From Pathology to Precision Medicine in Anaplastic Large Cell Lymphoma Expressing Anaplastic Lymphoma Kinase (ALK+ ALCL)
Abstract
:1. Anaplastic Large Cell Lymphoma in Historical Context
2. ALK Biology in the Crosshairs of Medicine
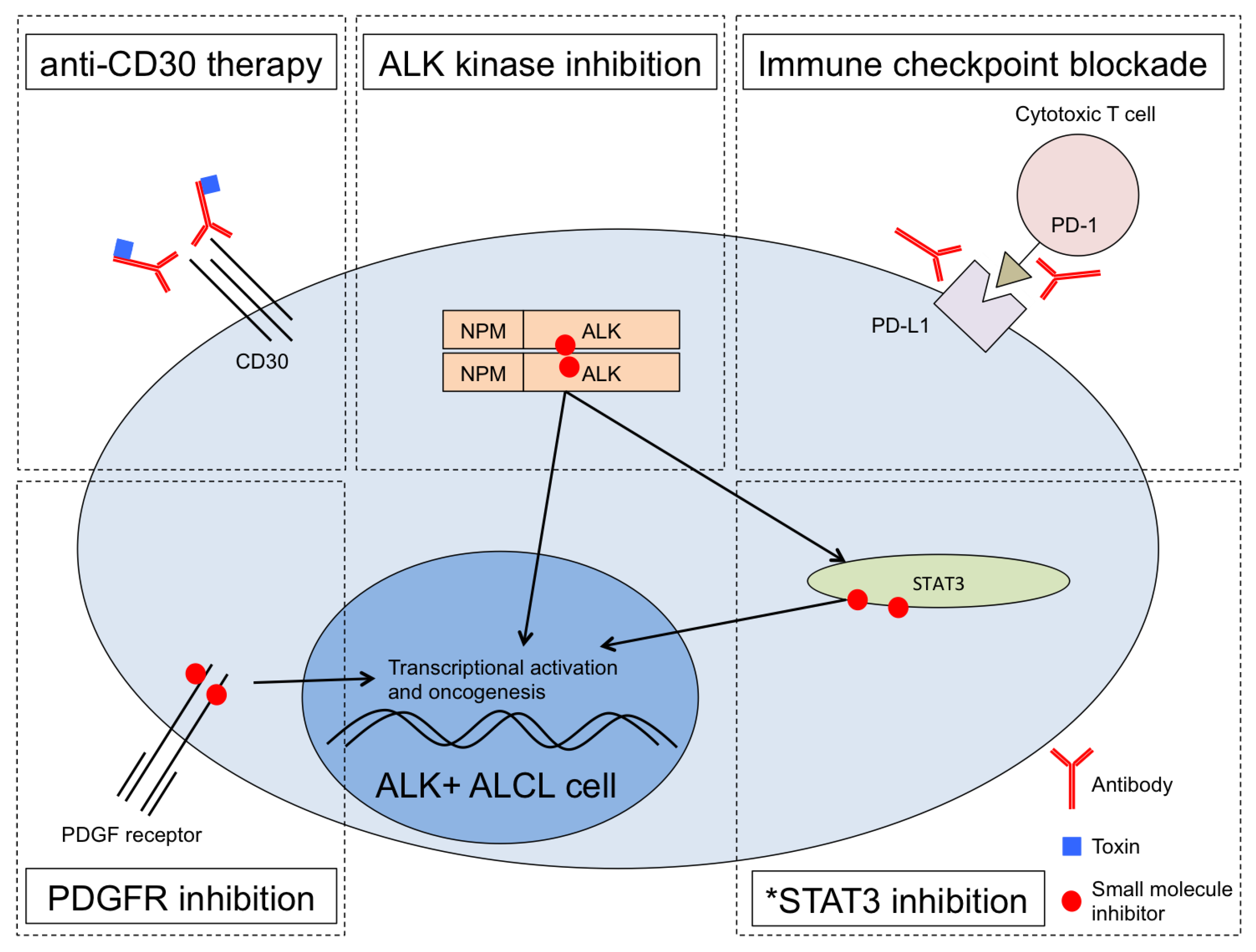
3. Targeting STAT3 in ALK+ ALCL
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Turner, S.D.; Lamant, L.; Kenner, L.; Brugieres, L. Anaplastic large cell lymphoma in paediatric and young adult patients. Br. J. Haematol. 2016, 173, 560–572. [Google Scholar] [CrossRef] [PubMed]
- Schwab, U.; Stein, H.; Gerdes, J.; Lemke, H.; Kirchner, H.; Schaadt, M.; Diehl, V. Production of a monoclonal antibody specific for hodgkin and sternberg-reed cells of Hodgkin’s disease and a subset of normal lymphoid cells. Nature 1982, 299, 65–67. [Google Scholar] [CrossRef] [PubMed]
- Stein, H.M.; Mason, D.Y.; Gerdes, J.; O’connor, N.; Wainscoat, J.; Pallesen, G.; Gatter, K.; Falini, B.; Delsol, G.; Lemke, H.; et al. The expression of the Hodgkin’s disease associated antigen Ki-1 in reactive and neoplastic lymphoid tissue: Evidence that Reed-Sternberg cells and histiocytic malignancies are derived from activated lymphoid cells. Blood 1985, 66, 848–858. [Google Scholar] [PubMed]
- Beverly, P. Activation antigens: New and previously defined clusters. In Leucocyte Typing III; McMichael, A., Beverly, P., Cobbold, S., Eds.; Oxford University Press: Oxford, UK, 1987; Volume 9, p. 516. [Google Scholar]
- Stansfeld, A.G.; Diebold, J.; Kapanci, Y.; Kelényi, G.; Lennert, K.; Mioduszewska, O.; Noel, H.; Rilke, F.; Sundstrom, C.; van Unnik, J.A.M.; et al. Updated kiel classification for lymphomas. Lancet 1988, 331, 292–293. [Google Scholar] [CrossRef]
- Rimokh, R.; Magaud, J.P.; Berger, F.; Samarut, J.; Coiffier, B.; Germain, D.; Mason, D.Y. A translocation involving a specific breakpoint (q35) on chromosome 5 is characteristic of anaplastic large cell lymphoma (‘Ki-1 lymphoma’). Br. J. Haematol. 1989, 71, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Morris, S.; Kirstein, M.; Valentine, M.; Dittmer, K.; Shapiro, D.; Saltman, D.; Look, A. Fusion of a kinase gene, alk, to a nucleolar protein gene, npm, in non-Hodgkin’s lymphoma. Science 1994, 263, 1281–1284. [Google Scholar] [CrossRef] [PubMed]
- Morris, S.W.; Naeve, C.; Mathew, P.; James, P.L.; Kirstein, M.N.; Cui, X.; Witte, D.P. ALK, the chromosome 2 gene locus altered by the t(2;5) in non-Hodgkin’s lymphoma, encodes a novel neural receptor tyrosine kinase that is highly related to leukocyte tyrosine kinase (LTK). Oncogene 1997, 14, 2175–2188. [Google Scholar] [CrossRef] [PubMed]
