMTS-2) develop fragile skin and male sterility. Biochem. J. 2001, 355, 271–278. [Google Scholar] [CrossRef]
- Tortorella, M.; Burn, T.; Pratta, M.; Abbaszade, I.; Hollis, J.; Liu, R.; Rosenfeld, S.; Copeland, R.; Decicco, C.; Wynn, R.; et al. Purification and cloning of aggrecanase-1: A member of the ADAMTS family of proteins. Science 1999, 284, 1664–1666. [Google Scholar] [CrossRef]
- Naito, S.; Shiomi, T.; Okada, A.; Kimura, T.; Chijiiwa, M.; Fujita, Y.; Yatabe, T.; Komiya, K.; Enomoto, H.; Fujikawa, K.; et al. Expression of ADAMTS4 (aggrecanase-1) in human osteoarthritic cartilage. Pathol. Int. 2007, 57, 703–711. [Google Scholar] [CrossRef]
- Matthews, R.; Gary, S.; Zerillo, C.; Pratta, M.; Solomon, K.; Arner, E.; Hockfield, S. Brain-enriched hyaluronan binding (BEHAB)/brevican cleavage in a glioma cell line is mediated by a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS) family member. J. Biol. Chem. 2000, 275, 22695–22703. [Google Scholar]
- Ehlen, H.W.; Sengle, G.; Klatt, A.R.; Talke, A.; Muller, S.; Paulsson, M.; Wagener, R. Proteolytic processing causes extensive heterogeneity of tissue matrilin forms. J. Biol. Chem. 2009, 284, 21545–21556. [Google Scholar]
- Weaver, M.S.; Workman, G.; Cardo-Vila, M.; Arap, W.; Pasqualini, R.; Sage, E.H. Processing of the matricellular protein hevin in mouse brain is dependent on ADAMTS4. J. Biol. Chem. 2010, 285, 5868–5877. [Google Scholar]
- Hisanaga, A.; Morishita, S.; Suzuki, K.; Sasaki, K.; Koie, M.; Kohno, T.; Hattori, M. A disintegrin and metalloproteinase with thrombospondin motifs 4 (ADAMTS-4) cleaves Reelin in an isoform-dependent manner. FEBS Lett. 2012, 586, 3349–3353. [Google Scholar]
- Glasson, S.S.; Askew, R.; Sheppard, B.; Carito, B.A.; Blanchet, T.; Ma, H.L.; Flannery, C.R.; Kanki, K.; Wang, E.; Peluso, D.; et al. Characterization of and osteoarthritis susceptibility in ADAMTS-4-knockout mice. Arthritis Rheum. 2004, 50, 2547–2558. [Google Scholar] [CrossRef]
- Abbaszade, I.; Liu, R.; Yang, F.; Rosenfeld, S.; Ross, O.H.; Link, J.R.; Ellis, D.M.; Tortorella, M.D.; Pratta, MA.; Hollis, J.M.; et al. Cloning and characterization of ADAMTS11, an aggrecanase from the ADAMTS family. J. Biol. Chem. 1999, 274, 23443–23450. [Google Scholar]
- Longpré, J.-M.; McCulloch, D.; Koo, B.-H.; Alexander, J.; Apte, S.; Leduc, R. Characterization of proADAMTS5 processing by proprotein convertases. Int. J. Biochem. Cell Biol. 2009, 41, 1116–1142. [Google Scholar]
- Nakada, M.; Miyamori, H.; Kita, D.; Takahashi, T.; Yamashita, J.; Sato, H.; Miura, R.; Yamaguchi, Y.; Okada, Y. Human glioblastomas overexpress ADAMTS-5 that degrades brevican. Acta Neuropathol. 2005, 110, 239–246. [Google Scholar] [CrossRef]
- Cross, N.; Chandrasekharan, S.; Jokonya, N.; Fowles, A.; Hamdy, F.; Buttle, D.; Eaton, C. The expression and regulation of ADAMTS-1, -4, -5, -9, and -15, and TIMP-3 by TGFbeta1 in prostate cells: Relevance to the accumulation of versican. Prostate 2005, 63, 269–275. [Google Scholar] [CrossRef]
- Glasson, S.; Askew, R.; Sheppard, B.; Carito, B.; Blanchet, T.; Ma, H.-L.; Flannery, C.; Peluso, D.; Kanki, K.; Yang, Z.; et al. Deletion of active ADAMTS5 prevents cartilage degradation in a murine model of osteoarthritis. Nature 2005, 434, 644–652. [Google Scholar]
- Stanton, H.; Rogerson, F.; East, C.; Golub, S.; Lawlor, K.; Meeker, C.; Little, C.; Last, K.; Farmer, P.; Campbell, I.; et al. ADAMTS5 is the major aggrecanase in mouse cartilage in vivo and in vitro. Nature 2005, 434, 648–700. [Google Scholar] [CrossRef]
- McCulloch, D.; Nelson, C.; Dixon, L.; Silver, D.; Wylie, J.; Lindner, V.; Sasaki, T.; Cooley, M.; Argraves, W.; Apte, S. ADAMTS metalloproteases generate active versican fragments that regulate interdigital web regression. Dev. Cell 2009, 17, 687–785. [Google Scholar]
- Velasco, J.; Li, J.; DiPietro, L.; Stepp, M.; Sandy, J.; Plaas, A. Adamts5 deletion blocks murine dermal repair through CD44-mediated aggrecan accumulation and modulation of transforming growth factor β1 (TGFβ1) signaling. J. Biol. Chem. 2011, 286, 26016–26043. [Google Scholar]
- Majumdar, M.; Askew, R.; Schelling, S.; Stedman, N.; Blanchet, T.; Hopkins, B.; Morris, E.; Glasson, S. Double-knockout of ADAMTS-4 and ADAMTS-5 in mice results in physiologically normal animals and prevents the progression of osteoarthritis. Arthritis Rheum. 2007, 56, 3670–3674. [Google Scholar] [CrossRef]
- Ilic, M.; East, C.; Rogerson, F.; Fosang, A.; Handley, C. Distinguishing aggrecan loss from aggrecan proteolysis in ADAMTS-4 and ADAMTS-5 single and double deficient mice. J. Biol. Chem. 2007, 282, 37420–37428. [Google Scholar] [CrossRef]
- Collins-Racie, L.; Flannery, C.; Zeng, W.; Corcoran, C.; Annis-Freeman, B.; Agostino, M.; Arai, M.; DiBlasio-Smith, E.; Dorner, A.; Georgiadis, K.; et al. ADAMTS-8 exhibits aggrecanase activity and is expressed in human articular cartilage. Matrix Biol. 2004, 23, 219–230. [Google Scholar] [CrossRef]
- Somerville, R.; Longpre, J.-M.; Jungers, K.; Engle, J.; Ross, M.; Evanko, S.; Wight, T.; Leduc, R.; Apte, S. Characterization of ADAMTS-9 and ADAMTS-20 as a distinct ADAMTS subfamily related to Caenorhabditis elegans GON-1. J. Biol. Chem. 2003, 278, 9503–9513. [Google Scholar]
- Enomoto, H.; Nelson, C.; Somerville, R.; Mielke, K.; Dixon, L.; Powell, K.; Apte, S. Cooperation of two ADAMTS metalloproteases in closure of the mouse palate identifies a requirement for versican proteolysis in regulating palatal mesenchyme proliferation. Development 2010, 137, 4029–4038. [Google Scholar]
- Koo, B.-H.; Coe, D.; Dixon, L.; Somerville, R.; Nelson, C.; Wang, L.; Young, M.; Lindner, D.; Apte, S. ADAMTS9 is a cell-autonomously acting, anti-angiogenic metalloprotease expressed by microvascular endothelial cells. Am. J. Pathol. 2010, 176, 1494–1504. [Google Scholar] [CrossRef]
