Roles of microRNAs and RNA-Binding Proteins in the Regulation of Colorectal Cancer Stem Cells
Abstract
:1. Introduction
2. Human Colorectal Cancer Stem Cells
2.1. Shared Molecular Properties between Colorectal CSCs and Normal Intestinal Stem Cells
2.2. Cell Surface Markers for Human Colorectal CSCs
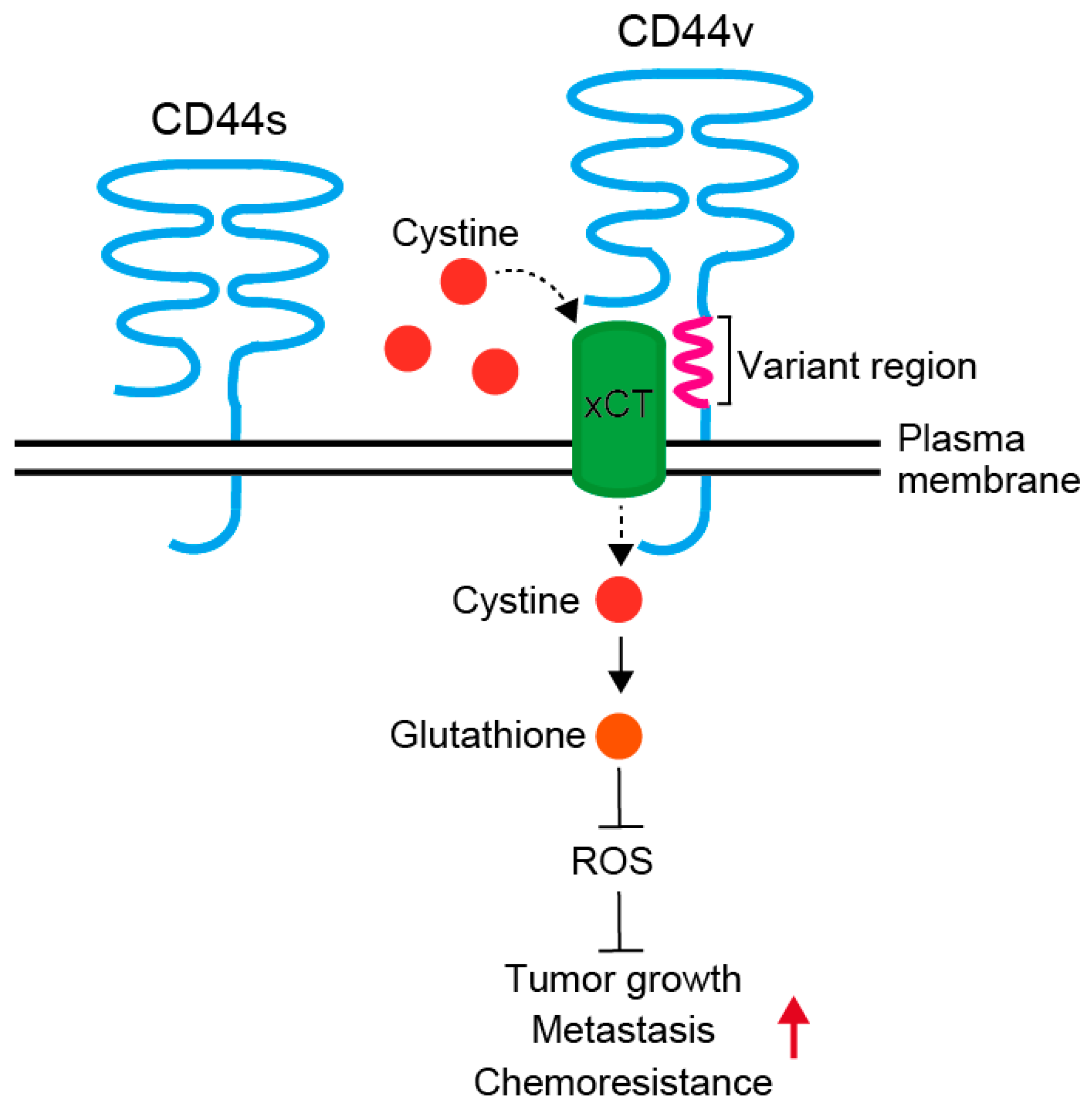
2.2.1. CD44 and Its Variants
2.2.2. CD133
2.2.3. CD166
2.2.4. LGR5
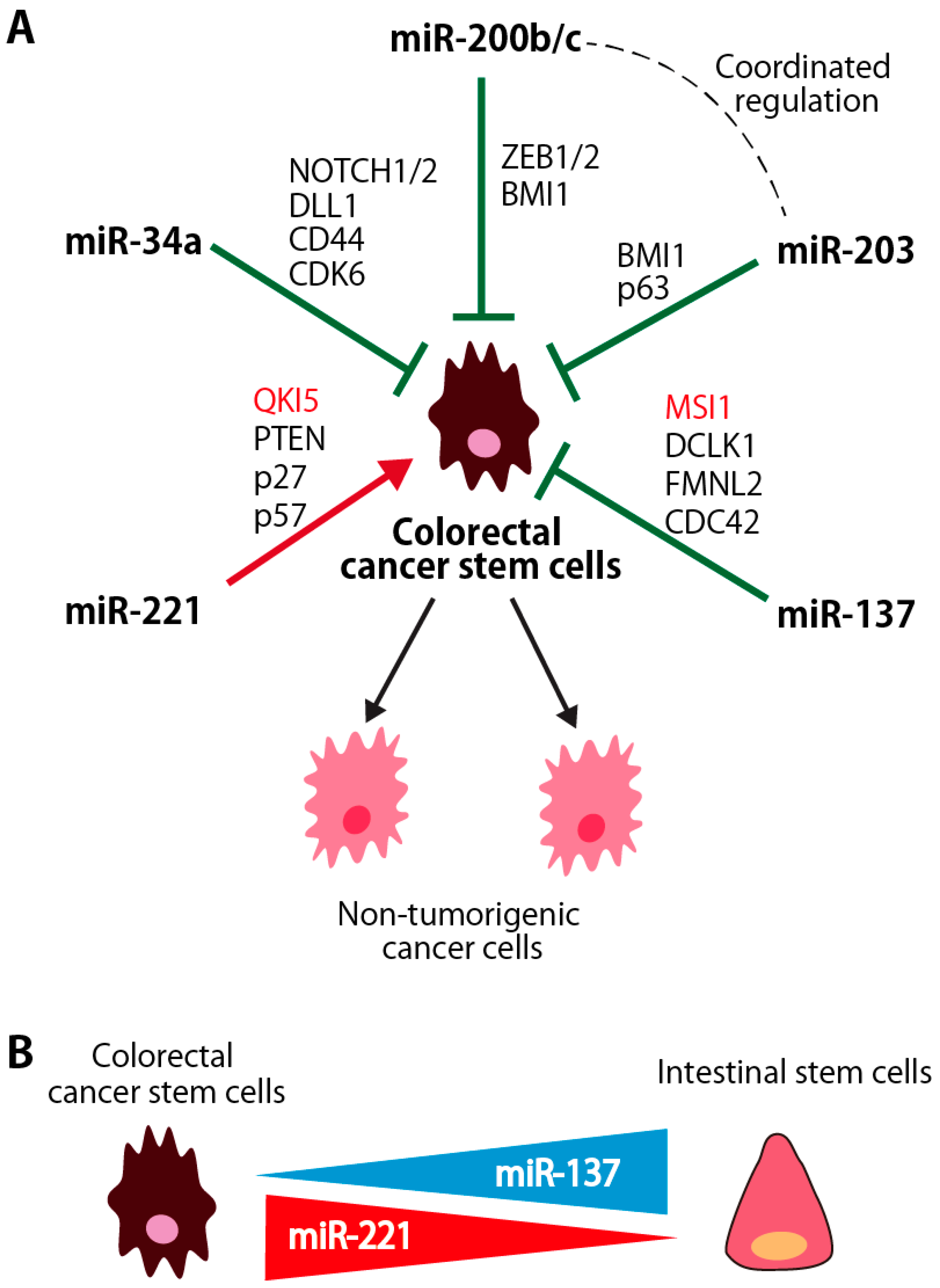
3. miRNAs for Stem Cell Regulation in the Human Colorectal CSCs
3.1. miR-200b/c and miR-203
3.1.1. Coordinated Regulation of miR-200 and miR-203 in Colorectal CSCs
3.1.2. miR-200 Family miRNAs
3.1.3. miR-203
3.1.4. miR-137
3.1.5. miR-221
3.1.6. miR-34a
4. Roles of RNA Binding Proteins in Colorectal CSCs
4.1. LIN28A/B
4.1.1. Molecular Characteristics
4.1.2. Roles in Stem Cell Regulation
4.1.3. Dysregulation in Cancer and CSCs
4.1.4. Molecular Functions for the Regulation of Stem Cells and CSCs
4.2. MSI1/2
4.2.1. Molecular Characteristics
4.2.2. Roles in Stem Cell Regulation
4.2.3. Dysregulation in Cancer and CSCs
4.2.4. Molecular Functions for the Regulation of Stem Cells and CSCs
4.3. QKI-5/6/7/7b
4.3.1. Molecular Characteristics
4.3.2. Roles in Stem Cell Maintenance and Differentiation
4.3.3. Dysregulation in Cancer and CSCs
4.3.4. Molecular Functions for the Regulation of Stem Cells and CSCs
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [PubMed]
- Zeuner, A.; Todaro, M.; Stassi, G.; De Maria, R. Colorectal cancer stem cells: From the crypt to the clinic. Cell Stem Cell 2014, 15, 692–705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Todaro, M.; Alea, M.P.; Di Stefano, A.B.; Cammareri, P.; Vermeulen, L.; Iovino, F.; Tripodo, C.; Russo, A.; Gulotta, G.; Medema, J.P.; et al. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell 2007, 1, 389–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricci-Vitiani, L.; Lombardi, D.G.; Pilozzi, E.; Biffoni, M.; Todaro, M.; Peschle, C.; De Maria, R. Identification and expansion of human colon-cancer-initiating cells. Nature 2007, 445, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Dalerba, P.; Dylla, S.J.; Park, I.K.; Liu, R.; Wang, X.; Cho, R.W.; Hoey, T.; Gurney, A.; Huang, E.H.; Simeone, D.M.; et al. Phenotypic characterization of human colorectal cancer stem cells. Proc. Natl. Acad. Sci. USA 2007, 104, 10158–10163. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, C.A.; Pollett, A.; Gallinger, S.; Dick, J.E. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 2007, 445, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Fearon, E.R.; Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 1990, 61, 759–767. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Shimono, Y.; Zabala, M.; Cho, R.W.; Lobo, N.; Dalerba, P.; Qian, D.; Diehn, M.; Liu, H.; Panula, S.P.; Chiao, E.; et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell 2009, 138, 592–603. [Google Scholar] [CrossRef] [PubMed]
- Isobe, T.; Hisamori, S.; Hogan, D.J.; Zabala, M.; Hendrickson, D.G.; Dalerba, P.; Cai, S.; Scheeren, F.; Kuo, A.H.; Sikandar, S.S.; et al. miR-142 regulates the tumorigenicity of human breast cancer stem cells through the canonical wnt signaling pathway. Elife 2014, 3. [Google Scholar] [CrossRef] [PubMed]
- Glisovic, T.; Bachorik, J.L.; Yong, J.; Dreyfuss, G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008, 582, 1977–1986. [Google Scholar] [CrossRef] [PubMed]
- Kedde, M.; Agami, R. Interplay between microRNAs and RNA-binding proteins determines developmental processes. Cell Cycle 2008, 7, 899–903. [Google Scholar] [CrossRef] [PubMed]
- Kemper, K.; Prasetyanti, P.R.; De Lau, W.; Rodermond, H.; Clevers, H.; Medema, J.P. Monoclonal antibodies against lgr5 identify human colorectal cancer stem cells. Stem Cells 2012, 30, 2378–2386. [Google Scholar] [CrossRef] [PubMed]
- Jung, P.; Sato, T.; Merlos-Suarez, A.; Barriga, F.M.; Iglesias, M.; Rossell, D.; Auer, H.; Gallardo, M.; Blasco, M.A.; Sancho, E.; et al. Isolation and in vitro expansion of human colonic stem cells. Nat. Med. 2011, 17, 1225–1227. [Google Scholar] [CrossRef] [PubMed]
- Huang, E.H.; Hynes, M.J.; Zhang, T.; Ginestier, C.; Dontu, G.; Appelman, H.; Fields, J.Z.; Wicha, M.S.; Boman, B.M. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (sc) and tracks sc overpopulation during colon tumorigenesis. Cancer Res. 2009, 69, 3382–3389. [Google Scholar] [CrossRef] [PubMed]
- Gemei, M.; Mirabelli, P.; Di Noto, R.; Corbo, C.; Iaccarino, A.; Zamboli, A.; Troncone, G.; Galizia, G.; Lieto, E.; Del Vecchio, L.; et al. Cd66c is a novel marker for colorectal cancer stem cell isolation, and its silencing halts tumor growth in vivo. Cancer 2013, 119, 729–738. [Google Scholar] [CrossRef] [PubMed]
- Todaro, M.; Gaggianesi, M.; Catalano, V.; Benfante, A.; Iovino, F.; Biffoni, M.; Apuzzo, T.; Sperduti, I.; Volpe, S.; Cocorullo, G.; et al. Cd44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell Stem Cell 2014, 14, 342–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barker, N. Adult intestinal stem cells: Critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol. 2014, 15, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Dalerba, P.; Kalisky, T.; Sahoo, D.; Rajendran, P.S.; Rothenberg, M.E.; Leyrat, A.A.; Sim, S.; Okamoto, J.; Johnston, D.M.; Qian, D.; et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat. Biotechnol. 2011, 29, 1120–1127. [Google Scholar] [CrossRef] [PubMed]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Heidt, D.G.; Dalerba, P.; Burant, C.F.; Zhang, L.; Adsay, V.; Wicha, M.; Clarke, M.F.; Simeone, D.M. Identification of pancreatic cancer stem cells. Cancer Res. 2007, 67, 1030–1037. [Google Scholar] [CrossRef] [PubMed]
- Collins, A.T.; Berry, P.A.; Hyde, C.; Stower, M.J.; Maitland, N.J. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005, 65, 10946–10951. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Wang, H.; He, L.; Zhang, J.; Ni, B.; Wang, X.; Jin, H.; Cahuzac, N.; Mehrpour, M.; Lu, Y.; et al. Cd44 is of functional importance for colorectal cancer stem cells. Clin. Cancer Res. 2008, 14, 6751–6760. [Google Scholar] [CrossRef] [PubMed]
- Sneath, R.J.; Mangham, D.C. The normal structure and function of cd44 and its role in neoplasia. Mol. Pathol. 1998, 51, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Wielenga, V.J.; Smits, R.; Korinek, V.; Smit, L.; Kielman, M.; Fodde, R.; Clevers, H.; Pals, S.T. Expression of cd44 in apc and tcf mutant mice implies regulation by the wnt pathway. Am. J. Pathol. 1999, 154, 515–523. [Google Scholar] [CrossRef]
- Screaton, G.R.; Bell, M.V.; Jackson, D.G.; Cornelis, F.B.; Gerth, U.; Bell, J.I. Genomic structure of DNA encoding the lymphocyte homing receptor cd44 reveals at least 12 alternatively spliced exons. Proc. Natl. Acad. Sci. USA 1992, 89, 12160–12164. [Google Scholar] [CrossRef] [PubMed]
- Iczkowski, K.A. Cell adhesion molecule CD44: Its functional roles in prostate cancer. Am. J. Transl. Res. 2010, 3, 1–7. [Google Scholar] [PubMed]
