Genomic Destabilization Triggered by Replication Stress during Senescence
Abstract
:1. Introduction
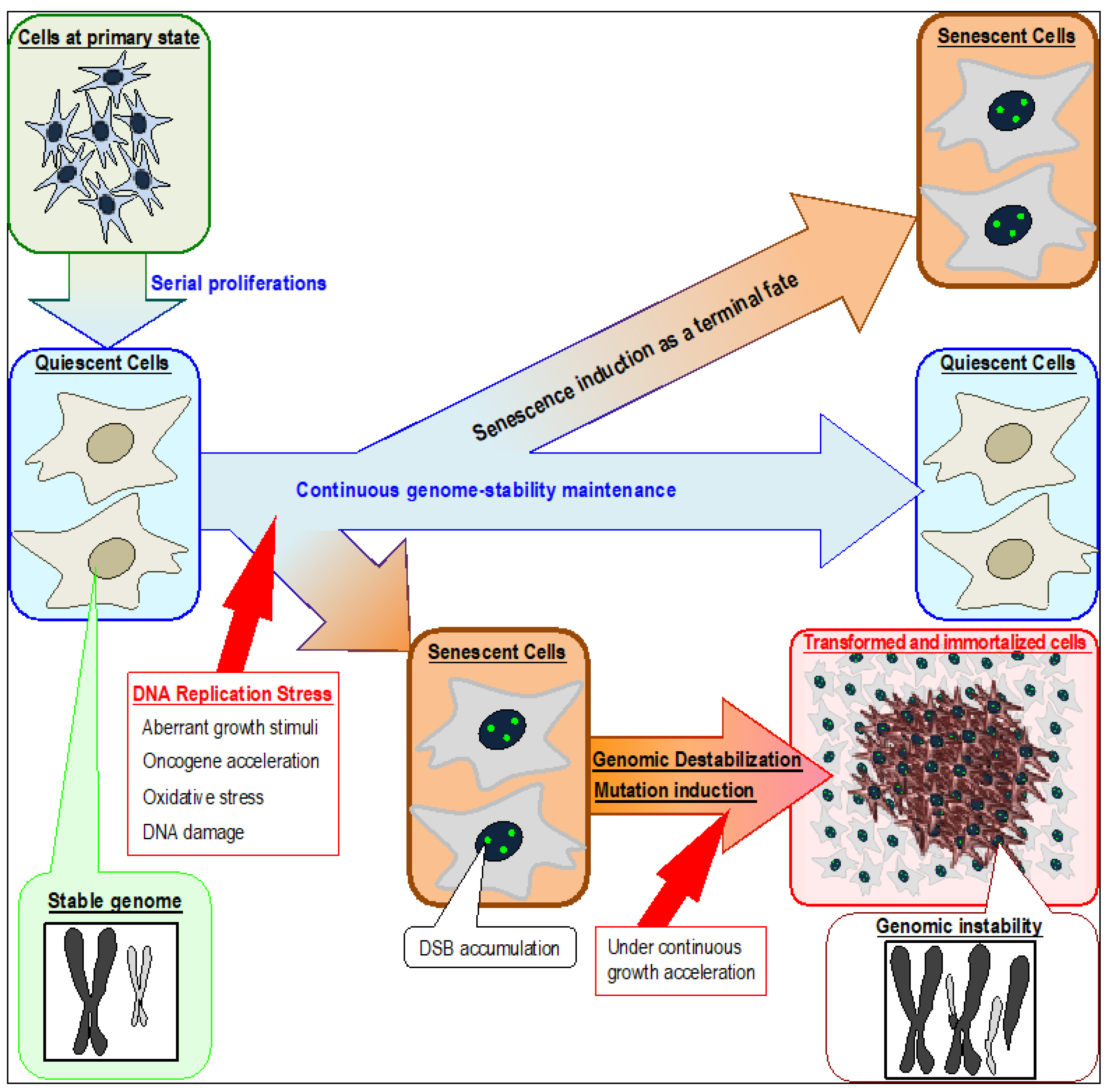
2. Genomic Instabilities in Cancer Cells and Cells Immortalized In Vitro
3. Genomic Destabilization and Replication Stress
4. Genomic Destabilization and Induction of Cancer-Driver Mutations
5. Oncogene Acceleration and Its Contribution to Cancer Development
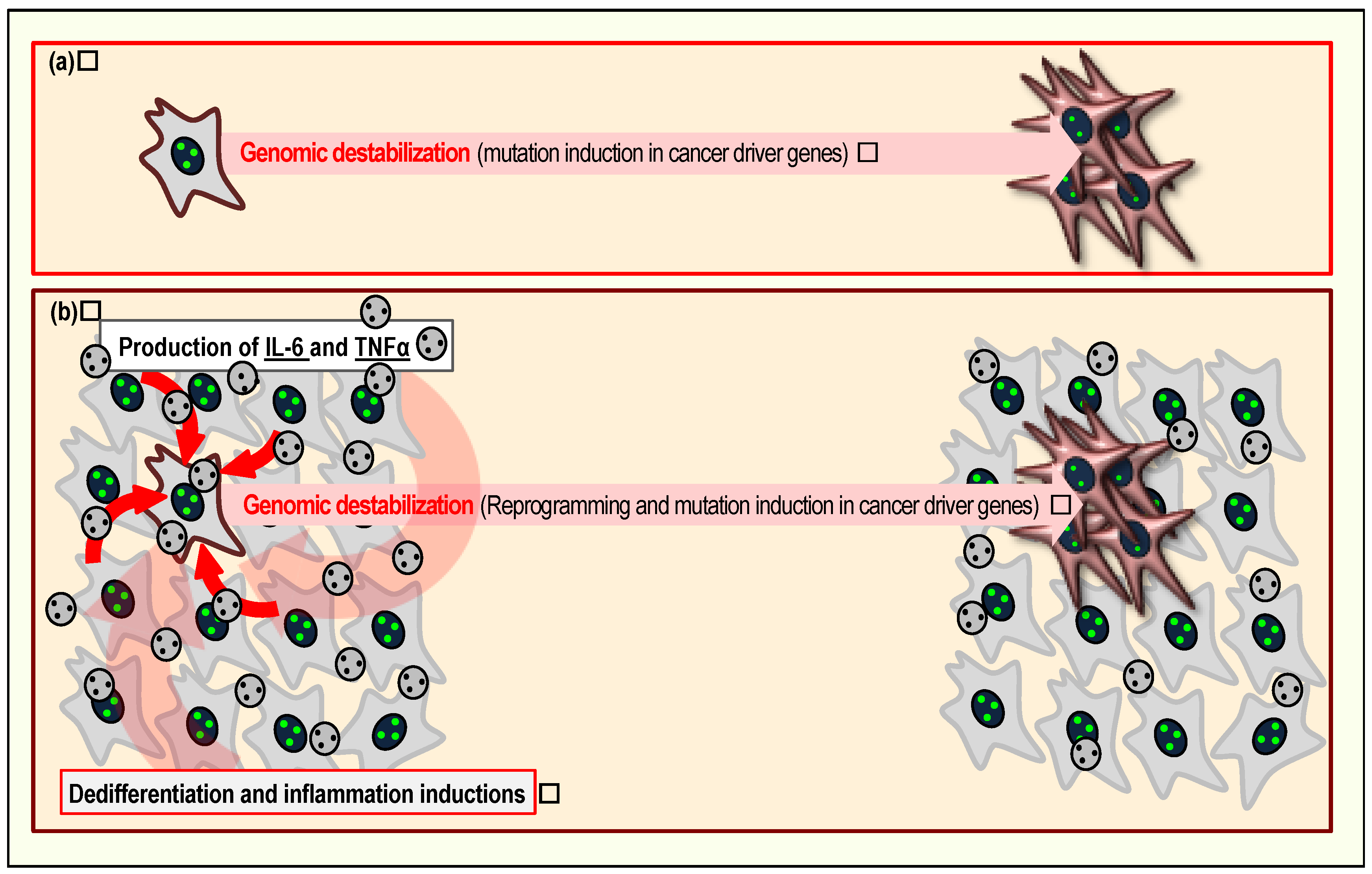
6. The Senescent Cellular Background and Its Contribution to Cancer Development
7. Age as a Risk Factor of Genomic Instability
8. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lengauer, C.; Kinzler, K.W.; Vogelstein, B. Genetic instability in colorectal cancers. Nature 1997, 386, 623–627. [Google Scholar] [CrossRef] [PubMed]
- Lengauer, C.; Kinzler, K.W.; Vogelstein, B. Genetic instabilities in human cancers. Nature 1998, 396, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Shih, I.M.; Zhou, W.; Goodman, S.N.; Lengauer, C.; Kinzler, K.W.; Vogelstein, B. Evidence that genetic instability occurs at an early stage of colorectal tumorigenesis. Cancer Res. 2001, 61, 818–822. [Google Scholar] [PubMed]
- Hveem, T.S.; Merok, M.A.; Pretorius, M.E.; Novelli, M.; Bævre, M.S.; Sjo, O.H.; Clinch, N.; Liestøl, K.; Svindland, A.; Lothe, R.A.; et al. Prognostic impact of genomic instability in colorectal cancer. Br. J. Cancer 2014, 110, 2159–2164. [Google Scholar] [CrossRef] [PubMed]
- Edwards, B.K.; Howe, H.L.; Ries, L.A.; Thun, M.J.; Rosenberg, H.M.; Yancik, R.; Wingo, P.A.; Jemal, A.; Feigal, E.G. Annual report to the nation on the status of cancer, 1973–1999, featuring implications of age and aging on U.S. cancer burden. Cancer 2002, 94, 2766–2792. [Google Scholar] [CrossRef] [PubMed]
- Peto, J. Cancer epidemiology in the last century and the next decade. Nature 2001, 411, 390–395. [Google Scholar] [CrossRef] [PubMed]
- Bartkova, J.; Horejsí, Z.; Koed, K.; Krämer, A.; Tort, F.; Zieger, K.; Guldberg, P.; Sehested, M.; Nesland, J.M.; Lukas, C.; et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 2005, 434, 864–870. [Google Scholar] [CrossRef] [PubMed]
- Gorgoulis, V.G.; Vassiliou, L.V.; Karakaidos, P.; Zacharatos, P.; Kotsinas, A.; Liloglou, T.; Venere, M.; Ditullio, R.A., Jr.; Kastrinakis, N.G.; Levy, B.; et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 2005, 434, 907–913. [Google Scholar] [CrossRef] [PubMed]
- Atsumi, Y.; Fujimori, H.; Fukuda, H.; Inase, A.; Shinohe, K.; Yoshioka, Y.; Shikanai, M.; Ichijima, Y.; Unno, J.; Mizutani, S.; et al. Onset of quiescence following p53 mediated down-regulation of H2AX in normal cells. PLoS ONE 2011, 6, e23432. [Google Scholar] [CrossRef] [PubMed]