- Kern, C.; Wessels, A.; McGarity, J.; Dixon, L.; Alston, E.; Argraves, W.; Geeting, D.; Nelson, C.; Menick, D.; Apte, S. Reduced versican cleavage due to Adamts9 haploinsufficiency is associated with cardiac and aortic anomalies. Matrix Biol. 2010, 29, 304–316. [Google Scholar]
- Liu, C.-J.; Kong, W.; Xu, K.; Luan, Y.; Ilalov, K.; Sehgal, B.; Yu, S.; Howell, R.; di Cesare, P. ADAMTS-12 associates with and degrades cartilage oligomeric matrix protein. J. Biol. Chem. 2006, 281, 15800–15808. [Google Scholar]
- Llamazares, M.; Obaya, A.; Moncada-Pazos, A.; Heljasvaara, R.; Espada, J.; López-Otín, C.; Cal, S. The ADAMTS12 metalloproteinase exhibits anti-tumorigenic properties through modulation of the Ras-dependent ERK signalling pathway. J. Cell Sci. 2007, 120, 3544–3552. [Google Scholar] [CrossRef]
- El Hour, M.; Moncada-Pazos, A.; Blacher, S.; Masset, A.; Cal, S.; Berndt, S.; Detilleux, J.; Host, L.; Obaya, A.; Maillard, C.; et al. Higher sensitivity of Adamts12-deficient mice to tumor growth and angiogenesis. Oncogene 2010, 29, 3025–3032. [Google Scholar] [CrossRef]
- Moncada-Pazos, A.; Obaya, A.; Llamazares, M.; Heljasvaara, R.; Suárez, M.; Colado, E.; Noël, A.; Cal, S.; López-Otin, C. ADAMTS-12 metalloprotease is necessary for normal inflammatory response. J. Biol. Chem. 2012, 287, 39554–39563. [Google Scholar]
- Fujikawa, K.; Suzuki, H.; McMullen, B.; Chung, D. Purification of human von Willebrand factor-cleaving protease and its identification as a new member of the metalloproteinase family. Blood 2001, 98, 1662–1666. [Google Scholar]
- Banno, F.; Kokame, K.; Okuda, T.; Honda, S.; Miyata, S.; Kato, H.; Tomiyama, Y.; Miyata, T. Complete deficiency in ADAMTS13 is prothrombotic, but it alone is not sufficient to cause thrombotic thrombocytopenic purpura. Blood 2006, 107, 3161–3166. [Google Scholar] [CrossRef]
- Motto, D.; Chauhan, A.; Zhu, G.; Homeister, J.; Lamb, C.; Desch, K.; Zhang, W.; Tsai, H.-M.; Wagner, D.; Ginsburg, D. Shigatoxin triggers thrombotic thrombocytopenic purpura in genetically susceptible ADAMTS13-deficient mice. J. Clin. Invest. 2005, 115, 2752–2761. [Google Scholar] [CrossRef]
- Yamaji, N.; Nishimura, K.; Abe, K.; Ohara, O.; Nagase, T.; Nomura, N. Novel metalloprotease having aggrecanase activity. EP1234875A1, 10 November 2000. [Google Scholar]
- Fraser, F.W.; Dancevic, C.M.; Stupka, N.S.; Fosang, A.J.; Rogerson, F.; Ward, A.C.; McCulloch, D.R. The biosynthesis and expression of ADAMTS15; a novel versican-cleaving proteoglycanase, Matrix Biology Society of Australia and New Zealand Annual Meeting, Gold Coast, Australia, 7 September 2012.
- Iruela-Arispe, M.; Carpizo, D.; Luque, A. ADAMTS1: A matrix metalloprotease with angioinhibitory properties. Ann. NY Acad. Sci. 2003, 995, 183–190. [Google Scholar]
- Fu, Y.; Nagy, J.; Brown, L.; Shih, S.-C.; Johnson, P.; Chan, C.; Dvorak, H.; Wight, T. Proteolytic cleavage of versican and involvement of ADAMTS-1 in VEGF-A/VPF-induced pathological angiogenesis. J. Histochem. Cytochem. 2011, 59, 463–473. [Google Scholar] [CrossRef]
- Obika, M.; Ogawa, H.; Takahashi, K.; Li, J.; Hatipoglu, O.; Cilek, M.; Miyoshi, T.; Inagaki, J.; Ohtsuki, T.; Kusachi, S.; et al. Tumor growth inhibitory effect of ADAMTS1 is accompanied by the inhibition of tumor angiogenesis. Cancer Sci. 2012, 103, 1889–1897. [Google Scholar] [Green Version]
- Liu, Y.J.; Xu, Y.; Yu, Q. Full-length ADAMTS-1 and the ADAMTS-1 fragments display pro- and antimetastatic activity, respectively. Oncogene 2006, 25, 2452–2467. [Google Scholar] [CrossRef]
- Nakamura, K.; Hirohata, S.; Murakami, T.; Miyoshi, T.; Demircan, K.; Oohashi, T.; Ogawa, H.; Koten, K.; Toeda, K.; Kusachi, S.; et al. Dynamic induction of ADAMTS1 gene in the early phase of acute myocardial infarction. J. Biochem. 2004, 136, 439–446. [Google Scholar] [CrossRef]
- Gustavsson, H.; Tesan, T.; Jennbacken, K.; Kuno, K.; Damber, J.-E.; Welén, K. ADAMTS1 alters blood vessel morphology and TSP1 levels in LNCaP and LNCaP-19 prostate tumors. BMC Cancer 2010, 10, 288. [Google Scholar] [CrossRef]
- Xu, Z.; Yu, Y.; Duh, E. Vascular endothelial growth factor upregulates expression of ADAMTS1 in endothelial cells through protein kinase C signaling. Invest. Ophthalmol. Vis. Sci. 2006, 47, 4059–4066. [Google Scholar]
- Casal, C.; Torres-Collado, A.; Plaza-Calonge, M.D.C.; Martino-Echarri, E.; Ramón Y Cajal, S.; Rojo, F.; Griffioen, A.; Rodríguez-Manzaneque, J. ADAMTS1 contributes to the acquisition of an endothelial-like phenotype in plastic tumor cells. Cancer Res. 2010, 70, 4676–4686. [Google Scholar]
- Dubail, J.; Kesteloot, F.; Deroanne, C.; Motte, P.; Lambert, V.; Rakic, J.M.; Lapière, C.; Nusgens, B.; Colige, A. ADAMTS-2 functions as anti-angiogenic and anti-tumoral molecule independently of its catalytic activity. Cell. Mol. Life Sci. 2010, 67, 4213–4232. [Google Scholar]
- Rao, N.; Ke, Z.; Liu, H.; Ho, C.-J.; Kumar, S.; Xiang, W.; Zhu, Y.; Ge, R. ADAMTS4 and Its Proteolytic Fragments Differentially Affect Melanoma Growth and Angiogenesis in Mice. Int. J. Cancer 2012. submitted. [Google Scholar]
- Karagiannis, E.D.; Popel, A.S. Anti-angiogenic peptides identified in thrombospondin type I domains. Biochem.Biophys. Res. Commun. 2007, 359, 63–69. [Google Scholar]
- Hsu, Y.P.; Staton, C.A.; Cross, N.; Buttle, D.J. Anti-angiogenic properties of ADAMTS-4 in vitro. Int. J. Exp. Pathol. 2012, 93, 70–77. [Google Scholar] [CrossRef]
- Kumar, S.; Sharghi-Namini, S.; Rao, N.; Ge, R. ADAMTS5 Functions as an Anti-Angiogenic and Anti-Tumorigenic Protein Independent of Its Proteoglycanase Activity. Am. J. Pathol. 2012, 181, 1056–1068. [Google Scholar]