- Underhill, C. Cd44: The hyaluronan receptor. J. Cell Sci. 1992, 103, 293–298. [Google Scholar] [PubMed]
- Goodison, S.; Urquidi, V.; Tarin, D. Cd44 cell adhesion molecules. MP 1999, 52, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Cooper, D.L.; Dougherty, G.J. To metastasize or not? Selection of cd44 splice sites. Nat. Med. 1995, 1, 635–637. [Google Scholar] [CrossRef] [PubMed]
- Tolg, C.; Hofmann, M.; Herrlich, P.; Ponta, H. Splicing choice from ten variant exons establishes cd44 variability. Nucleic Acids Res. 1993, 21, 1225–1229. [Google Scholar] [CrossRef] [PubMed]
- Nagano, O.; Okazaki, S.; Saya, H. Redox regulation in stem-like cancer cells by CD44 variant isoforms. Oncogene 2013, 32, 5191–5198. [Google Scholar] [CrossRef] [PubMed]
- Ishimoto, T.; Nagano, O.; Yae, T.; Tamada, M.; Motohara, T.; Oshima, H.; Oshima, M.; Ikeda, T.; Asaba, R.; Yagi, H.; et al. Cd44 variant regulates redox status in cancer cells by stabilizing the xct subunit of system xc(-) and thereby promotes tumor growth. Cancer Cell 2011, 19, 387–400. [Google Scholar] [CrossRef] [PubMed]
- Auvinen, P.; Tammi, R.; Tammi, M.; Johansson, R.; Kosma, V.M. Expression of CD 44 s, CD 44 v3 and CD 44 v6 in benign and malignant breast lesions: Correlation and colocalization with hyaluronan. Histopathology 2005, 47, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Yae, T.; Tsuchihashi, K.; Ishimoto, T.; Motohara, T.; Yoshikawa, M.; Yoshida, G.J.; Wada, T.; Masuko, T.; Mogushi, K.; Tanaka, H.; et al. Alternative splicing of cd44 mRNA by ESRP1 enhances lung colonization of metastatic cancer cell. Nat. Commun. 2012, 3, 883. [Google Scholar] [CrossRef] [PubMed]
- Shmelkov, S.V.; St Clair, R.; Lyden, D.; Rafii, S. Ac133/cd133/prominin-1. Int. J. Biochem. Cell Biol. 2005, 37, 715–719. [Google Scholar] [CrossRef] [PubMed]
- Mak, A.B.; Nixon, A.M.; Kittanakom, S.; Stewart, J.M.; Chen, G.I.; Curak, J.; Gingras, A.C.; Mazitschek, R.; Neel, B.G.; Stagljar, I.; et al. Regulation of cd133 by hdac6 promotes beta-catenin signaling to suppress cancer cell differentiation. Cell Rep. 2012, 2, 951–963. [Google Scholar] [CrossRef] [PubMed]
- Shimozato, O.; Waraya, M.; Nakashima, K.; Souda, H.; Takiguchi, N.; Yamamoto, H.; Takenobu, H.; Uehara, H.; Ikeda, E.; Matsushita, S.; et al. Receptor-type protein tyrosine phosphatase kappa directly dephosphorylates cd133 and regulates downstream akt activation. Oncogene 2015, 34, 1949–1960. [Google Scholar] [CrossRef] [PubMed]
- Yin, A.H.; Miraglia, S.; Zanjani, E.D.; Almeida-Porada, G.; Ogawa, M.; Leary, A.G.; Olweus, J.; Kearney, J.; Buck, D.W. Ac133, a novel marker for human hematopoietic stem and progenitor cells. Blood 1997, 90, 5002–5012. [Google Scholar] [PubMed]
- Miraglia, S.; Godfrey, W.; Yin, A.H.; Atkins, K.; Warnke, R.; Holden, J.T.; Bray, R.A.; Waller, E.K.; Buck, D.W. A novel five-transmembrane hematopoietic stem cell antigen: Isolation, characterization, and molecular cloning. Blood 1997, 90, 5013–5021. [Google Scholar] [PubMed]
- Singh, S.K.; Clarke, I.D.; Terasaki, M.; Bonn, V.E.; Hawkins, C.; Squire, J.; Dirks, P.B. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003, 63, 5821–5828. [Google Scholar] [PubMed]
- Singh, S.K.; Hawkins, C.; Clarke, I.D.; Squire, J.A.; Bayani, J.; Hide, T.; Henkelman, R.M.; Cusimano, M.D.; Dirks, P.B. Identification of human brain tumour initiating cells. Nature 2004, 432, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Suetsugu, A.; Nagaki, M.; Aoki, H.; Motohashi, T.; Kunisada, T.; Moriwaki, H. Characterization of cd133+ hepatocellular carcinoma cells as cancer stem/progenitor cells. Biochem. Biophys. Res. Commun. 2006, 351, 820–824. [Google Scholar] [CrossRef] [PubMed]
- Hermann, P.C.; Huber, S.L.; Herrler, T.; Aicher, A.; Ellwart, J.W.; Guba, M.; Bruns, C.J.; Heeschen, C. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell 2007, 1, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Eramo, A.; Lotti, F.; Sette, G.; Pilozzi, E.; Biffoni, M.; Di Virgilio, A.; Conticello, C.; Ruco, L.; Peschle, C.; De Maria, R. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008, 15, 504–514. [Google Scholar] [CrossRef] [PubMed]
- Ferrandina, G.; Bonanno, G.; Pierelli, L.; Perillo, A.; Procoli, A.; Mariotti, A.; Corallo, M.; Martinelli, E.; Rutella, S.; Paglia, A.; et al. Expression of cd133-1 and cd133-2 in ovarian cancer. Int. J. Gynecol. Cancer 2008, 18, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Uchida, N.; Buck, D.W.; He, D.; Reitsma, M.J.; Masek, M.; Phan, T.V.; Tsukamoto, A.S.; Gage, F.H.; Weissman, I.L. Direct isolation of human central nervous system stem cells. Proc. Natl. Acad. Sci. USA 2000, 97, 14720–14725. [Google Scholar] [CrossRef] [PubMed]
- Sagrinati, C.; Netti, G.S.; Mazzinghi, B.; Lazzeri, E.; Liotta, F.; Frosali, F.; Ronconi, E.; Meini, C.; Gacci, M.; Squecco, R.; et al. Isolation and characterization of multipotent progenitor cells from the bowman’s capsule of adult human kidneys. JASN 2006, 17, 2443–2456. [Google Scholar] [CrossRef] [PubMed]
- Oshima, Y.; Suzuki, A.; Kawashimo, K.; Ishikawa, M.; Ohkohchi, N.; Taniguchi, H. Isolation of mouse pancreatic ductal progenitor cells expressing CD133 and c-MET by flow cytometric cell sorting. Gastroenterology 2007, 132, 720–732. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Gibson, P.; Currle, D.S.; Tong, Y.; Richardson, R.J.; Bayazitov, I.T.; Poppleton, H.; Zakharenko, S.; Ellison, D.W.; Gilbertson, R.J. Prominin 1 marks intestinal stem cells that are susceptible to neoplastic transformation. Nature 2009, 457, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Horst, D.; Scheel, S.K.; Liebmann, S.; Neumann, J.; Maatz, S.; Kirchner, T.; Jung, A. The cancer stem cell marker cd133 has high prognostic impact but unknown functional relevance for the metastasis of human colon cancer. J. Clin. Pathol. 2009, 219, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Flint, A.; Dvorin, E.L.; Bischoff, J. Ac133–2, a novel isoform of human ac133 stem cell antigen. J. Biol. Chem. 2002, 277, 20711–20716. [Google Scholar] [CrossRef] [PubMed]
- Shmelkov, S.V.; Jun, L.; St Clair, R.; McGarrigle, D.; Derderian, C.A.; Usenko, J.K.; Costa, C.; Zhang, F.; Guo, X.; Rafii, S. Alternative promoters regulate transcription of the gene that encodes stem cell surface protein ac133. Blood 2004, 103, 2055–2061. [Google Scholar] [CrossRef] [PubMed]
- LaBarge, M.A.; Bissell, M.J. Is cd133 a marker of metastatic colon cancer stem cells? J. Clin. Investig. 2008, 118, 2021–2024. [Google Scholar] [CrossRef] [PubMed]
- Shmelkov, S.V.; Butler, J.M.; Hooper, A.T.; Hormigo, A.; Kushner, J.; Milde, T.; St Clair, R.; Baljevic, M.; White, I.; Jin, D.K.; et al. Cd133 expression is not restricted to stem cells, and both cd133+ and cd133− metastatic colon cancer cells initiate tumors. J. Clin. Investig. 2008, 118, 2111–2120. [Google Scholar] [CrossRef] [PubMed]