- Connor, F.; Bertwistle, D.; Mee, P.J.; Ross, G.M.; Swift, S.; Grigorieva, E.; Tybulewicz, V.L.; Ashworth, A. Tumorigenesis and a DNA repair defect in mice with a truncating Brca2 mutation. Nat. Genet. 1997, 17, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Moynahan, M.E.; Chiu, J.W.; Koller, B.H.; Jasin, M. BRCA1 controls homology-directed DNA repair. Mol. Cell 1999, 4, 511–518. [Google Scholar] [CrossRef]
- Campisi, J.; d’Adda di Fagagna, F. Cellular senescence: When bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 2007, 8, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Sharpless, N.E.; Sherr, C.J. Forging a signature of in vivo senescence. Nat. Rev. Cancer 2015, 15, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Terzi, M.Y.; Izmirli, M.; Gogebakan, B. The cell fate: Senescence or quiescence. Mol. Biol. Rep. 2016, 43, 1213–1220. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, K.; Atsumi, Y.; Fukuda, H.; Masutani, M.; Teraoka, H. The quiescent cellular state is Arf/p53-dependent and associated with H2AX downregulation and genome stability. Int. J. Mol. Sci. 2012, 13, 6492–6506. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, K.; Atsumi, Y.; Nakagama, H.; Teraoka, H. Development of cancer-initiating cells and immortalized cells with genomic instability. World J. Stem Cells 2015, 7, 483–489. [Google Scholar] [CrossRef] [PubMed]
- Grudzenski, S.; Raths, A.; Conrad, S.; Rübe, C.E.; Löbrich, M. Inducible response required for repair of low-dose radiation damage in human fibroblasts. Proc. Natl. Acad. Sci. USA 2010, 107, 14205–14210. [Google Scholar] [CrossRef] [PubMed]
- Sedelnikova, O.A.; Horikawa, I.; Zimonjic, D.B.; Popescu, N.C.; Bonner, W.M.; Barrett, J.C. Senescing human cells and ageing mice accumulate DNA lesions with unrepairable double-strand breaks. Nat. Cell Biol. 2004, 6, 168–170. [Google Scholar] [CrossRef] [PubMed]
- Campisi, J. Aging, cellular senescence, and cancer. Annu. Rev. Physiol. 2013, 75, 685–705. [Google Scholar] [CrossRef] [PubMed]
- Kulju, K.S.; Lehman, J.M. Increased p53 protein associated with aging in human-diploid fibroblasts. Exp. Cell Res. 1995, 217, 336–345. [Google Scholar] [CrossRef] [PubMed]
- Sabin, R.J.; Anderson, R.M. Cellular Senescence—Its role in cancer and the response to ionizing radiation. Genome Integr. 2011, 2, 7. [Google Scholar] [CrossRef] [PubMed]
- Sager, R. Senescence as a mode of tumor suppression. Environ. Health Perspect. 1991, 93, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Lecot, P.; Alimirah, F.; Desprez, P.Y.; Campisi, J.; Wiley, C. Context-dependent effects of cellular senescence in cancer development. Br. J. Cancer 2016, 114, 1180–1184. [Google Scholar] [CrossRef] [PubMed]