- Sharghi-Namini, S.; Fan, H.; Sulochana, K.; Potturi, P.; Xiang, W.; Chong, Y.-S.; Wang, Z.; Yang, H.; Ge, R. The first but not the second thrombospondin type 1 repeat of ADAMTS5 functions as an angiogenesis inhibitor. Biochem. Biophys. Res. Commun. 2008, 371, 215–219. [Google Scholar] [CrossRef]
- Lo, P.; Lung, H.; Cheung, A.; Apte, S.; Chan, K.; Kwong, F.; Ko, J.; Cheng, Y.; Law, S.; Srivastava, G.; et al. Extracellular protease ADAMTS9 suppresses esophageal and nasopharyngeal carcinoma tumor formation by inhibiting angiogenesis. Cancer Res. 2010, 70, 5567–5576. [Google Scholar] [CrossRef]
- Lee, N.; Sato, M.; Annis, D.; Loo, J.; Wu, L.; Mosher, D.; Iruela-Arispe, M. ADAMTS1 mediates the release of antiangiogenic polypeptides from TSP1 and 2. EMBO J. 2006, 25, 5270–5283. [Google Scholar] [CrossRef]
- Luque, A.; Carpizo, D.R.; Iruela-Arispe, M.L. ADAMTS1/METH1 inhibits endothelial cell proliferation by direct binding and sequestration of VEGF165. J. Biol. Chem. 2003, 278, 23656–23665. [Google Scholar]
- Hatipoglu, O.; Hirohata, S.; Cilek, M.; Ogawa, H.; Miyoshi, T.; Obika, M.; Demircan, K.; Shinohata, R.; Kusachi, S.; Ninomiya, Y. ADAMTS1 is a unique hypoxic early response gene expressed by endothelial cells. J. Biol. Chem. 2009, 284, 16325–16333. [Google Scholar]
- Cilek, M.; Hirohata, S.; Faruk Hatipoglu, O.; Ogawa, H.; Miyoshi, T.; Inagaki, J.; Ohtsuki, T.; Harada, H.; Kamikawa, S.; Kusachi, S.; et al. AHR, a novel acute hypoxia-response sequence, drives reporter gene expression under hypoxia in vitro and in vivo. Cell Biol. Int. 2011, 35, 1–8. [Google Scholar]
- Masui, T.; Hosotani, R.; Tsuji, S.; Miyamoto, Y.; Yasuda, S.; Ida, J.; Nakajima, S.; Kawaguchi, M.; Kobayashi, H.; Koizumi, M.; et al. Expression of METH-1 and METH-2 in pancreatic cancer. Clin. Cancer Res. 2001, 7, 3437–3443. [Google Scholar]
- Choi, J.; Kim, D.; Kim, E.; Chae, M.; Cha, S.; Kim, C.; Jheon, S.; Jung, T.; Park, J. Aberrant methylation of ADAMTS1 in non-small cell lung cancer. Cancer Genet. Cytogenet. 2008, 187, 80–84. [Google Scholar]
- Keightley, M.; Sales, K.; Jabbour, H. PGF2α-F-prostanoid receptor signalling via ADAMTS1 modulates epithelial cell invasion and endothelial cell function in endometrial cancer. BMC Cancer 2010, 10, 488. [Google Scholar]
- Ricciardelli, C.; Frewin, K.; Tan, I.; Williams, E.; Opeskin, K.; Pritchard, M.; Ingman, W.; Russell, D. The ADAMTS1 protease gene is required for mammary tumor growth and metastasis. Am. J. Pathol. 2011, 179, 3075–3085. [Google Scholar]
- Lu, X.; Wang, Q.; Hu, G.; van Poznak, C.; Fleisher, M.; Reiss, M.; Massagué, J.; Kang, Y. ADAMTS1 and MMP1 proteolytically engage EGF-like ligands in an osteolytic signaling cascade for bone metastasis. Genes Dev. 2009, 23, 1882–1894. [Google Scholar]
- Lee, Y.-J.; Koch, M.; Karl, D.; Torres-Collado, A.; Fernando, N.; Rothrock, C.; Kuruppu, D.; Ryeom, S.; Iruela-Arispe, M.; Yoon, S. Variable inhibition of thrombospondin 1 against liver and lung metastases through differential activation of metalloproteinase ADAMTS1. Cancer Res. 2010, 70, 948–956. [Google Scholar]
- Rocks, N.; Paulissen, G.; Quesada-Calvo, F.; Munaut, C.; Gonzalez, M.-L. A.; Gueders, M.; Hacha, J.; Gilles, C.; Foidart, J.-M.; Noel, A.; et al. ADAMTS-1 metalloproteinase promotes tumor development through the induction of a stromal reaction in vivo. Cancer Res. 2008, 68, 9541–9550. [Google Scholar] [CrossRef]
- Tyan, S.-W.; Hsu, C.-H.; Peng, K.-L.; Chen, C.-C.; Kuo, W.-H.; Lee, E.; Shew, J.-Y.; Chang, K.-J.; Juan, L.-J.; Lee, W.-H. Breast cancer cells induce stromal fibroblasts to secrete ADAMTS1 for cancer invasion through an epigenetic change. PLoS One 2012, 7, e35128. [Google Scholar]
- Kuno, K.; Bannai, K.; Hakozaki, M.; Matsushima, K.; Hirose, K. The carboxyl-terminal half region of ADAMTS-1 suppresses both tumorigenicity and experimental tumor metastatic potential. Biochem. Biophys. Res. Commun. 2004, 319, 1327–1333. [Google Scholar]
- Porter, S.; Scott, S.D.; Sassoon, E.M.; Williams, M.R.; Jones, J.L.; Girling, A.C.; Ball, R.Y.; Edwards, D.R. Dysregulated expression of adamalysin-thrombospondin genes in human breast carcinoma. Clin. Cancer Res. 2004, 10, 2429–2440. [Google Scholar]
- Demircan, K.; Gunduz, E.; Gunduz, M.; Beder, L.B.; Hirohata, S.; Nagatsuka, H.; Cengiz, B.; Cilek, M.Z.; Yamanaka, N.; Shimizu, K.; et al. Increased mRNA expression of ADAMTS metalloproteinases in metastatic foci of head and neck cancer. Head Neck 2009, 31, 793–801. [Google Scholar] [CrossRef]
- Held-Feindt, J.; Paredes, E.B.; Blomer, U.; Seidenbecher, C.; Stark, A.M.; Mehdorn, H.M.; Mentlein, R. Matrix-degrading proteases ADAMTS4 and ADAMTS5 (disintegrins and metalloproteinases with thrombospondin motifs 4 and 5) are expressed in human glioblastomas. Int. J. Cancer 2006, 118, 55–61. [Google Scholar]
- Minobe, K.; Ono, R.; Matsumine, A.; Shibata-Minoshima, F.; Izawa, K.; Oki, T.; Kitaura, J.; Iino, T.; Takita, J.; Iwamoto, S.; et al. Expression of ADAMTS4 in Ewing's sarcoma. Int. J. Oncol. 2010, 37, 569–581. [Google Scholar]
- Kim, Y.-H.; Lee, H.; Kim, S.-Y.; Yeom, Y.; Ryu, K.; Min, B.-H.; Kim, D.-H.; Son, H.; Rhee, P.-L.; Kim, J.; et al. Epigenomic analysis of aberrantly methylated genes in colorectal cancer identifies genes commonly affected by epigenetic alterations. Ann. Surg. Oncol. 2011, 18, 2338–2347. [Google Scholar] [CrossRef]
- Heighway, J.; Knapp, T.; Boyce, L.; Brennand, S.; Field, J.; Betticher, D.; Ratschiller, D.; Gugger, M.; Donovan, M.; Lasek, A.; et al. Expression profiling of primary non-small cell lung cancer for target identification. Oncogene 2002, 21, 7749–7763. [Google Scholar] [CrossRef]
- Dunn, J.; Panutsopulos, D.; Shaw, M.; Heighway, J.; Dormer, R.; Salmo, E.; Watson, S.; Field, J.; Liloglou, T. METH-2 silencing and promoter hypermethylation in NSCLC. Br. J. Cancer 2004, 91, 1149–1154. [Google Scholar]