- Kemper, K.; Sprick, M.R.; de Bree, M.; Scopelliti, A.; Vermeulen, L.; Hoek, M.; Zeilstra, J.; Pals, S.T.; Mehmet, H.; Stassi, G.; et al. The ac133 epitope, but not the cd133 protein, is lost upon cancer stem cell differentiation. Cancer Res. 2010, 70, 719–729. [Google Scholar] [CrossRef] [PubMed]
- Patel, D.D.; Wee, S.F.; Whichard, L.P.; Bowen, M.A.; Pesando, J.M.; Aruffo, A.; Haynes, B.F. Identification and characterization of a 100-kd ligand for cd6 on human thymic epithelial cells. J. Exp. Med. 1995, 181, 1563–1568. [Google Scholar] [CrossRef] [PubMed]
- Van Kempen, L.C.; van den Oord, J.J.; van Muijen, G.N.; Weidle, U.H.; Bloemers, H.P.; Swart, G.W. Activated leukocyte cell adhesion molecule/cd166, a marker of tumor progression in primary malignant melanoma of the skin. Am. J. Pathol. 2000, 156, 769–774. [Google Scholar] [CrossRef]
- Kristiansen, G.; Pilarsky, C.; Wissmann, C.; Stephan, C.; Weissbach, L.; Loy, V.; Loening, S.; Dietel, M.; Rosenthal, A. Alcam/cd166 is up-regulated in low-grade prostate cancer and progressively lost in high-grade lesions. Prostate 2003, 54, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Burkhardt, M.; Mayordomo, E.; Winzer, K.J.; Fritzsche, F.; Gansukh, T.; Pahl, S.; Weichert, W.; Denkert, C.; Guski, H.; Dietel, M.; et al. Cytoplasmic overexpression of alcam is prognostic of disease progression in breast cancer. J. Clin. Pathol. 2006, 59, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Mezzanzanica, D.; Fabbi, M.; Bagnoli, M.; Staurengo, S.; Losa, M.; Balladore, E.; Alberti, P.; Lusa, L.; Ditto, A.; Ferrini, S.; et al. Subcellular localization of activated leukocyte cell adhesion molecule is a molecular predictor of survival in ovarian carcinoma patients. Clin. Cancer Res. 2008, 14, 1726–1733. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Shukla, N.K.; Deo, S.V.; Gupta, S.D.; Ralhan, R. Memd/alcam: A potential marker for tumor invasion and nodal metastasis in esophageal squamous cell carcinoma. Oncology 2005, 68, 462–470. [Google Scholar] [CrossRef] [PubMed]
- Weichert, W.; Knosel, T.; Bellach, J.; Dietel, M.; Kristiansen, G. Alcam/cd166 is overexpressed in colorectal carcinoma and correlates with shortened patient survival. J. Clin. Pathol. 2004, 57, 1160–1164. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.C.; Shyh-Chang, N.; Yang, H.; Rai, A.; Umashankar, S.; Ma, S.; Soh, B.S.; Sun, L.L.; Tai, B.C.; Nga, M.E.; et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell 2012, 148, 259–272. [Google Scholar] [CrossRef] [PubMed]
- Rajasekhar, V.K.; Studer, L.; Gerald, W.; Socci, N.D.; Scher, H.I. Tumour-initiating stem-like cells in human prostate cancer exhibit increased nf-kappab signalling. Nat. Commun. 2011, 2, 162. [Google Scholar] [CrossRef] [PubMed]
- Degen, W.G.; van Kempen, L.C.; Gijzen, E.G.; van Groningen, J.J.; van Kooyk, Y.; Bloemers, H.P.; Swart, G.W. Memd, a new cell adhesion molecule in metastasizing human melanoma cell lines, is identical to alcam (activated leukocyte cell adhesion molecule). Am. J. Pathol. 1998, 152, 805–813. [Google Scholar] [PubMed]
- Cayrol, R.; Wosik, K.; Berard, J.L.; Dodelet-Devillers, A.; Ifergan, I.; Kebir, H.; Haqqani, A.S.; Kreymborg, K.; Krug, S.; Moumdjian, R.; et al. Activated leukocyte cell adhesion molecule promotes leukocyte trafficking into the central nervous system. Nat. Immunol. 2008, 9, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Ohneda, O.; Ohneda, K.; Arai, F.; Lee, J.; Miyamoto, T.; Fukushima, Y.; Dowbenko, D.; Lasky, L.A.; Suda, T. Alcam (cd166): Its role in hematopoietic and endothelial development. Blood 2001, 98, 2134–2142. [Google Scholar] [CrossRef] [PubMed]
- Levin, T.G.; Powell, A.E.; Davies, P.S.; Silk, A.D.; Dismuke, A.D.; Anderson, E.C.; Swain, J.R.; Wong, M.H. Characterization of the intestinal cancer stem cell marker cd166 in the human and mouse gastrointestinal tract. Gastroenterology 2010, 139, 2072–2082. [Google Scholar] [CrossRef] [PubMed]
- Smith, N.R.; Davies, P.S.; Levin, T.G.; Gallagher, A.C.; Keene, D.R.; Sengupta, S.K.; Wieghard, N.; El Rassi, E.; Wong, M.H. Cell adhesion molecule cd166/alcam functions within the crypt to orchestrate murine intestinal stem cell homeostasis. Cell. Mol. Gastroenterol. Hepatol. 2017, 3, 389–409. [Google Scholar] [CrossRef] [PubMed]
- Barker, N.; Ridgway, R.A.; van Es, J.H.; van de Wetering, M.; Begthel, H.; van den Born, M.; Danenberg, E.; Clarke, A.R.; Sansom, O.J.; Clevers, H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 2009, 457, 608–611. [Google Scholar] [CrossRef] [PubMed]
- Yui, S.; Nakamura, T.; Sato, T.; Nemoto, Y.; Mizutani, T.; Zheng, X.; Ichinose, S.; Nagaishi, T.; Okamoto, R.; Tsuchiya, K.; et al. Functional engraftment of colon epithelium expanded in vitro from a single adult lgr5(+) stem cell. Nat. Med. 2012, 18, 618–623. [Google Scholar] [CrossRef] [PubMed]
- Van der Flier, L.G.; Sabates-Bellver, J.; Oving, I.; Haegebarth, A.; De Palo, M.; Anti, M.; Van Gijn, M.E.; Suijkerbuijk, S.; Van de Wetering, M.; Marra, G.; et al. The intestinal WNT/TCF signature. Gastroenterology 2007, 132, 628–632. [Google Scholar] [CrossRef] [PubMed]
- De Lau, W.; Barker, N.; Low, T.Y.; Koo, B.K.; Li, V.S.; Teunissen, H.; Kujala, P.; Haegebarth, A.; Peters, P.J.; van de Wetering, M.; et al. Lgr5 homologues associate with WNT receptors and mediate r-spondin signalling. Nature 2011, 476, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Shimokawa, M.; Ohta, Y.; Nishikori, S.; Matano, M.; Takano, A.; Fujii, M.; Date, S.; Sugimoto, S.; Kanai, T.; Sato, T. Visualization and targeting of lgr5+ human colon cancer stem cells. Nature 2017, 545, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Melo, F.S.; Kurtova, A.V.; Harnoss, J.M.; Kljavin, N.; Hoeck, J.D.; Hung, J.; Anderson, J.E.; Storm, E.E.; Modrusan, Z.; Koeppen, H.; et al. A distinct role for lgr5+ stem cells in primary and metastatic colon cancer. Nature 2017, 543, 676–680. [Google Scholar] [CrossRef] [PubMed]
- Cantini, L.; Isella, C.; Petti, C.; Picco, G.; Chiola, S.; Ficarra, E.; Caselle, M.; Medico, E. microRNA-mRNA interactions underlying colorectal cancer molecular subtypes. Nat. Commun. 2015, 6, 8878. [Google Scholar] [CrossRef] [PubMed]
- Shimono, Y.; Mukohyama, J.; Nakamura, S.; Minami, H. microRNA regulation of human breast cancer stem cells. J. Clin. Med. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Brabletz, S.; Brabletz, T. The zeb/mir-200 feedback loop—A motor of cellular plasticity in development and cancer? EMBO Rep. 2010, 11, 670–677. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.I.; Young, R.A.; Sharp, P.A. Super-enhancer-mediated RNA processing revealed by integrative microRNA network analysis. Cell 2017, 168, 1000–1014. [Google Scholar] [CrossRef] [PubMed]
- Humphries, B.; Yang, C. The microRNA-200 family: Small molecules with novel roles in cancer development, progression and therapy. Oncotarget 2015, 6, 6472–6498. [Google Scholar] [CrossRef] [PubMed]