- Mosieniak, G.; Sikora, E. Polyploidy: The link between senescence and cancer. Curr. Pharm. Des. 2010, 16, 734–740. [Google Scholar] [CrossRef] [PubMed]
- Van Riggelen, J.; Felsher, D.W. Myc and a Cdk2 senescence switch. Nat. Cell Biol. 2010, 12, 7–9. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef]
- Poulogiannis, G.; Frayling, I.M.; Arends, M.J. DNA mismatch repair deficiency in sporadic colorectal cancer and Lynch syndrome. Histopathology 2010, 56, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Gologan, A.; Sepulveda, A.R. Microsatellite instability and DNA mismatch repair deficiency testing in hereditary and sporadic gastrointestinal cancers. Clin. Lab. Med. 2005, 25, 179–196. [Google Scholar] [CrossRef] [PubMed]
- Woerner, S.M.; Kloor, M.; von Knebel Doeberitz, M.; Gebert, J.F. Microsatellite instability in the development of DNA mismatch repair deficient tumors. Cancer Biomark. 2006, 2, 69–86. [Google Scholar] [CrossRef] [PubMed]
- Geigl, J.B.; Obenauf, A.C.; Schwarzbraun, T.; Speicher, M.R. Defining ‘chromosomal instability’. Trends Genet. 2008, 24, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Almoguera, C.; Shibata, D.; Forrester, K.; Martin, J.; Arnheim, N.; Perucho, M. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell 1988, 53, 549–554. [Google Scholar] [CrossRef] [Green Version]
- Williams, C.; Pontén, F.; Ahmadian, A.; Ren, Z.P.; Ling, G.; Rollman, O.; Ljung, A.; Jaspers, N.G.; Uhlén, M.; Lundeberg, J.; et al. Clones of normal keratinocytes and a variety of simultaneously present epidermal neoplastic lesions contain a multitude of p53 gene mutations in a xeroderma pigmentosum patient. Cancer Res. 1998, 58, 2449–2455. [Google Scholar] [PubMed]
- Nowell, P.C. Genetic alterations in leukemias and lymphomas: Impressive progress and continuing complexity. Cancer Genet. Cytogenet. 1997, 94, 13–19. [Google Scholar] [CrossRef]
- Thiagalingam, S.; Laken, S.; Willson, J.K.; Markowitz, S.D.; Kinzler, K.W.; Vogelstein, B.; Lengauer, C. Mechanisms underlying losses of heterozygosity in human colorectal cancers. Proc. Natl. Acad. Sci. USA 2001, 98, 2698–2702. [Google Scholar] [CrossRef] [PubMed]
- Baca, S.C.; Prandi, D.; Lawrence, M.S.; Mosquera, J.M.; Romanel, A.; Drier, Y.; Park, K.; Kitabayashi, N.; MacDonald, T.Y.; Ghandi, M.; et al. Punctuated evolution of prostate cancer genomes. Cell 2013, 153, 666–677. [Google Scholar] [CrossRef] [PubMed]