- Dunn, J.; Reed, J.; du Plessis, D.; Shaw, E.; Reeves, P.; Gee, A.; Warnke, P.; Walker, C. Expression of ADAMTS-8, a secreted protease with antiangiogenic properties, is downregulated in brain tumours. Br. J. Cancer 2006, 94, 1186–1193. [Google Scholar] [Green Version]
- Lo, P.; Leung, A.C.; Kwok, C.; Cheung, W.; Ko, J.; Yang, L.; Law, S.; Wang, L.; Li, J.; Stanbridge, E.; et al. Identification of a tumor suppressive critical region mapping to 3p14.2 in esophageal squamous cell carcinoma and studies of a candidate tumor suppressor gene, ADAMTS9. Oncogene 2007, 26, 148–157. [Google Scholar]
- Lung, H.; Lo, P.; Xie, D.; Apte, S.; Cheung, A.; Cheng, Y.; Law, E.; Chua, D.; Zeng, Y.-X.; Tsao, S.; et al. Characterization of a novel epigenetically-silenced, growth-suppressive gene, ADAMTS9, and its association with lymph node metastases in nasopharyngeal carcinoma. Int. J. Cancer 2008, 123, 401–408. [Google Scholar]
- Zhang, C.; Shao, Y.; Zhang, W.; Wu, Q.; Yang, H.; Zhong, Q.; Zhang, J.; Guan, M.; Yu, B.; Wan, J. High-resolution melting analysis of ADAMTS9 methylation levels in gastric, colorectal, and pancreatic cancers. Cancer Genet. Cytogenet. 2010, 196, 38–44. [Google Scholar] [CrossRef]
- Du, W.; Wang, S.; Zhou, Q.; Li, X.; Chu, J.; Chang, Z.; Tao, Q.; Ng, E.; Fang, J.; Sung, J.; et al. ADAMTS9 is a functional tumor suppressor through inhibiting AKT/mTOR pathway and associated with poor survival in gastric cancer. Oncogene 2012. [Google Scholar] [CrossRef]
- Moncada-Pazos, A.; Obaya, A.; Fraga, M.; Viloria, C.; Capellá, G.; Gausachs, M.; Esteller, M.; López-Otín, C.; Cal, S. The ADAMTS12 metalloprotease gene is epigenetically silenced in tumor cells and transcriptionally activated in the stroma during progression of colon cancer. J. Cell. Sci. 2009, 122, 2906–2913. [Google Scholar]
- Oleksowicz, L.; Bhagwati, N.; DeLeon-Fernandez, M. Deficient activity of von Willebrand’s factor-cleaving protease in patients with disseminated malignancies. Cancer Res. 1999, 59, 2244–2250. [Google Scholar]
- Koo, B.-H.; Oh, D.; Chung, S.; Kim, N.; Park, S.; Jang, Y.; Chung, K.-H. Deficiency of von Willebrand factor-cleaving protease activity in the plasma of malignant patients. Thromb. Res. 2002, 105, 471–476. [Google Scholar] [CrossRef]
- Böhm, M.; Gerlach, R.; Beecken, W.-D.; Scheuer, T.; Stier-Brück, I.; Scharrer, I. ADAMTS-13 activity in patients with brain and prostate tumors is mildly reduced, but not correlated to stage of malignancy and metastasis. Thromb. Res. 2003, 111, 33–37. [Google Scholar]
- Viloria, C.; Obaya, A.; Moncada-Pazos, A.; Llamazares, M.; Astudillo, A.; Capellá, G.; Cal, S.; López-Otín, C. Genetic inactivation of ADAMTS15 metalloprotease in human colorectal cancer. Cancer Res. 2009, 69, 4926–4934. [Google Scholar]
- Jones, S.; Zhang, X.; Parsons, D.; Lin, J.; Leary, R.; Angenendt, P.; Mankoo, P.; Carter, H.; Kamiyama, H.; Jimeno, A.; et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 2008, 321, 1801–1806. [Google Scholar]
- Connolly, K.; Gabra, H.; Millwater, C.; Taylor, K.; Rabiasz, G.; Watson, J.; Smyth, J.; Wyllie, A.; Jodrell, D. Identification of a region of frequent loss of heterozygosity at 11q24 in colorectal cancer. Cancer Res. 1999, 59, 2806–2809. [Google Scholar]
- Li, Z.; Zhang, W.; Shao, Y.; Zhang, C.; Wu, Q.; Yang, H.; Wan, X.; Zhang, J.; Guan, M.; Wan, J.; et al. High-resolution melting analysis of ADAMTS18 methylation levels in gastric, colorectal and pancreatic cancers. Med. Oncol. 2010, 27, 998–1004. [Google Scholar]
- Wood, L.D.; Parsons, D.W.; Jones, S.; Lin, J.; Sjoblom, T.; Leary, R.J.; Shen, D.; Boca, S.M.; Barber, T.; Ptak, J.; et al. The genomic landscapes of human breast and colorectal cancers. Science 2007, 318, 1108–1113. [Google Scholar]
- Wei, X.; Prickett, T.D.; Viloria, C.G.; Molinolo, A.; Lin, J.C.; Cardenas-Navia, I.; Cruz, P.; Rosenberg, S.A.; Davies, M.A.; Gershenwald, J.E.; et al. Mutational and functional analysis reveals ADAMTS18 metalloproteinase as a novel driver in melanoma. Mol. Cancer Res. 2010, 8, 1513–1525. [Google Scholar]
- Lapière, C.; Lenaers, A.; Kohn, L. Procollagen peptidase: An enzyme excising the coordination peptides of procollagen. Proc. Natl. Acad. Sci. USA 1971, 68, 3054–3058. [Google Scholar]
- Hojima, Y.; van der Rest, M.; Prockop, D. Type I procollagen carboxyl-terminal proteinase from chick embryo tendons. Purification and characterization. J. Biol. Chem. 1985, 260, 15996–16003. [Google Scholar]
- Hanset, R.; Ansay, M. Dermatosparaxie (peau déchirée) chez le veau: Un défaut général du tissu conjonctif, de nature héréditaire. Ann. Med. Vet. 1967, 7, 451–470. [Google Scholar]
- Smith, L.; Wertelecki, W.; Milstone, L.; Petty, E.; Seashore, M.; Braverman, I.; Jenkins, T.; Byers, P. Human dermatosparaxis: A form of Ehlers-Danlos syndrome that results from failure to remove the amino-terminal propeptide of type I procollagen. Am. J. Hum. Genet. 1992, 51, 235–244. [Google Scholar]
- Nusgens, B.; Verellen-Dumoulin, C.; Hermanns-Lê, T.; de Paepe, A.; Nuytinck, L.; Piérard, G.; Lapière, C. Evidence for a relationship between Ehlers-Danlos type VII C in humans and bovine dermatosparaxis. Nat. Genet. 1992, 1, 214–217. [Google Scholar]
- Colige, A.; Li, S.; Sieron, A.; Nusgens, B.; Prockop, D.; Lapière, C. cDNA cloning and expression of bovine procollagen I N-proteinase: A new member of the superfamily of zinc-metalloproteinases with binding sites for cells and other matrix components. Proc. Natl. Acad. Sci. USA 1997, 94, 2374–2379. [Google Scholar] [CrossRef]
- Nardi, J.; Martos, R.; Walden, K.; Lampe, D.; Robertson, H. Expression of lacunin, a large multidomain extracellular matrix protein, accompanies morphogenesis of epithelial monolayers in Manduca sexta. Insect Biochem. Mol. Biol. 1999, 29, 883–897. [Google Scholar] [CrossRef]