- Wellner, U.; Schubert, J.; Burk, U.C.; Schmalhofer, O.; Zhu, F.; Sonntag, A.; Waldvogel, B.; Vannier, C.; Darling, D.; zur Hausen, A.; et al. The EMT-activator zeb1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat. Cell Biol. 2009, 11, 1487–1495. [Google Scholar] [CrossRef] [PubMed]
- Park, I.K.; Qian, D.; Kiel, M.; Becker, M.W.; Pihalja, M.; Weissman, I.L.; Morrison, S.J.; Clarke, M.F. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 2003, 423, 302–305. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, B.; Li, W.; Fu, L.; Fu, L.; Zhu, Z.; Dong, J.T. Epigenetic silencing of miR-203 upregulates snai2 and contributes to the invasiveness of malignant breast cancer cells. Genes Cancer 2011, 2, 782–791. [Google Scholar] [CrossRef] [PubMed]
- Moes, M.; Le Bechec, A.; Crespo, I.; Laurini, C.; Halavatyi, A.; Vetter, G.; Del Sol, A.; Friederich, E. A novel network integrating a miRNA-203/snai1 feedback loop which regulates epithelial to mesenchymal transition. PLoS ONE 2012, 7, e35440. [Google Scholar] [CrossRef] [PubMed]
- Marisetty, A.L.; Singh, S.K.; Nguyen, T.N.; Coarfa, C.; Liu, B.; Majumder, S. Rest represses mir-124 and mir-203 to regulate distinct oncogenic properties of glioblastoma stem cells. Neuro. Oncol. 2017, 19, 514–523. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.P.; Tang, G.B.; Xu, Y.J.; Zeng, Y.Q.; Zhang, S.F.; Du, H.Z.; Teng, Z.Q.; Liu, C.M. miR-203 interplays with polycomb repressive complexes to regulate the proliferation of neural stem/progenitor cells. Stem Cell Rep. 2017, 9, 190–202. [Google Scholar] [CrossRef] [PubMed]
- Yi, R.; Poy, M.N.; Stoffel, M.; Fuchs, E. A skin microRNA promotes differentiation by repressing ‘stemness’. Nature 2008, 452, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Lena, A.M.; Shalom-Feuerstein, R.; Rivetti di Val Cervo, P.; Aberdam, D.; Knight, R.A.; Melino, G.; Candi, E. Mir-203 represses ‘stemness’ by repressing deltanp63. Cell Death Differ. 2008, 15, 1187–1195. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Jiang, X.; Li, H.; Guo, L.; Jiang, W.; Lu, S.H. Mir-203 inhibits the proliferation and self-renewal of esophageal cancer stem-like cells by suppressing stem renewal factor bmi-1. Stem Cells Dev. 2014, 23, 576–585. [Google Scholar] [CrossRef] [PubMed]
- Viticchie, G.; Lena, A.M.; Latina, A.; Formosa, A.; Gregersen, L.H.; Lund, A.H.; Bernardini, S.; Mauriello, A.; Miano, R.; Spagnoli, L.G.; et al. miR-203 controls proliferation, migration and invasive potential of prostate cancer cell lines. Cell Cycle 2011, 10, 1121–1131. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Park, S.I.; McCauley, L.K.; Wang, C.Y. Signaling between transforming growth factor beta (tgf-beta) and transcription factor snai2 represses expression of microRNA miR-203 to promote epithelial-mesenchymal transition and tumor metastasis. J. Biol. Chem. 2013, 288, 10241–10253. [Google Scholar] [CrossRef] [PubMed]
- Balaguer, F.; Link, A.; Lozano, J.J.; Cuatrecasas, M.; Nagasaka, T.; Boland, C.R.; Goel, A. Epigenetic silencing of miR-137 is an early event in colorectal carcinogenesis. Cancer Res. 2010, 70, 6609–6618. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Chen, X.; Zhang, M.; Fan, Q.; Luo, S.; Cao, X. miR-137 is frequently down-regulated in gastric cancer and is a negative regulator of cdc42. Dig. Dis. Sci. 2011, 56, 2009–2016. [Google Scholar] [CrossRef] [PubMed]
- Silber, J.; Lim, D.A.; Petritsch, C.; Persson, A.I.; Maunakea, A.K.; Yu, M.; Vandenberg, S.R.; Ginzinger, D.G.; James, C.D.; Costello, J.F.; et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008, 6, 14. [Google Scholar] [CrossRef] [PubMed]
- Bemis, L.T.; Chen, R.; Amato, C.M.; Classen, E.H.; Robinson, S.E.; Coffey, D.G.; Erickson, P.F.; Shellman, Y.G.; Robinson, W.A. MicroRNA-137 targets microphthalmia-associated transcription factor in melanoma cell lines. Cancer Res. 2008, 68, 1362–1368. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Li, X.; Zhang, X.; Lv, Z.; He, G.; Zhao, W.; Ren, X.; Li, Y.; Bian, X.; Liao, W.; et al. microRNA-137, an hmga1 target, suppresses colorectal cancer cell invasion and metastasis in mice by directly targeting FMNL2. Gastroenterology 2013, 144, 624–635. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.R.; Marquez, R.T.; Tsao, W.C.; Pathak, S.; Roy, A.; Ping, J.; Wilkerson, B.; Lan, L.; Meng, W.; Neufeld, K.L.; et al. Tumor suppressive microRNA-137 negatively regulates musashi-1 and colorectal cancer progression. Oncotarget 2015, 6, 12558–12573. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Lang, N.; Qiu, M.; Xu, F.; Li, Q.; Tang, Q.; Chen, J.; Chen, X.; Zhang, S.; Liu, Z.; et al. Mir-137 targets cdc42 expression, induces cell cycle g1 arrest and inhibits invasion in colorectal cancer cells. Int. J. Cancer 2011, 128, 1269–1279. [Google Scholar] [CrossRef] [PubMed]
- Jiang, K.; Ren, C.; Nair, V.D. MicroRNA-137 represses klf4 and tbx3 during differentiation of mouse embryonic stem cells. Stem Cell Res. 2013, 11, 1299–1313. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, Y.; Seno, H.; Fukuoka, A.; Ueo, T.; Yamaga, Y.; Maruno, T.; Nakanishi, N.; Kanda, K.; Komekado, H.; Kawada, M.; et al. DCLK1 distinguishes between tumor and normal stem cells in the intestine. Nat. Genet. 2013, 45, 98–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakaguchi, M.; Hisamori, S.; Oshima, N.; Sato, F.; Shimono, Y.; Sakai, Y. Mir-137 regulates the tumorigenicity of colon cancer stem cells through the inhibition of dclk1. Mol. Cancer Res. 2016, 14, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, M.; Quintavalle, C.; Romano, G.; Croce, C.M.; Condorelli, G. miR221/222 in cancer: Their role in tumor progression and response to therapy. Curr. Mol. Med. 2012, 12, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Sun, X.; Wang, M.; Hou, Y.; Zhan, Y.; Jiang, Y.; Liu, Z.; Cao, X.; Chen, P.; Liu, Z.; et al. A microRNA 221- and 222-mediated feedback loop maintains constitutive activation of NFKAPPAB and STAT3 in colorectal cancer cells. Gastroenterology 2014, 147, 847–859. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Li, G.; Fan, C.; Diao, Y.; Wu, B.; Li, J. Increased expression of microRNA-221 in gastric cancer and its clinical significance. J. Int. Med. Res. 2012, 40, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Gramantieri, L.; Fornari, F.; Ferracin, M.; Veronese, A.; Sabbioni, S.; Calin, G.A.; Grazi, G.L.; Croce, C.M.; Bolondi, L.; Negrini, M. microRNA-221 targets bmf in hepatocellular carcinoma and correlates with tumor multifocality. Clin. Cancer Res. 2009, 15, 5073–5081. [Google Scholar] [CrossRef] [PubMed]
- Fornari, F.; Gramantieri, L.; Ferracin, M.; Veronese, A.; Sabbioni, S.; Calin, G.A.; Grazi, G.L.; Giovannini, C.; Croce, C.M.; Bolondi, L.; et al. miR-221 controls cdkn1c/p57 and cdkn1b/p27 expression in human hepatocellular carcinoma. Oncogene 2008, 27, 5651–5661. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Dubaybo, H.; Ali, S.; Goncalves, P.; Kollepara, S.L.; Sethi, S.; Philip, P.A.; Li, Y. Down-regulation of mir-221 inhibits proliferation of pancreatic cancer cells through up-regulation of pten, p27(kip1), p57(kip2), and puma. Am. J. Cancer Res. 2013, 3, 465–477. [Google Scholar] [PubMed]