- Iwahara, T.; Fujimoto, J.; Wen, D.; Cupples, R.; Bucay, N.; Arakawa, T.; Mori, S.; Ratzkin, B.; Yamamoto, T. Molecular characterization of ALK, a receptor tyrosine kinase expressed specifically in the nervous system. Oncogene 1997, 14, 439–449. [Google Scholar] [CrossRef] [PubMed]
- Shiota, M.; Fujimoto, J.; Semba, T.; Satoh, H.; Yamamoto, T.; Mori, S. Hyperphosphorylation of a novel 80 kDa protein-tyrosine kinase similar to Ltk in a human Ki-1 lymphoma cell line, AMS3. Oncogene 1994, 9, 1567–1574. [Google Scholar] [PubMed]
- Fujimoto, J.; Shiota, M.; Iwahara, T.; Seki, N.; Satoh, H.; Mori, S.; Yamamoto, T. Characterization of the transforming activity of p80, a hyperphosphorylated protein in a ki-1 lymphoma cell line with chromosomal translocation t(2;5). Proc. Natl. Acad. Sci. USA 1996, 93, 4181–4186. [Google Scholar] [CrossRef] [PubMed]
- Bischof, D.; Pulford, K.; Mason, D.Y.; Morris, S.W. Role of the nucleophosmin (NPM) portion of the non-Hodgkin’s lymphoma-associated npm-anaplastic lymphoma kinase fusion protein in oncogenesis. Mol. Cell. Biol. 1997, 17, 2312–2325. [Google Scholar] [CrossRef] [PubMed]
- Wasik, M.A.; Zhang, Q.; Marzec, M.; Kasprzycka, M.; Wang, H.Y.; Liu, X. Anaplastic lymphoma kinase (ALK)-induced malignancies: Novel mechanisms of cell transformation and potential therapeutic approaches. Semin. Oncol. 2009, 36, S27–S35. [Google Scholar] [CrossRef] [PubMed]
- Chiarle, R.; Voena, C.; Ambrogio, C.; Piva, R.; Inghirami, G. The anaplastic lymphoma kinase in the pathogenesis of cancer. Nat. Rev. Cancer 2008, 8, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Swerdlow, S.H.; Campo, E.; Harris, N.L.; Jaffe, E.S.; Pileri, S.A.; Stein, H.; Thiele, J.; Vardiman, J.W. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues; International Agency for Research on Cancer: Lyon, France, 2008. [Google Scholar]
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016, 127, 2375–2390. [Google Scholar] [CrossRef] [PubMed]
- Mathas, S.; Kreher, S.; Meaburn, K.J.; Johrens, K.; Lamprecht, B.; Assaf, C.; Sterry, W.; Kadin, M.E.; Daibata, M.; Joos, S.; et al. Gene deregulation and spatial genome reorganization near breakpoints prior to formation of translocations in anaplastic large cell lymphoma. Proc. Natl. Acad. Sci. USA 2009, 106, 5831–5836. [Google Scholar] [CrossRef] [PubMed]