- Colige, A.; Nuytinck, L.; Hausser, I.; van Essen, A.; Thiry, M.; Herens, C.; Adès, L.; Malfait, F.; Paepe, A.; Franck, P.; et al. Novel types of mutation responsible for the dermatosparactic type of Ehlers-Danlos syndrome (Type VIIC) and common polymorphisms in the ADAMTS2 gene. J. Invest. Dermatol. 2004, 123, 656–663. [Google Scholar] [CrossRef]
- De Coster, P.; Cornelissen, M.; de Paepe, A.; Martens, L.; Vral, A. Abnormal dentin structure in two novel gene mutations [COL1A1, Arg134Cys] and [ADAMTS2, Trp795-to-ter] causing rare type I collagen disorders. Arch. Oral Biol. 2007, 52, 101–109. [Google Scholar] [CrossRef]
- Zhou, H.; Hickford, J.; Fang, Q. A premature stop codon in the ADAMTS2 gene is likely to be responsible for dermatosparaxis in Dorper sheep. Anim. Genet. 2012, 43, 471–473. [Google Scholar]
- Tolsma, S.; Volpert, O.; Good, D.; Frazier, W.; Polverini, P.; Bouck, N. Peptides derived from two separate domains of the matrix protein thrombospondin-1 have anti-angiogenic activity. J. Cell Biol. 1993, 122, 497–511. [Google Scholar]
- Arner, E.C.; Pratta, M.A.; Trzaskos, J.M.; Decicco, C.P.; Tortorella, M.D. Generation and characterization of aggrecanase. A soluble, cartilage-derived aggrecan-degrading activity. J. Biol. Chem. 1999, 274, 6594–6601. [Google Scholar]
- Wang, P.; Tortorella, M.; England, K.; Malfait, A.M.; Thomas, G.; Arner, E.C.; Pei, D. Proprotein convertase furin interacts with and cleaves pro-ADAMTS4 (Aggrecanase-1) in the trans-Golgi network. J. Biol. Chem. 2004, 279, 15434–15440. [Google Scholar]
- Flannery, C.R.; Zeng, W.; Corcoran, C.; Collins-Racie, L.A.; Chockalingam, P.S.; Hebert, T.; Mackie, S.A.; McDonagh, T.; Crawford, T.K.; Tomkinson, K.N.; et al. Autocatalytic cleavage of ADAMTS-4 (Aggrecanase-1) reveals multiple glycosaminoglycan-binding sites. J. Biol. Chem. 2002, 277, 42775–42780. [Google Scholar]
- Gao, G.; Plaas, A.; Thompson, V.P.; Jin, S.; Zuo, F.; Sandy, J.D. ADAMTS4 (aggrecanase-1) activation on the cell surface involves C-terminal cleavage by glycosylphosphatidyl inositol-anchored membrane type 4-matrix metalloproteinase and binding of the activated proteinase to chondroitin sulfate and heparan sulfate on syndecan-1. J. Biol. Chem. 2004, 279, 10042–10051. [Google Scholar]
- Boerboom, D.; Lafond, J.-F.; Zheng, X.; Lapointe, E.; Mittaz, L.; Boyer, A.; Pritchard, M.; DeMayo, F.; Mort, J.; Drolet, R.; et al. Partially redundant functions of Adamts1 and Adamts4 in the perinatal development of the renal medulla. Dev. Dyn. 2011, 240, 1806–1814. [Google Scholar]
- Kashiwagi, M.; Tortorella, M.; Nagase, H.; Brew, K. TIMP-3 is a potent inhibitor of aggrecanase 1 (ADAM-TS4) and aggrecanase 2 (ADAM-TS5). J. Biol. Chem. 2001, 276, 12501–12504. [Google Scholar]
- Hashimoto, G.; Aoki, T.; Nakamura, H.; Tanzawa, K.; Okada, Y. Inhibition of ADAMTS4 (aggrecanase-1) by tissue inhibitors of metalloproteinases (TIMP-1, 2, 3 and 4). FEBS Lett. 2001, 494, 192–195. [Google Scholar] [CrossRef]
- Wayne, G.; Deng, S.-J.; Amour, A.; Borman, S.; Matico, R.; Carter, H.; Murphy, G. TIMP-3 inhibition of ADAMTS-4 (Aggrecanase-1) is modulated by interactions between aggrecan and the C-terminal domain of ADAMTS-4. J. Biol. Chem. 2007, 282, 20991–20998. [Google Scholar]
- Pratta, M.A.; Scherle, P.A.; Yang, G.; Liu, R.Q.; Newton, R.C. Induction of aggrecanase 1 (ADAM-TS4) by interleukin-1 occurs through activation of constitutively produced protein. Arthritis Rheum. 2003, 48, 119–133. [Google Scholar]
- Caterson, B.; Flannery, C.R.; Hughes, C.E.; Little, C.B. Mechanisms involved in cartilage proteoglycan catabolism. Matrix Biol. 2000, 19, 333–344. [Google Scholar] [CrossRef]
- Thirunavukkarasu, K.; Pei, Y.; Moore, T.; Wang, H.; Yu, X.-P.; Geiser, A.; Chandrasekhar, S. Regulation of the human ADAMTS-4 promoter by transcription factors and cytokines. Biochem. Biophys. Res. Commun. 2006, 345, 197–204. [Google Scholar] [CrossRef]
- Wågsäter, D.; Björk, H.; Zhu, C.; Björkegren, J.; Valen, G.; Hamsten, A.; Eriksson, P. ADAMTS-4 and -8 are inflammatory regulated enzymes expressed in macrophage-rich areas of human atherosclerotic plaques. Atherosclerosis 2008, 196, 514–522. [Google Scholar] [CrossRef]
- Wainwright, S.; Bondeson, J.; Hughes, C. An alternative spliced transcript of ADAMTS4 is present in human synovium from OA patients. Matrix Biol. 2006, 25, 317–320. [Google Scholar]
- Richards, J.; Hernandez-Gonzalez, I.; Gonzalez-Robayna, I.; Teuling, E.; Lo, Y.; Boerboom, D.; Falender, A.; Doyle, K.; LeBaron, R.; Thompson, V.; et al. Regulated expression of ADAMTS family members in follicles and cumulus oocyte complexes: Evidence for specific and redundant patterns during ovulation. Biol. Reprod. 2005, 72, 1241–1255. [Google Scholar] [CrossRef]
- Hamel, M.; Ajmo, J.; Leonardo, C.; Zuo, F.; Sandy, J.; Gottschall, P. Multimodal signaling by the ADAMTSs (a disintegrin and metalloproteinase with thrombospondin motifs) promotes neurite extension. Exp. Neurol. 2008, 210, 428–440. [Google Scholar]
- Tauchi, R.; Imagama, S.; Natori, T.; Ohgomori, T.; Muramoto, A.; Shinjo, R.; Matsuyama, Y.; Ishiguro, N.; Kadomatsu, K. The endogenous proteoglycan-degrading enzyme ADAMTS-4 promotes functional recovery after spinal cord injury. J. Neuroinflammation 2012, 9, 53. [Google Scholar]
- Tortorella, M.D.; Malfait, A.M.; Deccico, C.; Arner, E. The role of ADAM-TS4 (aggrecanase-1) and ADAM-TS5 (aggrecanase-2) in a model of cartilage degradation. Osteoarthr. Cartil. 2001, 9, 539–552. [Google Scholar]
- Tortorella, M.; Pratta, M.; Liu, R.; Austin, J.; Ross, O.; Abbaszade, I.; Burn, T.; Arner, E. Sites of aggrecan cleavage by recombinant human aggrecanase-1 (ADAMTS-4). J. Biol. Chem. 2000, 275, 18566–18573. [Google Scholar]