- Stephens, P.J.; Greenman, C.D.; Fu, B.; Yang, F.; Bignell, G.R.; Mudie, L.J.; Pleasance, E.D.; Lau, K.W.; Beare, D.; Stebbings, L.A.; et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 2011, 144, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Ichijima, Y.; Yoshioka, K.; Yoshioka, Y.; Shinohe, K.; Fujimori, H.; Unno, J.; Takagi, M.; Goto, H.; Inagaki, M.; Mizutani, S.; et al. DNA lesions induced by replication stress trigger mitotic aberration and tetraploidy development. PLoS ONE 2010, 5, e8821. [Google Scholar] [CrossRef] [PubMed]
- Fujimori, H.; Shikanai, M.; Teraoka, H.; Masutani, M.; Yoshioka, K. Induction of cancerous stem cells during embryonic stem cell differentiation. J. Biol. Chem. 2012, 287, 36777–36791. [Google Scholar] [CrossRef] [PubMed]
- Osawa, T.; Atsumi, Y.; Sugihara, E.; Saya, H.; Kanno, M.; Tashiro, F.; Masutani, M.; Yoshioka, K. Arf and p53 act as guardians of a quiescent cellular state by protecting against immortalization of cells with stable genomes. Biochem. Biophys. Res. Commun. 2013, 432, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Hoeijmakers, J.H. DNA Damage, Aging, and Cancer. N. Engl. J. Med. 2009, 361, 1475–1485. [Google Scholar] [CrossRef] [PubMed]
- Sebastian, R.; Raghavan, S.C. Induction of DNA damage and erroneous repair can explain genomic instability caused by endosulfan. Carcinogenesis 2016, 37, 929–940. [Google Scholar] [CrossRef] [PubMed]
- Ciccia, A.; Elledge, S.J. The DNA damage response: Making it safe to play with knives. Mol. Cell 2010, 40, 179–204. [Google Scholar] [CrossRef] [PubMed]
- Roos, W.P.; Kaina, B. DNA damage-induced cell death by apoptosis. Trends Mol. Med. 2006, 12, 440–450. [Google Scholar] [CrossRef] [PubMed]
- Stucki, M.; Jackson, S.P. gamma H2AX and MDC1: Anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair 2006, 5, 534–543. [Google Scholar] [CrossRef] [PubMed]
- Bonner, W.M.; Redon, C.E.; Dickey, J.S.; Nakamura, A.J.; Sedelnikova, O.A.; Solier, S.; Pommier, Y. gamma H2AX and cancer. Nat. Rev. Cancer 2008, 8, 957–967. [Google Scholar] [CrossRef] [PubMed]
- Atsumi, Y.; Minakawa, Y.; Ono, M.; Dobashi, S.; Shinohe, K.; Shinohara, A.; Takeda, S.; Takagi, M.; Takamatsu, N.; Nakagama, H.; et al. ATM and SIRT6/SNF2H mediate transient H2AX stabilization when DSBs form by blocking HUWE1 to allow efficient gamma H2AX Foci formation. Cell Rep. 2015, 13, 2728–2740. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, A.; Garcia-Muse, T. Causes of genome instability. Annu. Rev. Genet. 2013, 47, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Gaillard, H.; Garcia-Muse, T.; Aguilera, A. Replication stress and cancer. Nat. Rev. Cancer 2015, 15, 276–289. [Google Scholar] [CrossRef] [PubMed]