- Feldman, A.L.; Law, M.; Remstein, E.D.; Macon, W.R.; Erickson, L.A.; Grogg, K.L.; Kurtin, P.J.; Dogan, A. Recurrent translocations involving the IRF4 oncogene locus in peripheral T-cell lymphomas. Leukemia 2008, 23, 574–580. [Google Scholar] [CrossRef] [PubMed]
- Feldman, A.L.; Dogan, A.; Smith, D.I.; Law, M.E.; Ansell, S.M.; Johnson, S.H.; Porcher, J.C.; Ozsan, N.; Wieben, E.D.; Eckloff, B.W.; et al. Discovery of recurrent t(6;7)(p25.3;q32.3) translocations in ALK-negative anaplastic large cell lymphomas by massively parallel genomic sequencing. Blood 2011, 117, 915–919. [Google Scholar] [CrossRef] [PubMed]
- Vasmatzis, G.; Johnson, S.H.; Knudson, R.A.; Ketterling, R.P.; Braggio, E.; Fonseca, R.; Viswanatha, D.S.; Law, M.E.; Kip, N.S.; Ozsan, N.; et al. Genome-wide analysis reveals recurrent structural abnormalities of TP63 and other p53-related genes in peripheral T-cell lymphomas. Blood 2012, 120, 2280–2289. [Google Scholar] [CrossRef] [PubMed]
- Crescenzo, R.; Abate, F.; Lasorsa, E.; Tabbo’, F.; Gaudiano, M.; Chiesa, N.; Di Giacomo, F.; Spaccarotella, E.; Barbarossa, L.; Ercole, E.; et al. Convergent mutations and kinase fusions lead to oncogenic STAT3 activation in anaplastic large cell lymphoma. Cancer Cell 2015, 27, 516–532. [Google Scholar] [CrossRef] [PubMed]
- Hsu, F.Y.-Y.; Johnston, P.B.; Burke, K.A.; Zhao, Y. The expression of CD30 in anaplastic large cell lymphoma is regulated by nucleophosmin-anaplastic lymphoma kinase-mediated junb level in a cell type-specific manner. Cancer Res. 2006, 66, 9002–9008. [Google Scholar] [CrossRef] [PubMed]
- Horwitz, S.M.; Shustov, A.R.; Forero-Torres, A.; Bartlett, N.L.; Advani, R.; Pro, B.; Chen, R.; Davies, A.J.; Illidge, T.; Huebner, D.; et al. Frontline treatment of CD30+ Peripheral T-cell lymphomas with brentuximab vedotin in combination with CHP: 3-year durability and survival follow-up. Blood 2015, 126, 1537. [Google Scholar]
- Werner, M.T.; Zhao, C.; Zhang, Q.; Wasik, M.A. Nucleophosmin-anaplastic lymphoma kinase: The ultimate oncogene and therapeutic target. Blood 2016, 129, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Raghunath, P.N.; Xue, L.; Majewski, M.; Carpentieri, D.F.; Odum, N.; Morris, S.; Skorski, T.; Wasik, M.A. Multilevel dysregulation of STAT3 activation in anaplastic lymphoma kinase-positive t/null-cell lymphoma. J. Immunol. 2002, 168, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Chiarle, R.; Simmons, W.J.; Cai, H.; Dhall, G.; Zamo, A.; Raz, R.; Karras, J.G.; Levy, D.E.; Inghirami, G. Stat3 is required for ALK-mediated lymphomagenesis and provides a possible therapeutic target. Nat. Med. 2005, 11, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Peters, S.; Camidge, D.R.; Shaw, A.T.; Gadgeel, S.; Ahn, J.S.; Kim, D.-W.; Ou, S.-H.I.; Pérol, M.; Dziadziuszko, R.; Rosell, R.; et al. Alectinib versus crizotinib in untreated ALK-positive non-small-cell lung cancer. N. Engl. J. Med. 2017. [Google Scholar] [CrossRef] [PubMed]