- Xu, Q.; Li, P.; Chen, X.; Zong, L.; Jiang, Z.; Nan, L.; Lei, J.; Duan, W.; Zhang, D.; Li, X.; et al. Mir-221/222 induces pancreatic cancer progression through the regulation of matrix metalloproteinases. Oncotarget 2015, 6, 14153–14164. [Google Scholar] [CrossRef] [PubMed]
- Miller, T.E.; Ghoshal, K.; Ramaswamy, B.; Roy, S.; Datta, J.; Shapiro, C.L.; Jacob, S.; Majumder, S. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27kip1. J. Biol. Chem. 2008, 283, 29897–29903. [Google Scholar] [CrossRef] [PubMed]
- Quintavalle, C.; Garofalo, M.; Zanca, C.; Romano, G.; Iaboni, M.; del Basso De Caro, M.; Martinez-Montero, J.C.; Incoronato, M.; Nuovo, G.; Croce, C.M.; et al. Mir-221/222 overexpession in human glioblastoma increases invasiveness by targeting the protein phosphate ptpmu. Oncogene 2012, 31, 858–868. [Google Scholar] [CrossRef] [PubMed]
- Tao, K.; Yang, J.; Guo, Z.; Hu, Y.; Sheng, H.; Gao, H.; Yu, H. Prognostic value of miR-221-3p, miR-342-3p and mir-491-5p expression in colon cancer. Am. J. Transl. Res. 2014, 6, 391–401. [Google Scholar] [PubMed]
- Sun, K.; Wang, W.; Zeng, J.J.; Wu, C.T.; Lei, S.T.; Li, G.X. MicroRNA-221 inhibits cdkn1c/p57 expression in human colorectal carcinoma. Acta Pharmacol. Sin. 2011, 32, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, M.; Di Leva, G.; Romano, G.; Nuovo, G.; Suh, S.S.; Ngankeu, A.; Taccioli, C.; Pichiorri, F.; Alder, H.; Secchiero, P.; et al. Mir-221&222 regulate trail resistance and enhance tumorigenicity through pten and timp3 downregulation. Cancer Cell 2009, 16, 498–509. [Google Scholar] [PubMed]
- Zhao, Y.; Zhao, L.; Ischenko, I.; Bao, Q.; Schwarz, B.; Niess, H.; Wang, Y.; Renner, A.; Mysliwietz, J.; Jauch, K.W.; et al. Antisense inhibition of microRNA-21 and microRNA-221 in tumor-initiating stem-like cells modulates tumorigenesis, metastasis, and chemotherapy resistance in pancreatic cancer. Target. Oncol. 2015, 10, 535–548. [Google Scholar] [CrossRef] [PubMed]
- Aldaz, B.; Sagardoy, A.; Nogueira, L.; Guruceaga, E.; Grande, L.; Huse, J.T.; Aznar, M.A.; Diez-Valle, R.; Tejada-Solis, S.; Alonso, M.M.; et al. Involvement of miRNAs in the differentiation of human glioblastoma multiforme stem-like cells. PLoS ONE 2013, 8, e77098. [Google Scholar] [CrossRef] [PubMed]
- Howe, E.N.; Cochrane, D.R.; Richer, J.K. The mir-200 and mir-221/222 microRNA families: Opposing effects on epithelial identity. J. Mammary Gland Biol. Neoplasia 2012, 17, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Hermeking, H. P53 enters the microRNA world. Cancer Cell 2007, 12, 414–418. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Guessous, F.; Zhang, Y.; Dipierro, C.; Kefas, B.; Johnson, E.; Marcinkiewicz, L.; Jiang, J.; Yang, Y.; Schmittgen, T.D.; et al. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 2009, 69, 7569–7576. [Google Scholar] [CrossRef] [PubMed]
- Bommer, G.T.; Gerin, I.; Feng, Y.; Kaczorowski, A.J.; Kuick, R.; Love, R.E.; Zhai, Y.; Giordano, T.J.; Qin, Z.S.; Moore, B.B.; et al. P53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr. Biol. 2007, 17, 1298–1307. [Google Scholar] [CrossRef] [PubMed]
- He, L.; He, X.; Lowe, S.W.; Hannon, G.J. MicroRNAs join the p53 network—Another piece in the tumour-suppression puzzle. Nat. Rev. Cancer 2007, 7, 819–822. [Google Scholar] [CrossRef] [PubMed]
- He, L.; He, X.; Lim, L.P.; de Stanchina, E.; Xuan, Z.; Liang, Y.; Xue, W.; Zender, L.; Magnus, J.; Ridzon, D.; et al. A microRNA component of the p53 tumour suppressor network. Nature 2007, 447, 1130–1134. [Google Scholar] [CrossRef] [PubMed]
- Corney, D.C.; Flesken-Nikitin, A.; Godwin, A.K.; Wang, W.; Nikitin, A.Y. MicroRNA-34b and microRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007, 67, 8433–8438. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Lane, D.; Levine, A.J. Surfing the p53 network. Nature 2000, 408, 307–310. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Kelnar, K.; Liu, B.; Chen, X.; Calhoun-Davis, T.; Li, H.; Patrawala, L.; Yan, H.; Jeter, C.; Honorio, S.; et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat. Med. 2011, 17, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Hermeking, H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010, 17, 193–199. [Google Scholar] [CrossRef] [PubMed]
- De Antonellis, P.; Medaglia, C.; Cusanelli, E.; Andolfo, I.; Liguori, L.; De Vita, G.; Carotenuto, M.; Bello, A.; Formiggini, F.; Galeone, A.; et al. miR-34a targeting of notch ligand delta-like 1 impairs CD15+/CD133+ tumor-propagating cells and supports neural differentiation in medulloblastoma. PLoS ONE 2011, 6, e24584. [Google Scholar] [CrossRef] [PubMed]
- Tarantino, C.; Paolella, G.; Cozzuto, L.; Minopoli, G.; Pastore, L.; Parisi, S.; Russo, T. miRNA 34a, 100, and 137 modulate differentiation of mouse embryonic stem cells. FASEB J. 2010, 24, 3255–3263. [Google Scholar] [CrossRef] [PubMed]
- Bu, P.; Chen, K.Y.; Chen, J.H.; Wang, L.; Walters, J.; Shin, Y.J.; Goerger, J.P.; Sun, J.; Witherspoon, M.; Rakhilin, N.; et al. A microRNA miR-34a-regulated bimodal switch targets notch in colon cancer stem cells. Cell Stem Cell 2013, 12, 602–615. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Hermeking, H. miR-34a and miR-34b/c suppress intestinal tumorigenesis. Cancer Res. 2017, 77, 2746–2758. [Google Scholar] [CrossRef] [PubMed]
- Bouchie, A. First microRNA mimic enters clinic. Nat. Biotechnol. 2013, 31, 577. [Google Scholar] [CrossRef] [PubMed]
- Bader, A.G. miR-34—A microRNA replacement therapy is headed to the clinic. Front. Genet. 2012, 3, 120. [Google Scholar] [CrossRef] [PubMed]
- Keene, J.D. RNA regulons: Coordination of post-transcriptional events. Nat. Rev. Genet. 2007, 8, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Gerstberger, S.; Hafner, M.; Tuschl, T. A census of human RNA-binding proteins. Nat. Rev. Genet. 2014, 15, 829–845. [Google Scholar] [CrossRef] [PubMed]
- Lunde, B.M.; Moore, C.; Varani, G. RNA-binding proteins: Modular design for efficient function. Nat. Rev. Mol. Cell Biol. 2007, 8, 479–490. [Google Scholar] [CrossRef] [PubMed]
- Lukong, K.E.; Chang, K.W.; Khandjian, E.W.; Richard, S. RNA-binding proteins in human genetic disease. Trends Genet. 2008, 24, 416–425. [Google Scholar] [CrossRef] [PubMed]
- Ambros, V.; Horvitz, H.R. Heterochronic mutants of the nematode caenorhabditis elegans. Science 1984, 226, 409–416. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Chen, Y.; Ito, H.; Watanabe, A.; Ge, X.; Kodama, T.; Aburatani, H. Identification and characterization of lin-28 homolog b (lin28b) in human hepatocellular carcinoma. Gene 2006, 384, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, S.R.; Daley, G.Q. Lin28: A microRNA regulator with a macro role. Cell 2010, 140, 445–449. [Google Scholar] [CrossRef] [PubMed]