- Mazouzi, A.; Velimezi, G.; Loizou, J.I. DNA replication stress: Causes, resolution and disease. Exp. Cell Res. 2014, 329, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Atsumi, Y.; Inase, A.; Osawa, T.; Sugihara, E.; Sakasai, R.; Fujimori, H.; Teraoka, H.; Saya, H.; Kanno, M.; Tashiro, F.; et al. The Arf/p53 protein module, which induces apoptosis, down-regulates histone H2AX to allow normal cells to survive in the presence of anti-cancer drugs. J. Biol. Chem. 2013, 288, 13269–13277. [Google Scholar] [CrossRef] [PubMed]
- Anderson, G.R.; Brenner, B.M.; Swede, H.; Chen, N.; Henry, W.M.; Conroy, J.M.; Karpenko, M.J.; Issa, J.P.; Bartos, J.D.; Brunelle, J.K.; et al. Intrachromosomal genomic instability in human sporadic colorectal cancer measured by genome-wide allelotyping and inter-(simple sequence repeat) PCR. Cancer Res. 2001, 61, 8274–8283. [Google Scholar] [PubMed]
- Zeman, M.K.; Cimprich, K.A. Causes and consequences of replication stress. Nat. Cell Biol. 2014, 16, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Abbas, T.; Keaton, M.A.; Dutta, A. Genomic instability in cancer. Cold Spring Harb. Perspect. Biol. 2013, 3, a012914. [Google Scholar] [CrossRef] [PubMed]
- Sedelnikova, O.A.; Horikawa, I.; Redon, C.; Nakamura, A.; Zimonjic, D.B.; Popescu, N.C.; Bonner, W.M. Delayed kinetics of DNA double-strand break processing in normal and pathological aging. Aging Cell 2008, 7, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Minakawa, Y.; Atsumi, Y.; Shinohara, A.; Murakami, Y.; Yoshioka, K. Gamma-irradiated quiescent cells repair directly induced double-strand breaks but accumulate persistent double-strand breaks during subsequent DNA replication. Genes Cells 2016, 21, 789–797. [Google Scholar] [CrossRef] [PubMed]
- Rao, C.V.; Yamada, H.Y. Genomic instability and colon carcinogenesis: From the perspective of genes. Front. Oncol. 2013, 3, 130. [Google Scholar] [CrossRef] [PubMed]
- Blokzijl, F.; de Ligt, J.; Jager, M.; Sasselli, V.; Roerink, S.; Sasaki, N.; Huch, M.; Boymans, S.; Kuijk, E.; Prins, P.; et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature 2016, 538, 260–264. [Google Scholar] [CrossRef] [PubMed]
- Martincorena, I.; Campbell, P.J. Somatic mutation in cancer and normal cells. Science 2015, 349, 1483–1489. [Google Scholar] [CrossRef] [PubMed]
- Tomasetti, C.; Vogelstein, B. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science 2015, 347, 78–81. [Google Scholar] [CrossRef] [PubMed]
- Kane, D.P.; Shcherbakova, P.V. A common cancer-associated DNA polymerase epsilon mutation causes an exceptionally strong mutator phenotype, indicating fidelity defects distinct from loss of proofreading. Cancer Res. 2014, 74, 1895–1901. [Google Scholar] [CrossRef] [PubMed]