- Nagase, H.; Kashiwagi, M. Aggrecanases and cartilage matrix degradation. Arthritis Res. Ther. 2003, 5, 94–103. [Google Scholar]
- Iruela-Arispe, M.L.; Vazquez, F.; Ortega, M.A. Antiangiogenic domains shared by thrombospondins and metallospondins, a new family of angiogenic inhibitors. Ann. NY Acad. Sci. 1999, 886, 58–66. [Google Scholar]
- Kahn, J.; Mehraban, F.; Ingle, G.; Xin, X.; Bryant, J.; Vehar, G.; Schoenfeld, J.; Grimaldi, C.; Peale, F.; Draksharapu, A.; et al. Gene expression profiling in an in vitro model of angiogenesis. Am. J. Pathol. 2000, 156, 1887–1900. [Google Scholar]
- Kintakas, C.; McCulloch, D. Emerging roles for ADAMTS5 during development and disease. Matrix Biol. 2011, 30, 311–318. [Google Scholar]
- Fosang, A.; Rogerson, F.; East, C.J.; Stanton, H. ADAMTS-5: The story so far. Eur. Cell Mater. 2008, 15, 11–26. [Google Scholar]
- Mosyak, L.; Georgiadis, K.; Shane, T.; Svenson, K.; Hebert, T.; McDonagh, T.; Mackie, S.; Olland, S.; Lin, L.; Zhong, X.; et al. Crystal structures of the two major aggrecan degrading enzymes, ADAMTS4 and ADAMTS5. Protein Sci. 2008, 17, 16–37. [Google Scholar] [CrossRef]
- Shieh, H.-S.; Mathis, K.; Williams, J.; Hills, R.; Wiese, J.; Benson, T.; Kiefer, J.; Marino, M.; Carroll, J.; Leone, J.; et al. High resolution crystal structure of the catalytic domain of ADAMTS-5 (aggrecanase-2). J. Biol. Chem. 2008, 283, 1501–1508. [Google Scholar]
- Maingot, L.; Leroux, F.; Landry, V.; Dumont, J.; Nagase, H.; Villoutreix, B.; Sperandio, O.; Deprez-Poulain, R.; Deprez, B. New non-hydroxamic ADAMTS-5 inhibitors based on the 1,2,4-triazole-3-thiol scaffold. Bioorg. Med. Chem. Lett. 2010, 20, 6213–6219. [Google Scholar]
- Longpré, J.-M.; Leduc, R. Identification of prodomain determinants involved in ADAMTS-1 biosynthesis. J. Biol. Chem. 2004, 279, 33237–33282. [Google Scholar]
- Koo, B.-H.; Longpré, J.-M.; Somerville, R.; Alexander, J.; Leduc, R.; Apte, S. Cell-surface processing of pro-ADAMTS9 by furin. J. Biol. Chem. 2006, 281, 12485–12494. [Google Scholar]
- Zeng, W.; Corcoran, C.; Collins-Racie, L.; Lavallie, E.; Morris, E.; Flannery, C. Glycosaminoglycan-binding properties and aggrecanase activities of truncated ADAMTSs: Comparative analyses with ADAMTS-5, -9, -16 and -18. Biochim. Biophys. Acta 2006, 1760, 517–541. [Google Scholar] [CrossRef]
- Gendron, C.; Kashiwagi, M.; Lim, N.; Enghild, J.; Thøgersen, I.; Hughes, C.; Caterson, B.; Nagase, H. Proteolytic activities of human ADAMTS-5: Comparative studies with ADAMTS-4. J. Biol. Chem. 2007, 282, 18294–18600. [Google Scholar]
- McCulloch, D.; Le Goff, C.; Bhatt, S.; Dixon, L.; Sandy, J.; Apte, S. Adamts5, the gene encoding a proteoglycan-degrading metalloprotease, is expressed by specific cell lineages during mouse embryonic development and in adult tissues. Gene Expr. Patterns 2009, 9, 314–337. [Google Scholar] [CrossRef]
- Fosang, A.; Rogerson, F. Identifying the human aggrecanase. Osteoarthr. Cartil. 2010, 18, 1109–1116. [Google Scholar]
- Koshy, P.; Lundy, C.; Rowan, A.; Porter, S.; Edwards, D.; Hogan, A.; Clark, I.; Cawston, T. The modulation of matrix metalloproteinase and ADAM gene expression in human chondrocytes by interleukin-1 and oncostatin M: A time-course study using real-time quantitative reverse transcription-polymerase chain reaction. Arthritis Rheum. 2002, 46, 961–967. [Google Scholar]
- Cortial, D.; Gouttenoire, J.; Rousseau, C.; Ronzière, M.C.; Piccardi, N.; Msika, P.; Herbage, D.; Mallein-Gerin, F.; Freyria, A.M. Activation by IL-1 of bovine articular chondrocytes in culture within a 3D collagen-based scaffold. An in vitro model to address the effect of compounds with therapeutic potential in osteoarthritis. Osteoarthr. Cartil. 2006, 14, 631–640. [Google Scholar]
- Arai, M.; Anderson, D.; Kurdi, Y.; Annis-Freeman, B.; Shields, K.; Collins-Racie, L.; Corcoran, C.; DiBlasio-Smith, E.; Pittman, D.; Dorner, A.; et al. Effect of adenovirus-mediated overexpression of bovine ADAMTS-4 and human ADAMTS-5 in primary bovine articular chondrocyte pellet culture system. Osteoarthr. Cartil. 2004, 12, 599–613. [Google Scholar]
- Bondeson, J.; Lauder, S.; Wainwright, S.; Amos, N.; Evans, A.; Hughes, C.; Feldmann, M.; Caterson, B. Adenoviral gene transfer of the endogenous inhibitor IkappaBalpha into human osteoarthritis synovial fibroblasts demonstrates that several matrix metalloproteinases and aggrecanases are nuclear factor-kappaB-dependent. J. Rheumatol. 2007, 34, 523–533. [Google Scholar]
- Moulharat, N.; Lesur, C.; Thomas, M.; Rolland-Valognes, G.; Pastoureau, P.; Anract, P.; de Ceuninck, F.; Sabatini, M. Effects of transforming growth factor-beta on aggrecanase production and proteoglycan degradation by human chondrocytes in vitro. Osteoarthr. Cartil. 2004, 12, 296–305. [Google Scholar]
- Yamanishi, Y.; Boyle, D.; Clark, M.; Maki, R.; Tortorella, M.; Arner, E.; Firestein, G. Expression and regulation of aggrecanase in arthritis: The role of TGF-beta. J. Immunol. 2002, 168, 1405–1412. [Google Scholar]
- Didangelos, A.; Mayr, U.; Monaco, C.; Mayr, M. Novel role of ADAMTS-5 protein in proteoglycan turnover and lipoprotein retention in atherosclerosis. J. Biol. Chem. 2012, 287, 19341–19346. [Google Scholar]
- Chia, S.-L.; Sawaji, Y.; Burleigh, A.; McLean, C.; Inglis, J.; Saklatvala, J.; Vincent, T. Fibroblast growth factor 2 is an intrinsic chondroprotective agent that suppresses ADAMTS-5 and delays cartilage degradation in murine osteoarthritis. Arthritis Rheum. 2009, 60, 2019–2027. [Google Scholar]
- Yamamoto, K.; Troeberg, L.; Scilabra, S.; Pelosi, M.; Murphy, C.; Strickland, D.; Nagase, H. LRP-1-mediated endocytosis regulates extracellular activity of ADAMTS-5 in articular cartilage. FASEB J. 2012. [Google Scholar] [CrossRef]