- Shinoda, G.; Shyh-Chang, N.; Soysa, T.Y.; Zhu, H.; Seligson, M.T.; Shah, S.P.; Abo-Sido, N.; Yabuuchi, A.; Hagan, J.P.; Gregory, R.I.; et al. Fetal deficiency of lin28 programs life-long aberrations in growth and glucose metabolism. Stem Cells 2013, 31, 1563–1573. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007, 318, 1917–1920. [Google Scholar] [CrossRef] [PubMed]
- Hanna, J.; Saha, K.; Pando, B.; van Zon, J.; Lengner, C.J.; Creyghton, M.P.; van Oudenaarden, A.; Jaenisch, R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature 2009, 462, 595–601. [Google Scholar] [CrossRef] [PubMed]
- Gunsalus, K.T.; Wagoner, M.P.; Meyer, K.; Potter, W.B.; Schoenike, B.; Kim, S.; Alexander, C.M.; Friedl, A.; Roopra, A. Induction of the RNA regulator lin28a is required for the growth and pathogenesis of restless breast tumors. Cancer Res. 2012, 72, 3207–3216. [Google Scholar] [CrossRef] [PubMed]
- Hamano, R.; Miyata, H.; Yamasaki, M.; Sugimura, K.; Tanaka, K.; Kurokawa, Y.; Nakajima, K.; Takiguchi, S.; Fujiwara, Y.; Mori, M.; et al. High expression of lin28 is associated with tumour aggressiveness and poor prognosis of patients in oesophagus cancer. Br. J. Cancer 2012, 106, 1415–1423. [Google Scholar] [CrossRef] [PubMed]
- King, C.E.; Cuatrecasas, M.; Castells, A.; Sepulveda, A.R.; Lee, J.S.; Rustgi, A.K. Lin28b promotes colon cancer progression and metastasis. Cancer Res. 2011, 71, 4260–4268. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Lin, X.; Zhong, X.; Kaur, S.; Li, N.; Liang, S.; Lassus, H.; Wang, L.; Katsaros, D.; Montone, K.; et al. Double-negative feedback loop between reprogramming factor lin28 and microRNA let-7 regulates aldehyde dehydrogenase 1-positive cancer stem cells. Cancer Res. 2010, 70, 9463–9472. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Maihle, N.J.; Huang, Y. Pluripotency factors lin28 and oct4 identify a sub-population of stem cell-like cells in ovarian cancer. Oncogene 2010, 29, 2153–2159. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Cao, F.; Bai, L.; Liu, Y.; Xie, J.; Wang, W.; Si, Q.; Yang, J.; Chang, A.; Liu, D.; et al. Ikkbeta enforces a lin28b/tcf7l2 positive feedback loop that promotes cancer cell stemness and metastasis. Cancer Res. 2015, 75, 1725–1735. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.Y.; Wei, T.Z.; Luo, Q.C.; Wu, Q.W.; Liu, Q.F.; Yang, M.; Ye, G.D.; Wu, J.F.; Chen, Y.Y.; Sun, G.B.; et al. The wnt-beta-catenin pathway represses let-7 microRNA expression through transactivation of lin28 to augment breast cancer stem cell expansion. J. Cell Sci. 2013, 126, 2877–2889. [Google Scholar] [CrossRef] [PubMed]
- Lightfoot, H.L.; Miska, E.A.; Balasubramanian, S. Identification of small molecule inhibitors of the lin28-mediated blockage of pre-let-7g processing. Org. Biomol. Chem. 2016, 14, 10208–10216. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Sevignani, C.; Dumitru, C.D.; Hyslop, T.; Noch, E.; Yendamuri, S.; Shimizu, M.; Rattan, S.; Bullrich, F.; Negrini, M.; et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA 2004, 101, 2999–3004. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, S.R.; Daley, G.Q.; Gregory, R.I. Selective blockade of microRNA processing by lin28. Science 2008, 320, 97–100. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, S.R.; Powers, J.T.; Einhorn, W.; Hoshida, Y.; Ng, T.L.; Toffanin, S.; O’Sullivan, M.; Lu, J.; Phillips, L.A.; Lockhart, V.L.; et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat. Genet. 2009, 41, 843–848. [Google Scholar] [CrossRef] [PubMed]
- Iliopoulos, D.; Hirsch, H.A.; Struhl, K. An epigenetic switch involving nf-kappab, lin28, let-7 microRNA, and il6 links inflammation to cell transformation. Cell 2009, 139, 693–706. [Google Scholar] [CrossRef] [PubMed]
- Sampson, V.B.; Rong, N.H.; Han, J.; Yang, Q.; Aris, V.; Soteropoulos, P.; Petrelli, N.J.; Dunn, S.P.; Krueger, L.J. MicroRNA let-7a down-regulates myc and reverts myc-induced growth in burkitt lymphoma cells. Cancer Res. 2007, 67, 9762–9770. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.M.; Grosshans, H.; Shingara, J.; Byrom, M.; Jarvis, R.; Cheng, A.; Labourier, E.; Reinert, K.L.; Brown, D.; Slack, F.J. Ras is regulated by the let-7 microRNA family. Cell 2005, 120, 635–647. [Google Scholar] [CrossRef] [PubMed]
- Mayr, C.; Hemann, M.T.; Bartel, D.P. Disrupting the pairing between let-7 and hmga2 enhances oncogenic transformation. Science 2007, 315, 1576–1579. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Zhao, J.; Hu, W.; Yang, G.; Yu, H.; Wang, R.; Wang, L.; Zhang, G.; Fu, W.; Dai, L.; et al. Disturbance of the let-7/lin28 double-negative feedback loop is associated with radio- and chemo-resistance in non-small cell lung cancer. PLoS ONE 2017, 12, e0172787. [Google Scholar] [CrossRef] [PubMed]
- Qiu, C.; Ma, Y.; Wang, J.; Peng, S.; Huang, Y. Lin28-mediated post-transcriptional regulation of oct4 expression in human embryonic stem cells. Nucleic Acids Res. 2010, 38, 1240–1248. [Google Scholar] [CrossRef] [PubMed]
- Polesskaya, A.; Cuvellier, S.; Naguibneva, I.; Duquet, A.; Moss, E.G.; Harel-Bellan, A. Lin-28 binds igf-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev. 2007, 21, 1125–1138. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Chen, L.L.; Lei, X.X.; Yang, L.; Lin, H.; Carmichael, G.G.; Huang, Y. Genome-wide studies reveal that lin28 enhances the translation of genes important for growth and survival of human embryonic stem cells. Stem Cells 2011, 29, 496–504. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.X.; Xu, J.; Ma, W.; Qiao, C.; Newman, M.A.; Hammond, S.M.; Huang, Y. Determinants of mRNA recognition and translation regulation by lin28. Nucleic Acids Res. 2012, 40, 3574–3584. [Google Scholar] [CrossRef] [PubMed]
- Tsanov, K.M.; Pearson, D.S.; Wu, Z.; Han, A.; Triboulet, R.; Seligson, M.T.; Powers, J.T.; Osborne, J.K.; Kane, S.; Gygi, S.P.; et al. Lin28 phosphorylation by mapk/erk couples signalling to the post-transcriptional control of pluripotency. Nat. Cell Biol. 2017, 19, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Okano, H.; Blendy, J.A.; Montell, C. Musashi, a neural RNA-binding protein required for drosophila adult external sensory organ development. Neuron 1994, 13, 67–81. [Google Scholar] [CrossRef]
- Sakakibara, S.; Nakamura, Y.; Satoh, H.; Okano, H. RNA-binding protein musashi2: Developmentally regulated expression in neural precursor cells and subpopulations of neurons in mammalian CNS. J. Neurosci. 2001, 21, 8091–8107. [Google Scholar] [PubMed]
- Kurihara, Y.; Nagata, T.; Imai, T.; Hiwatashi, A.; Horiuchi, M.; Sakakibara, S.; Katahira, M.; Okano, H.; Uesugi, S. Structural properties and RNA-binding activities of two RNA recognition motifs of a mouse neural RNA-binding protein, mouse-musashi-1. Gene 1997, 186, 21–27. [Google Scholar] [CrossRef]
- Akasaka, Y.; Saikawa, Y.; Fujita, K.; Kubota, T.; Ishikawa, Y.; Fujimoto, A.; Ishii, T.; Okano, H.; Kitajima, M. Expression of a candidate marker for progenitor cells, musashi-1, in the proliferative regions of human antrum and its decreased expression in intestinal metaplasia. Histopathology 2005, 47, 348–356. [Google Scholar] [CrossRef] [PubMed]