- Gambacorti-Passerini, C.; Messa, C.; Pogliani, E.M. Crizotinib in anaplastic large-cell lymphoma. N. Engl. J. Med. 2011, 364, 775–776. [Google Scholar] [CrossRef] [PubMed]
- Mossé, Y.P.; Lim, M.S.; Voss, S.D.; Wilner, K.; Ruffner, K.; Laliberte, J.; Rolland, D.; Balis, F.M.; Maris, J.M.; Weigel, B.J.; et al. Safety and activity of crizotinib for paediatric patients with refractory solid tumours or anaplastic large-cell lymphoma: A children’s oncology group phase 1 consortium study. Lancet Oncol. 2013, 14, 472–480. [Google Scholar] [CrossRef]
- Gambacorti Passerini, C.; Farina, F.; Stasia, A.; Redaelli, S.; Ceccon, M.; Mologni, L.; Messa, C.; Guerra, L.; Giudici, G.; Sala, E.; et al. Crizotinib in advanced, chemoresistant anaplastic lymphoma kinase-positive lymphoma patients. J. Natl. Cancer Inst. 2014, 106. [Google Scholar] [CrossRef] [PubMed]
- Mosse, Y.P.; Voss, S.D.; Lim, M.S.; Rolland, D.; Minard, C.G.; Fox, E.; Adamson, P.; Wilner, K.; Blaney, S.M.; Weigel, B.J. Targeting ALK with crizotinib in pediatric anaplastic large cell lymphoma and inflammatory myofibroblastic tumor: A children’s oncology group study. J. Clin. Oncol. 2017, 35, 3215–3221. [Google Scholar] [CrossRef] [PubMed]
- Zdzalik, D.; Dymek, B.; Grygielewicz, P.; Gunerka, P.; Bujak, A.; Lamparska-Przybysz, M.; Wieczorek, M.; Dzwonek, K. Activating mutations in alk kinase domain confer resistance to structurally unrelated ALK inhibitors in NPM-ALK-positive anaplastic large-cell lymphoma. J. Cancer Res. Clin. Oncol. 2014, 140, 589–598. [Google Scholar] [CrossRef] [PubMed]
- Amin, A.D.; Rajan, S.S.; Liang, W.S.; Pongtornpipat, P.; Groysman, M.J.; Tapia, E.O.; Peters, T.L.; Cuyugan, L.; Adkins, J.; Rimsza, L.M.; et al. Evidence suggesting that discontinuous dosing of ALK kinase inhibitors may prolong control of ALK+ tumors. Cancer Res. 2015, 75, 2916–2927. [Google Scholar] [CrossRef] [PubMed]
- Ceccon, M.; Mologni, L.; Bisson, W.; Scapozza, L.; Gambacorti-Passerini, C. Crizotinib-resistant NPM-ALK mutants confer differential sensitivity to unrelated alk inhibitors. Mol. Cancer Res. 2012, 11, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Hebart, H.; Lang, P.; Woessmann, W. Nivolumab for refractory anaplastic large cell lymphoma: A case report. Ann. Intern. Med. 2016, 165, 607. [Google Scholar] [CrossRef] [PubMed]