- Hattori, N.; Carrino, D.; Lauer, M.; Vasanji, A.; Wylie, J.; Nelson, C.; Apte, S. Pericellular versican regulates the fibroblast-myofibroblast transition: A role for ADAMTS5 protease-mediated proteolysis. J. Biol. Chem. 2011, 286, 34298–34310. [Google Scholar]
- Nissinen, L.; Kähäri, V.-M. ADAMTS5: A New Player in the Vascular Field. Am. J. Pathol. 2012, 181, 743–745. [Google Scholar] [CrossRef]
- Frischknecht, R.; Seidenbecher, C. Brevican: A key proteoglycan in the perisynaptic extracellular matrix of the brain. Int. J. Biochem. Cell Biol. 2012, 44, 1051–1054. [Google Scholar]
- Georgiadis, K.; Hirohata, S.; Seldin, M.; Apte, S. ADAM-TS8, a novel metalloprotease of the ADAM-TS family located on mouse chromosome 9 and human chromosome 11. Genomics 1999, 62, 312–315. [Google Scholar]
- Porter, S.; Span, P.; Sweep, F.; Tjan-Heijnen, V.; Pennington, C.; Pedersen, T.; Johnsen, M.; Lund, L.; Rømer, J.; Edwards, D. ADAMTS8 and ADAMTS15 expression predicts survival in human breast carcinoma. Int. J. Cancer 2006, 118, 1241–1247. [Google Scholar]
- Clark, M.; Kelner, G.; Turbeville, L.; Boyer, A.; Arden, K.; Maki, R. ADAMTS9, a novel member of the ADAM-TS/ metallospondin gene family. Genomics 2000, 67, 343–350. [Google Scholar]
- Blelloch, R.; Kimble, J. Control of organ shape by a secreted metalloprotease in the nematode Caenorhabditis elegans. Nature 1999, 399, 586–590. [Google Scholar]
- Jungers, K.; Le Goff, C.; Somerville, R.; Apte, S. Adamts9 is widely expressed during mouse embryo development. Gene Expr. Patterns 2005, 5, 609–617. [Google Scholar] [CrossRef]
- Demircan, K.; Hirohata, S.; Nishida, K.; Hatipoglu, O.; Oohashi, T.; Yonezawa, T.; Apte, S.; Ninomiya, Y. ADAMTS-9 is synergistically induced by interleukin-1beta and tumor necrosis factor alpha in OUMS-27 chondrosarcoma cells and in human chondrocytes. Arthritis Rheum. 2005, 52, 1451–1460. [Google Scholar]
- Yaykasli, K.; Oohashi, T.; Hirohata, S.; Hatipoglu, O.; Inagawa, K.; Demircan, K.; Ninomiya, Y. ADAMTS9 activation by interleukin 1 beta via NFATc1 in OUMS-27 chondrosarcoma cells and in human chondrocytes. Mol. Cell. Biochem. 2009, 323, 69–79. [Google Scholar] [CrossRef]
- Bevitt, D.; Mohamed, J.; Catterall, J.; Li, Z.; Arris, C.; Hiscott, P.; Sheridan, C.; Langton, K.; Barker, M.; Clarke, M.; McKie, N. Expression of ADAMTS metalloproteinases in the retinal pigment epithelium derived cell line ARPE-19: Transcriptional regulation by TNFalpha. Biochim. Biophys. Acta 2003, 1626, 83–91. [Google Scholar]
- Silver, D.; Hou, L.; Somerville, R.; Young, M.; Apte, S.; Pavan, W. The secreted metalloprotease ADAMTS20 is required for melanoblast survival. PLoS Genet. 2008, 4, e1000003. [Google Scholar]
- Coughlan, T.; Crawford, A.; Goldring, M.; Hatton, P.; Barker, M. Lentiviral shRNA knock-down of ADAMTS-5 and -9 restores matrix deposition in 3D chondrocyte culture. J. Tissue Eng. Regen. Med. 2010, 4, 611–618. [Google Scholar] [CrossRef]
- Cal, S.; Arguelles, J.; Fernandez, P.; López-Otín, C. Identification, characterization, and intracellular processing of ADAM-TS12, a novel human disintegrin with a complex structural organization involving multiple thrombospondin-1 repeats. J. Biol. Chem. 2001, 276, 17932–17940. [Google Scholar]
- Beristain, A.; Zhu, H.; Leung, P. Regulated expression of ADAMTS-12 in human trophoblastic cells: A role for ADAMTS-12 in epithelial cell invasion? PLoS One 2011, 6, e18473. [Google Scholar] [CrossRef]
- Tseng, S.; Reddi, A.; di Cesare, P. Cartilage Oligomeric Matrix Protein (COMP): A Biomarker of Arthritis. Biomark. Insights 2009, 4, 33–44. [Google Scholar]
- Luan, Y.; Kong, L.; Howell, D.; Ilalov, K.; Fajardo, M.; Bai, X.H.; di Cesare, P.; Goldring, M.; Abramson, S.; Liu, C.J. Inhibition of ADAMTS-7 and ADAMTS-12 degradation of cartilage oligomeric matrix protein by alpha-2-macroglobulin. Osteoarthr. Cartil. 2008, 16, 1413–1420. [Google Scholar]
- Guo, F.; Lai, Y.; Tian, Q.; Lin, E.; Kong, L.; Liu, C. Granulin-epithelin precursor binds directly to ADAMTS-7 and ADAMTS-12 and inhibits their degradation of cartilage oligomeric matrix protein. Arthritis Rheum. 2010, 62, 2023–2036. [Google Scholar]
- Denis, C.; Lenting, P. von Willebrand factor: At the crossroads of bleeding and thrombosis. Int. J. Hematol. 2012, 95, 353–361. [Google Scholar]
- Levy, G.; Nichols, W.; Lian, E.; Foroud, T.; McClintick, J.; McGee, B.; Yang, A.; Siemieniak, D.; Stark, K.; Gruppo, R.; et al. Mutations in a member of the ADAMTS gene family cause thrombotic thrombocytopenic purpura. Nature 2001, 413, 488–494. [Google Scholar]
- Sadler, J. Von Willebrand factor, ADAMTS13, and thrombotic thrombocytopenic purpura. Blood 2008, 112, 11–18. [Google Scholar] [CrossRef]
- Zheng, X.; Chung, D.; Takayama, T.; Majerus, E.; Sadler, J.; Fujikawa, K. Structure of von Willebrand factor-cleaving protease (ADAMTS13), a metalloprotease involved in thrombotic thrombocytopenic purpura. J. Biol. Chem. 2001, 276, 41059–41063. [Google Scholar]
- Shomron, N.; Hamasaki-Katagiri, N.; Hunt, R.; Hershko, K.; Pommier, E.; Geetha, S.; Blaisdell, A.; Dobkin, A.; Marple, A.; Roma, I.; et al. A splice variant of ADAMTS13 is expressed in human hepatic stellate cells and cancerous tissues. Thromb. Haemost. 2010, 104, 531–535. [Google Scholar] [CrossRef]
- Majerus, E.; Anderson, P.; Sadler, J. Binding of ADAMTS13 to von Willebrand factor. J. Biol. Chem. 2005, 280, 21773–21778. [Google Scholar]
- Gao, W.; Zhu, J.; Westfield, L.; Tuley, E.; Anderson, P.; Sadler, J. Rearranging Exosites in Noncatalytic Domains Can Redirect the Substrate Specificity of ADAMTS Proteases. J. Biol. Chem. 2012, 287, 26944–26952. [Google Scholar]
- Gerritsen, H.; Robles, R.; Lämmle, B.; Furlan, M. Partial amino acid sequence of purified von Willebrand factor-cleaving protease. Blood 2001, 98, 1654–1661. [Google Scholar]