- Kayahara, T.; Sawada, M.; Takaishi, S.; Fukui, H.; Seno, H.; Fukuzawa, H.; Suzuki, K.; Hiai, H.; Kageyama, R.; Okano, H.; et al. Candidate markers for stem and early progenitor cells, musashi-1 and hes1, are expressed in crypt base columnar cells of mouse small intestine. FEBS Lett. 2003, 535, 131–135. [Google Scholar] [CrossRef]
- Potten, C.S.; Booth, C.; Tudor, G.L.; Booth, D.; Brady, G.; Hurley, P.; Ashton, G.; Clarke, R.; Sakakibara, S.; Okano, H. Identification of a putative intestinal stem cell and early lineage marker; musashi-1. Differentiation 2003, 71, 28–41. [Google Scholar] [CrossRef] [PubMed]
- Okano, H.; Kawahara, H.; Toriya, M.; Nakao, K.; Shibata, S.; Imai, T. Function of RNA-binding protein musashi-1 in stem cells. Exp. Cell Res. 2005, 306, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama-Nakagiri, Y.; Akiyama, M.; Shibata, S.; Okano, H.; Shimizu, H. Expression of RNA-binding protein musashi in hair follicle development and hair cycle progression. Am. J. Pathol. 2006, 168, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Sakakibara, S.; Imai, T.; Hamaguchi, K.; Okabe, M.; Aruga, J.; Nakajima, K.; Yasutomi, D.; Nagata, T.; Kurihara, Y.; Uesugi, S.; et al. Mouse-musashi-1, a neural RNA-binding protein highly enriched in the mammalian cns stem cell. Dev. Biol. 1996, 176, 230–242. [Google Scholar] [CrossRef] [PubMed]
- Kharas, M.G.; Lengner, C.J.; Al-Shahrour, F.; Bullinger, L.; Ball, B.; Zaidi, S.; Morgan, K.; Tam, W.; Paktinat, M.; Okabe, R.; et al. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat. Med. 2010, 16, 903–908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kudinov, A.E.; Karanicolas, J.; Golemis, E.A.; Boumber, Y. Musashi RNA-binding proteins as cancer drivers and novel therapeutic targets. Clin. Cancer Res. 2017, 23, 2143–2153. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Peng, X.; Yan, D.; Tang, H.; Huang, F.; Yang, Y.; Peng, Z. Msi-1 is a predictor of survival and a novel therapeutic target in colon cancer. Ann. Surg. Oncol. 2011, 18, 2074–2083. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.F.; Dong, W.G.; Jiang, C.Q.; Xia, D.; Liao, F.; Yu, Q.F. Expression of putative stem cell genes musashi-1 and beta1-integrin in human colorectal adenomas and adenocarcinomas. Int. J. Colorectal Dis. 2010, 25, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Bobryshev, Y.V.; Freeman, A.K.; Botelho, N.K.; Tran, D.; Levert-Mignon, A.J.; Lord, R.V. Expression of the putative stem cell marker musashi-1 in barrett’s esophagus and esophageal adenocarcinoma. Dis. Esophagus 2010, 23, 580–589. [Google Scholar] [CrossRef] [PubMed]
- Nikpour, P.; Baygi, M.E.; Steinhoff, C.; Hader, C.; Luca, A.C.; Mowla, S.J.; Schulz, W.A. The RNA binding protein musashi1 regulates apoptosis, gene expression and stress granule formation in urothelial carcinoma cells. J. Cell. Mol. Med. 2011, 15, 1210–1224. [Google Scholar] [CrossRef] [PubMed]
- Moreira, A.L.; Gonen, M.; Rekhtman, N.; Downey, R.J. Progenitor stem cell marker expression by pulmonary carcinomas. Mod. Pathol. 2010, 23, 889–895. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Y.; Penalva, L.O.; Yuan, H.; Linnoila, R.I.; Lu, J.; Okano, H.; Glazer, R.I. Musashi1 regulates breast tumor cell proliferation and is a prognostic indicator of poor survival. Mol. Cancer 2010, 9, 221. [Google Scholar] [CrossRef] [PubMed]
- Ye, F.; Zhou, C.; Cheng, Q.; Shen, J.; Chen, H. Stem-cell-abundant proteins nanog, nucleostemin and musashi1 are highly expressed in malignant cervical epithelial cells. BMC Cancer 2008, 8, 108. [Google Scholar] [CrossRef] [PubMed]
- Gotte, M.; Wolf, M.; Staebler, A.; Buchweitz, O.; Kelsch, R.; Schuring, A.N.; Kiesel, L. Increased expression of the adult stem cell marker musashi-1 in endometriosis and endometrial carcinoma. J. Pathol. 2008, 215, 317–329. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Diaz, P.C.; Burton, T.L.; Burns, S.C.; Hung, J.Y.; Penalva, L.O. Musashi1 modulates cell proliferation genes in the medulloblastoma cell line daoy. BMC Cancer 2008, 8, 280. [Google Scholar] [CrossRef] [PubMed]
- Seigel, G.M.; Hackam, A.S.; Ganguly, A.; Mandell, L.M.; Gonzalez-Fernandez, F. Human embryonic and neuronal stem cell markers in retinoblastoma. Mol. Vis. 2007, 13, 823–832. [Google Scholar] [PubMed]
- Toda, M.; Iizuka, Y.; Yu, W.; Imai, T.; Ikeda, E.; Yoshida, K.; Kawase, T.; Kawakami, Y.; Okano, H.; Uyemura, K. Expression of the neural RNA-binding protein musashi1 in human gliomas. Glia 2001, 34, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Kwon, H.Y.; Zimdahl, B.; Congdon, K.L.; Blum, J.; Lento, W.E.; Zhao, C.; Lagoo, A.; Gerrard, G.; Foroni, L.; et al. Regulation of myeloid leukaemia by the cell-fate determinant musashi. Nature 2010, 466, 765–768. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, N.; Yousefi, M.; Nakauka-Ddamba, A.; Li, F.; Parada, K.; Rao, S.; Minuesa, G.; Katz, Y.; Gregory, B.D.; et al. Transformation of the intestinal epithelium by the msi2 RNA-binding protein. Nat. Commun. 2015, 6, 6517. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Yousefi, M.; Nakauka-Ddamba, A.; Li, F.; Vandivier, L.; Parada, K.; Woo, D.H.; Wang, S.; Naqvi, A.S.; Rao, S.; et al. The msi family of RNA-binding proteins function redundantly as intestinal oncoproteins. Cell Rep. 2015, 13, 2440–2455. [Google Scholar] [CrossRef] [PubMed]
- Battelli, C.; Nikopoulos, G.N.; Mitchell, J.G.; Verdi, J.M. The RNA-binding protein musashi-1 regulates neural development through the translational repression of p21waf-1. Mol. Cell. Neurosci. 2006, 31, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Imai, T.; Tokunaga, A.; Yoshida, T.; Hashimoto, M.; Mikoshiba, K.; Weinmaster, G.; Nakafuku, M.; Okano, H. The neural RNA-binding protein musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol. Cell. Biol. 2001, 21, 3888–3900. [Google Scholar] [CrossRef] [PubMed]
- Spears, E.; Neufeld, K.L. Novel double-negative feedback loop between adenomatous polyposis coli and musashi1 in colon epithelia. J. Biol. Chem. 2011, 286, 4946–4950. [Google Scholar] [CrossRef] [PubMed]
- Kondo, T.; Furuta, T.; Mitsunaga, K.; Ebersole, T.A.; Shichiri, M.; Wu, J.; Artzt, K.; Yamamura, K.; Abe, K. Genomic organization and expression analysis of the mouse QKI locus. Mamm. Genome 1999, 10, 662–669. [Google Scholar] [PubMed]
- Darbelli, L.; Richard, S. Emerging functions of the quaking RNA-binding proteins and link to human diseases. Wiley Interdiscip. Rev. RNA 2016, 7, 399–412. [Google Scholar] [CrossRef] [PubMed]