- Laimer, D.; Dolznig, H.; Kollmann, K.; Vesely, P.W.; Schlederer, M.; Merkel, O.; Schiefer, A.-I.; Hassler, M.R.; Heider, S.; Amenitsch, L.; et al. PDGFR blockade is a rational and effective therapy for NPM-ALK-driven lymphomas. Nat. Med. 2012, 18, 1699–1704. [Google Scholar] [CrossRef] [PubMed]
- Giuriato, S.; Turner, S.D. Twenty years of modelling NPM-ALK-induced lymphomagenesis. Front. Biosci. (Sch. Ed.) 2014, 7, 236–247. [Google Scholar]
- Murga-Zamalloa, C.A.; Mendoza-Reinoso, V.; Sahasrabuddhe, A.A.; Rolland, D.; Hwang, S.R.; McDonnell, S.R.P.; Sciallis, A.P.; Wilcox, R.A.; Bashur, V.; Elenitoba-Johnson, K.; et al. NPM-ALK phosphorylates WASp Y102 and contributes to oncogenesis of anaplastic large cell lymphoma. Oncogene 2017, 36, 2085–2094. [Google Scholar] [CrossRef] [PubMed]
- Peterson, J.R.; Bickford, L.C.; Morgan, D.; Kim, A.S.; Ouerfelli, O.; Kirschner, M.W.; Rosen, M.K. Chemical inhibition of N-WASP by stabilization of a native autoinhibited conformation. Nat. Struct. Mol. Biol. 2004, 11, 747–755. [Google Scholar] [CrossRef] [PubMed]
- Rolland, D.C.; Basrur, V.; Jeon, Y.K.; McNeil-Schwalm, C.; Fermin, D.; Conlon, K.P.; Zhou, Y.; Ng, S.Y.; Tsou, C.C.; Brown, N.A.; et al. Functional proteogenomics reveals biomarkers and therapeutic targets in lymphomas. Proc. Natl. Acad. Sci. USA 2017, 114, 6581–6586. [Google Scholar] [CrossRef] [PubMed]
- Ohmatsu, H.; Sugaya, M.; Suga, H.; Morimura, S.; Miyagaki, T.; Kai, H.; Kagami, S.; Fujita, H.; Asano, Y.; Tada, Y.; et al. Serum IL-31 levels are increased in patients with cutaneous T-cell lymphoma. Acta Derm. Venereol. 2012, 92, 282–283. [Google Scholar] [CrossRef] [PubMed]
- Singer, E.M.; Shin, D.B.; Nattkemper, L.A.; Benoit, B.M.; Klein, R.S.; Didigu, C.A.; Loren, A.W.; Dentchev, T.; Wysocka, M.; Yosipovitch, G.; et al. IL-31 is produced by the malignant T-cell population in cutaneous T-Cell lymphoma and correlates with CTCL pruritus. J. Invest. Dermatol. 2013, 133, 2783–2785. [Google Scholar] [CrossRef] [PubMed]
- Ferretti, E.; Tripodo, C.; Pagnan, G.; Guarnotta, C.; Marimpietri, D.; Corrias, M.V.; Ribatti, D.; Zupo, S.; Fraternali-Orcioni, G.; Ravetti, J.L.; et al. The interleukin (IL)-31/IL-31R axis contributes to tumor growth in human follicular lymphoma. Leukemia 2015, 29, 958–967. [Google Scholar] [CrossRef] [PubMed]