- Uemura, M.; Tatsumi, K.; Matsumoto, M.; Fujimoto, M.; Matsuyama, T.; Ishikawa, M.; Iwamoto, T.-A.; Mori, T.; Wanaka, A.; Fukui, H.; et al. Localization of ADAMTS13 to the stellate cells of human liver. Blood 2005, 106, 922–924. [Google Scholar]
- Zhou, W.; Inada, M.; Lee, T.-P.; Benten, D.; Lyubsky, S.; Bouhassira, E.; Gupta, S.; Tsai, H.-M. ADAMTS13 is expressed in hepatic stellate cells. Lab. Invest. 2005, 85, 780–788. [Google Scholar]
- Plaimauer, B.; Zimmermann, K.; Völkel, D.; Antoine, G.; Kerschbaumer, R.; Jenab, P.; Furlan, M.; Gerritsen, H.; Lämmle, B.; Schwarz, H.; et al. Cloning, expression, and functional characterization of the von Willebrand factor-cleaving protease (ADAMTS13). Blood 2002, 100, 3626–3632. [Google Scholar] [CrossRef]
- Liu, L.; Choi, H.; Bernardo, A.; Bergeron, A.; Nolasco, L.; Ruan, C.; Moake, J.; Dong, J.F. Platelet-derived VWF-cleaving metalloprotease ADAMTS-13. J. Thromb. Haemost. 2005, 3, 2536–2544. [Google Scholar]
- Tauchi, R.; Imagama, S.; Ohgomori, T.; Natori, T.; Shinjo, R.; Ishiguro, N.; Kadomatsu, K. ADAMTS-13 is produced by glial cells and upregulated after spinal cord injury. Neurosci. Lett. 2012, 517, 1–6. [Google Scholar]
- Turner, N.; Nolasco, L.; Tao, Z.; Dong, J.F.; Moake, J. Human endothelial cells synthesize and release ADAMTS‐13. J. Thromb. Haemost. 2006, 4, 1396–1404. [Google Scholar]
- Ruggeri, Z. The role of von Willebrand factor in thrombus formation. Thromb. Res. 2007, 120, 9. [Google Scholar]
- De Meyer, S.; Savchenko, A.; Haas, M.; Schatzberg, D.; Carroll, M.; Schiviz, A.; Dietrich, B.; Rottensteiner, H.; Scheiflinger, F.; Wagner, D. Protective anti-inflammatory effect of ADAMTS13 on myocardial ischemia/reperfusion injury in mice. Blood 2012. PMID:22915644. [Google Scholar]
- Gandhi, C.; Motto, D.; Jensen, M.; Lentz, S.; Chauhan, A. ADAMTS13 deficiency exacerbates VWF-dependent acute myocardial ischemia/reperfusion injury in mice. Blood 2012. [Google Scholar] [CrossRef]
- Starke, R.; Ferraro, F.; Paschalaki, K.; Dryden, N.; McKinnon, T.; Sutton, R.; Payne, E.; Haskard, D.; Hughes, A.; Cutler, D.; et al. Endothelial von Willebrand factor regulates angiogenesis. Blood 2011, 117, 1071–1080. [Google Scholar]
- Cal, S.; Obaya, A.; Llamazares, M.; Garabaya, C.; Quesada, V.; López-Otín, C. Cloning, expression analysis, and structural characterization of seven novel human ADAMTSs, a family of metalloproteinases with disintegrin and thrombospondin-1 domains. Gene 2002, 283, 49–62. [Google Scholar]
- Molokwu, C.; Adeniji, O.; Chandrasekharan, S.; Hamdy, F.; Buttle, D. Androgen regulates ADAMTS15 gene expression in prostate cancer cells. Cancer Invest. 2010, 28, 698–710. [Google Scholar]
- Sjoblom, T.; Jones, S.; Wood, L.D.; Parsons, D.W.; Lin, J.; Barber, T.D.; Mandelker, D.; Leary, R.J.; Ptak, J.; Silliman, N.; et al. The consensus coding sequences of human breast and colorectal cancers. Science 2006, 314, 268–274. [Google Scholar] [CrossRef]
- Wagstaff, L.; Kelwick, R.; Decock, J.; Pennington, C.; Jaworski, D.; Edwards, D. ADAMTS15 Metalloproteinase Inhibits Breast Cacer Cell Migration. Breast Cancer Res. 2010, 12, P15. [Google Scholar]
- Jin, H.; Wang, X.; Ying, J.; Wong, A.; Li, H.; Lee, K.; Srivastava, G.; Chan, A.; Yeo, W.; Ma, B.; et al. Epigenetic identification of ADAMTS18 as a novel 16q23.1 tumor suppressor frequently silenced in esophageal, nasopharyngeal and multiple other carcinomas. Oncogene 2007, 26, 7490–7498. [Google Scholar]
- Wang, J.; Zhang, W.; Yi, Z.; Wang, S.; Li, Z. Identification of a thrombin cleavage site and a short form of ADAMTS-18. Biochem. Biophys. Res. Commun. 2012, 419, 692–697. [Google Scholar]
- Li, Z.; Nardi, M.A.; Li, Y.S.; Zhang, W.; Pan, R.; Dang, S.; Yee, H.; Quartermain, D.; Jonas, S.; Karpatkin, S. C-terminal ADAMTS-18 fragment induces oxidative platelet fragmentation, dissolves platelet aggregates, and protects against carotid artery occlusion and cerebral stroke. Blood 2009, 113, 6051–6060. [Google Scholar] [CrossRef]
- Aldahmesh, M.A.; Khan, A.O.; Mohamed, J.Y.; Alkuraya, H.; Ahmed, H.; Bobis, S.; Al-Mesfer, S.; Alkuraya, F.S. Identification of ADAMTS18 as a gene mutated in Knobloch syndrome. J. Med. Genet. 2011, 48, 597–601. [Google Scholar]
- Balsara, B.; Pei, J.; de Rienzo, A.; Simon, D.; Tosolini, A.; Lu, Y.; Shen, F.; Fan, X.; Lin, W.; Buetow, K.; et al. Human hepatocellular carcinoma is characterized by a highly consistent pattern of genomic imbalances, including frequent loss of 16q23.1-24.1. Genes Chromosomes Cancer 2001, 30, 245–253. [Google Scholar] [CrossRef]
- Lo, K.; Teo, P.; Hui, A.; To, K.; Tsang, Y.; Chan, S.; Mak, K.; Lee, J.; Huang, D. High resolution allelotype of microdissected primary nasopharyngeal carcinoma. Cancer Res. 2000, 60, 3348–3353. [Google Scholar]
- Mori, Y.; Matsunaga, M.; Abe, T.; Fukushige, S.; Miura, K.; Sunamura, M.; Shiiba, K.; Sato, M.; Nukiwa, T.; Horii, A. Chromosome band 16q24 is frequently deleted in human gastric cancer. Br. J. Cancer 1999, 80, 556–562. [Google Scholar] [CrossRef]
- Riegman, P.; Vissers, K.; Alers, J.; Geelen, E.; Hop, W.; Tilanus, H.; van Dekken, H. Genomic alterations in malignant transformation of Barrett’s esophagus. Cancer Res. 2001, 61, 3164–3170. [Google Scholar]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Kumar, S.; Rao, N.; Ge, R. Emerging Roles of ADAMTSs in Angiogenesis and Cancer. Cancers 2012, 4, 1252-1299. https://doi.org/10.3390/cancers4041252
Kumar S, Rao N, Ge R. Emerging Roles of ADAMTSs in Angiogenesis and Cancer. Cancers. 2012; 4(4):1252-1299. https://doi.org/10.3390/cancers4041252
Chicago/Turabian StyleKumar, Saran, Nithya Rao, and Ruowen Ge. 2012. "Emerging Roles of ADAMTSs in Angiogenesis and Cancer" Cancers 4, no. 4: 1252-1299. https://doi.org/10.3390/cancers4041252