- Pilotte, J.; Larocque, D.; Richard, S. Nuclear translocation controlled by alternatively spliced isoforms inactivates the quaking apoptotic inducer. Genes Dev. 2001, 15, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.P.; Nagel, R.J.; Fagg, W.S.; Shiue, L.; Cline, M.S.; Perriman, R.J.; Donohue, J.P.; Ares, M., Jr. Quaking and ptb control overlapping splicing regulatory networks during muscle cell differentiation. RNA 2013, 19, 627–638. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.I.; Reed, R.B.; Grabowski, P.J.; Artzt, K. Function of quaking in myelination: Regulation of alternative splicing. Proc. Natl. Acad. Sci. USA 2002, 99, 4233–4238. [Google Scholar] [CrossRef] [PubMed]
- Larocque, D.; Galarneau, A.; Liu, H.N.; Scott, M.; Almazan, G.; Richard, S. Protection of p27(kip1) mRNA by quaking RNA binding proteins promotes oligodendrocyte differentiation. Nat. Neurosci. 2005, 8, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Vogel, G.; Yu, Z.; Richard, S. The QKI-5 and QKI-6 RNA binding proteins regulate the expression of microRNA 7 in glial cells. Mol. Cell. Biol. 2013, 33, 1233–1243. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Song, W.; Zhao, H.; Ma, Y.; Li, Y.; Zhai, D.; Pi, J.; Si, Y.; Xu, J.; Dong, L.; et al. The RNA-binding protein QKI5 regulates primary mir-124-1 processing via a distal RNA motif during erythropoiesis. Cell Res. 2017, 27, 416–439. [Google Scholar] [CrossRef] [PubMed]
- Conn, S.J.; Pillman, K.A.; Toubia, J.; Conn, V.M.; Salmanidis, M.; Phillips, C.A.; Roslan, S.; Schreiber, A.W.; Gregory, P.A.; Goodall, G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015, 160, 1125–1134. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Richard, S. Structure-function analysis of qk1: A lethal point mutation in mouse quaking prevents homodimerization. Mol. Cell. Biol. 1998, 18, 4863–4871. [Google Scholar] [CrossRef] [PubMed]
- Vernet, C.; Artzt, K. Star, a gene family involved in signal transduction and activation of RNA. Trends Genet. 1997, 13, 479–484. [Google Scholar] [CrossRef]
- Noveroske, J.K.; Lai, L.; Gaussin, V.; Northrop, J.L.; Nakamura, H.; Hirschi, K.K.; Justice, M.J. Quaking is essential for blood vessel development. Genesis 2002, 32, 218–230. [Google Scholar] [CrossRef] [PubMed]
- Darbelli, L.; Vogel, G.; Almazan, G.; Richard, S. Quaking regulates neurofascin 155 expression for myelin and axoglial junction maintenance. J. Neurosci. 2016, 36, 4106–4120. [Google Scholar] [CrossRef] [PubMed]
- Cochrane, A.; Kelaini, S.; Tsifaki, M.; Bojdo, J.; Vila-Gonzalez, M.; Drehmer, D.; Caines, R.; Magee, C.; Eleftheriadou, M.; Hu, Y.; et al. Quaking is a key regulator of endothelial cell differentiation, neovascularization, and angiogenesis. Stem Cells 2017, 35, 952–966. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.J.; Paik, J.H.; Zhang, H.; Shukla, S.A.; Mortensen, R.; Hu, J.; Ying, H.; Hu, B.; Hurt, J.; Farny, N.; et al. Star RNA-binding protein quaking suppresses cancer via stabilization of specific miRNA. Genes Dev. 2012, 26, 1459–1472. [Google Scholar] [CrossRef] [PubMed]
- Bandopadhayay, P.; Ramkissoon, L.A.; Jain, P.; Bergthold, G.; Wala, J.; Zeid, R.; Schumacher, S.E.; Urbanski, L.; O’Rourke, R.; Gibson, W.J.; et al. Myb-QKI rearrangements in angiocentric glioma drive tumorigenicity through a tripartite mechanism. Nat. Genet. 2016, 48, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Feng, F.; Xu, J.; Lu, X.; Wang, S.; Wang, L.; Lu, H.; Wei, M.; Yang, G.; Wang, L.; et al. QKI impairs self-renewal and tumorigenicity of oral cancer cells via repression of sox2. Cancer Biol. Ther. 2014, 15, 1174–1184. [Google Scholar] [CrossRef] [PubMed]
- Novikov, L.; Park, J.W.; Chen, H.; Klerman, H.; Jalloh, A.S.; Gamble, M.J. QKI-mediated alternative splicing of the histone variant macroh2a1 regulates cancer cell proliferation. Mol. Cell. Biol. 2011, 31, 4244–4255. [Google Scholar] [CrossRef] [PubMed]
- Bian, Y.; Wang, L.; Lu, H.; Yang, G.; Zhang, Z.; Fu, H.; Lu, X.; Wei, M.; Sun, J.; Zhao, Q.; et al. Downregulation of tumor suppressor QKI in gastric cancer and its implication in cancer prognosis. Biochem. Biophys. Res. Commun. 2012, 422, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Ji, S.; Ye, G.; Zhang, J.; Wang, L.; Wang, T.; Wang, Z.; Zhang, T.; Wang, G.; Guo, Z.; Luo, Y.; et al. miR-574-5p negatively regulates QKI6/7 to impact beta-catenin/Wnt signalling and the development of colorectal cancer. Gut 2013, 62, 716–726. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Fu, H.; Zhang, J.; Lu, X.; Yu, F.; Jin, L.; Bai, L.; Huang, B.; Shen, L.; Feng, Y.; et al. RNA-binding protein quaking, a critical regulator of colon epithelial differentiation and a suppressor of colon cancer. Gastroenterology 2010, 138, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Zong, F.Y.; Fu, X.; Wei, W.J.; Luo, Y.G.; Heiner, M.; Cao, L.J.; Fang, Z.; Fang, R.; Lu, D.; Ji, H.; et al. The RNA-binding protein QKI suppresses cancer-associated aberrant splicing. PLoS Genet. 2014, 10, e1004289. [Google Scholar] [CrossRef] [PubMed]
- De Miguel, F.J.; Pajares, M.J.; Martinez-Terroba, E.; Ajona, D.; Morales, X.; Sharma, R.D.; Pardo, F.J.; Rouzaut, A.; Rubio, A.; Montuenga, L.M.; et al. A large-scale analysis of alternative splicing reveals a key role of QKI in lung cancer. Mol. Oncol. 2016, 10, 1437–1449. [Google Scholar] [CrossRef] [PubMed]
- Shingu, T.; Ho, A.L.; Yuan, L.; Zhou, X.; Dai, C.; Zheng, S.; Wang, Q.; Zhong, Y.; Chang, Q.; Horner, J.W.; et al. QKI deficiency maintains stemness of glioma stem cells in suboptimal environment by downregulating endolysosomal degradation. Nat. Genet. 2017, 49, 75–86. [Google Scholar] [CrossRef] [PubMed]
- De Bruin, R.G.; Shiue, L.; Prins, J.; de Boer, H.C.; Singh, A.; Fagg, W.S.; van Gils, J.M.; Duijs, J.M.; Katzman, S.; Kraaijeveld, A.O.; et al. Quaking promotes monocyte differentiation into pro-atherogenic macrophages by controlling pre-mRNA splicing and gene expression. Nat. Commun. 2016, 7, 10846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nilsen, T.W.; Graveley, B.R. Expansion of the eukaryotic proteome by alternative splicing. Nature 2010, 463, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Stoss, O.; Olbrich, M.; Hartmann, A.M.; Konig, H.; Memmott, J.; Andreadis, A.; Stamm, S. The star/gsg family protein rslm-2 regulates the selection of alternative splice sites. J. Biol. Chem. 2001, 276, 8665–8673. [Google Scholar] [CrossRef] [PubMed]
- You, J.S.; Jones, P.A. Cancer genetics and epigenetics: Two sides of the same coin? Cancer Cell 2012, 22, 9–20. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mukohyama, J.; Shimono, Y.; Minami, H.; Kakeji, Y.; Suzuki, A. Roles of microRNAs and RNA-Binding Proteins in the Regulation of Colorectal Cancer Stem Cells. Cancers 2017, 9, 143. https://doi.org/10.3390/cancers9100143
Mukohyama J, Shimono Y, Minami H, Kakeji Y, Suzuki A. Roles of microRNAs and RNA-Binding Proteins in the Regulation of Colorectal Cancer Stem Cells. Cancers. 2017; 9(10):143. https://doi.org/10.3390/cancers9100143
Chicago/Turabian StyleMukohyama, Junko, Yohei Shimono, Hironobu Minami, Yoshihiro Kakeji, and Akira Suzuki. 2017. "Roles of microRNAs and RNA-Binding Proteins in the Regulation of Colorectal Cancer Stem Cells" Cancers 9, no. 10: 143. https://doi.org/10.3390/cancers9100143