- Rayner, E.; van Gool, I.C.; Palles, C.; Kearsey, S.E.; Bosse, T.; Tomlinson, I.; Church, D.N. A panoply of errors: Polymerase proofreading domain mutations in cancer. Nat. Rev. Cancer 2016, 16, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Caldwell, C.M.; Green, R.A.; Kaplan, K.B. APC mutations lead to cytokinetic failures in vitro and tetraploid genotypes in Min mice. J. Cell Biol. 2007, 178, 1109–1120. [Google Scholar] [CrossRef] [PubMed]
- Ceol, C.J.; Pellman, D.; Zon, L.I. APC and colon cancer: Two hits for one. Nat. Med. 2007, 13, 1286–1287. [Google Scholar] [CrossRef] [PubMed]
- Dikovskaya, D.; Schiffmann, D.; Newton, I.P.; Oakley, A.; Kroboth, K.; Sansom, O.; Jamieson, T.J.; Meniel, V.; Clarke, A.; Näthke, I.S. Loss of APC induces polyploidy as a result of a combination of defects in mitosis and apoptosis. J. Cell Biol. 2007, 176, 183–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nik-Zainal, S.; Alexandrov, L.B.; Wedge, D.C.; Van Loo, P.; Greenman, C.D.; Raine, K.; Jones, D.; Hinton, J.; Marshall, J.; Stebbings, L.A.; et al. Mutational processes molding the genomes of 21 breast cancers. Cell 2012, 149, 979–993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartek, J.; Lukas, J. Pathways governing G1/S transition and their response to DNA damage. FEBS Lett. 2001, 490, 117–122. [Google Scholar] [CrossRef]
- Sasaki, R.; Narisawa-Saito, M.; Yugawa, T.; Fujita, M.; Tashiro, H.; Katabuchi, H.; Kiyono, T. Oncogenic transformation of human ovarian surface epithelial cells with defined cellular oncogenes. Carcinogenesis 2009, 30, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Bass, A.J.; Watanabe, H.; Mermel, C.H.; Yu, S.; Perner, S.; Verhaak, R.G.; Kim, S.Y.; Wardwell, L.; Tamayo, P.; Gat-Viks, I.; et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat. Genet. 2009, 41, 1238–1242. [Google Scholar] [CrossRef] [PubMed]
- Felsher, D.W.; Bishop, J.M. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol. Cell 1999, 4, 199–207. [Google Scholar] [CrossRef]
- Gidekel, S.; Pizov, G.; Bergman, Y.; Pikarsky, E. Oct-3/4 is a dose-dependent oncogenic fate determinant. Cancer Cell 2003, 4, 361–370. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keath, E.J.; Caimi, P.G.; Cole, M.D. Fibroblast lines expressing activated c-myc oncogenes are tumorigenic in nude mice and syngeneic animals. Cell 1984, 39, 339–348. [Google Scholar] [CrossRef]
- D’Cruz, C.M.; Gunther, E.J.; Boxer, R.B.; Hartman, J.L.; Sintasath, L.; Moody, S.E.; Cox, J.D.; Ha, S.I.; Belka, G.K.; Golant, A.; et al. c-MYC induces mammary tumorigenesis by means of a preferred pathway involving spontaneous Kras2 mutations. Nat. Med. 2001, 7, 235–239. [Google Scholar] [CrossRef] [PubMed]