- Marzec, M.; Halasa, K.; Liu, X.; Wang, H.Y.; Cheng, M.; Baldwin, D.; Tobias, J.W.; Schuster, S.J.; Woetmann, A.; Zhang, Q.; et al. Malignant transformation of CD4+ T lymphocytes mediated by oncogenic kinase NPM/ALK recapitulates IL-2-induced cell signaling and gene expression reprogramming. J. Immunol. 2013, 191, 6200–6207. [Google Scholar] [CrossRef] [PubMed]
- Furtek, S.L.; Backos, D.S.; Matheson, C.J.; Reigan, P. Strategies and approaches of targeting STAT3 for cancer treatment. ACS Chem. Biol. 2016, 11, 308–318. [Google Scholar] [CrossRef] [PubMed]
- Amin, H.M.; Medeiros, L.J.; Ma, Y.; Feretzaki, M.; Das, P.; Leventaki, V.; Rassidakis, G.Z.; O’Connor, S.L.; McDonnell, T.J.; Lai, R.; et al. Inhibition of Jak3 induces apoptosis and decreases anaplastic lymphoma kinase activity in anaplastic large cell lymphoma. Oncogene 2003, 22, 5399–5407. [Google Scholar] [CrossRef] [PubMed]
- Crockett, D.K.; Lin, Z.; Elenitoba-Johnson, K.S.J.; Lim, M.S. Identification of NPM-ALK interacting proteins by tandem mass spectrometry. Oncogene 2004, 23, 2617–2629. [Google Scholar] [CrossRef] [PubMed]
- Lai, R.; Rassidakis, G.; Lin, Q.; Atwell, C.; Medeiros, L.; Amin, H. Jak3 activation is significantly associated with ALK expression in anaplastic large cell lymphoma. Hum. Pathol. 2005, 36, 939–944. [Google Scholar] [CrossRef] [PubMed]
- Zamo, A.; Chiarle, R.; Piva, R.; Howes, J.; Fan, Y.; Chilosi, M.; Levy, D.E.; Inghirami, G. Anaplastic lymphoma kinase (ALK) activates STAT3 and protects hematopoietic cells from cell death. Oncogene 2002, 21, 1038–1047. [Google Scholar] [CrossRef] [PubMed]
- Marzec, M.; Kasprzycka, M.; Ptasznik, A.; Wlodarski, P.; Zhang, Q.; Odum, N.; Wasik, M.A. Inhibition of ALK enzymatic activity in T-cell lymphoma cells induces apoptosis and suppresses proliferation and STAT3 phosphorylation independently of Jak3. Lab. Investig. 2005, 85, 1544–1554. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Werner, M.T.; Zhang, Q.; Wasik, M.A. From Pathology to Precision Medicine in Anaplastic Large Cell Lymphoma Expressing Anaplastic Lymphoma Kinase (ALK+ ALCL). Cancers 2017, 9, 138. https://doi.org/10.3390/cancers9100138
Werner MT, Zhang Q, Wasik MA. From Pathology to Precision Medicine in Anaplastic Large Cell Lymphoma Expressing Anaplastic Lymphoma Kinase (ALK+ ALCL). Cancers. 2017; 9(10):138. https://doi.org/10.3390/cancers9100138
Chicago/Turabian StyleWerner, Michael T., Qian Zhang, and Mariusz A. Wasik. 2017. "From Pathology to Precision Medicine in Anaplastic Large Cell Lymphoma Expressing Anaplastic Lymphoma Kinase (ALK+ ALCL)" Cancers 9, no. 10: 138. https://doi.org/10.3390/cancers9100138