- Lavialle, C.; Modjtahedi, N.; Cassingena, R.; Brison, O. High c-myc amplification level contributes to the tumorigenic phenotype of the human breast carcinoma cell line SW 613-S. Oncogene 1988, 3, 335–339. [Google Scholar] [PubMed]
- Pu, H.; Zheng, Q.; Li, H.; Wu, M.; An, J.; Gui, X.; Li, T.; Lu, D. CUDR promotes liver cancer stem cell growth through upregulating TERT and C-Myc. Oncotarget 2015, 6, 40775–40798. [Google Scholar] [CrossRef] [PubMed]
- Narisawa-Saito, M.; Handa, K.; Yugawa, T.; Ohno, S.; Fujita, M.; Kiyono, T. HPV16 E6-mediated stabilization of ErbB2 in neoplastic transformation of human cervical keratinocytes. Oncogene 2007, 26, 2988–2996. [Google Scholar] [CrossRef] [PubMed]
- Zindy, F.; Eischen, C.M.; Randle, D.H.; Kamijo, T.; Cleveland, J.L.; Sherr, C.J.; Roussel, M.F. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998, 12, 2424–2433. [Google Scholar] [CrossRef] [PubMed]
- Lasry, A.; Ben-Neriah, Y. Senescence-associated inflammatory responses: Aging and cancer perspectives. Trends Immunol. 2015, 36, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Collado, M.; Serrano, M. Senescence in tumours: Evidence from mice and humans. Nat. Rev. Cancer 2010, 10, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Lamm, N.; Ben-David, U.; Golan-Lev, T.; Storchová, Z.; Benvenisty, N.; Kerem, B. Genomic instability in human pluripotent stem cells arises from replicative stress and chromosome condensation defects. Cell Stem Cell 2016, 18, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Courtois-Cox, S.; Jones, S.L.; Cichowski, K. Many roads lead to oncogene-induced senescence. Oncogene 2008, 27, 2801–2809. [Google Scholar] [CrossRef] [PubMed]
- Galanos, P.; Vougas, K.; Walter, D.; Polyzos, A.; Maya-Mendoza, A.; Haagensen, E.J.; Kokkalis, A.; Roumelioti, F.M.; Gagos, S.; Tzetis, M.; et al. Chronic p53-independent p21 expression causes genomic instability by deregulating replication licensing. Nat. Cell Biol. 2016, 18, 777–789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knoepfler, P.S. Deconstructing stem cell tumorigenicity: A roadmap to safe regenerative medicine. Stem Cells 2009, 27, 1050–1056. [Google Scholar] [CrossRef] [PubMed]
- Mosteiro, L.; Pantoja, C.; Alcazar, N.; Marión, R.M.; Chondronasiou, D.; Rovira, M.; Fernandez-Marcos, P.J.; Muñoz-Martin, M.; Blanco-Aparicio, C.; Pastor, J.; et al. Tissue damage and senescence provide critical signals for cellular reprogramming in vivo. Science 2016, 354. [Google Scholar] [CrossRef] [PubMed]
- Cooks, T.; Pateras, I.S.; Tarcic, O.; Solomon, H.; Schetter, A.J.; Wilder, S.; Lozano, G.; Pikarsky, E.; Forshew, T.; Rosenfeld, N.; et al. Mutant p53 prolongs NF-kappaB activation and promotes chronic inflammation and inflammation-associated colorectal cancer. Cancer Cell 2013, 23, 634–646. [Google Scholar] [CrossRef] [PubMed]
- Grivennikov, S.I.; Karin, M. Inflammatory cytokines in cancer: Tumour necrosis factor and interleukin 6 take the stage. Ann. Rheum. Dis. 2011, 70, I104–I108. [Google Scholar] [CrossRef] [PubMed]
- Sethi, G.; Sung, B.; Aggarwal, B.B. TNF: A master switch for inflammation to cancer. Front. Biosci. 2008, 13, 5094–5107. [Google Scholar] [CrossRef] [PubMed]
- Vartanian, J.P.; Henry, M.; Marchio, A.; Suspène, R.; Aynaud, M.M.; Guétard, D.; Cervantes-Gonzalez, M.; Battiston, C.; Mazzaferro, V.; Pineau, P.; et al. Massive APOBEC3 editing of hepatitis B viral DNA in cirrhosis. PLoS Pathog. 2010, 6, e1000928. [Google Scholar] [CrossRef] [PubMed]
- Taylor, B.J.; Nik-Zainal, S.; Wu, Y.L.; Stebbings, L.A.; Raine, K.; Campbell, P.J.; Rada, C.; Stratton, M.R.; Neuberger, M.S. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. Elife 2013, 16, e00534. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; La Rosa, S.; Hagos, E.G. Oxidative DNA damage causes premature senescence in mouse embryonic fibroblasts deficient for Krüppel-like factor 4. Mol. Carcinog. 2015, 54, 889–899. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.; Xu, Q.; Martin, T.D.; Li, M.Z.; Demaria, M.; Aron, L.; Lu, T.; Yankner, B.A.; Campisi, J.; Elledge, S.J. The DNA damage response induces inflammation and senescence by inhibiting autophagy of GATA4. Science 2015, 349. [Google Scholar] [CrossRef] [PubMed]
- White, R.R.; Vijg, J. Do DNA double-strand breaks drive aging? Mol. Cell 2016, 63, 729–738. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhang, X.; Zeng, M.; Yuan, J.; Liu, M.; Yin, Y.; Wu, X.; Keefe, D.L.; Liu, L. Increased DNA damage and repair deficiency in granulosa cells are associated with ovarian aging in rhesus monkey. J. Assist. Reprod. Genet. 2015, 32, 1069–1078. [Google Scholar] [CrossRef] [PubMed]
- Gorbunova, V.; Seluanov, A. DNA double strand break repair, aging and the chromatin connection. Mutat. Res. 2016, 788, 2–6. [Google Scholar] [CrossRef] [PubMed]
- Giblin, W.; Skinner, M.E.; Lombard, D.B. Sirtuins: Guardians of mammalian healthspan. Trends Genet. 2014, 30, 271–286. [Google Scholar] [CrossRef] [PubMed]
- Haigis, M.C.; Sinclair, D.A. Mammalian sirtuins: Biological insights and disease relevance. Annu. Rev. Pathol. 2010, 5, 253–295. [Google Scholar] [CrossRef] [PubMed]
- Utani, K.; Fu, H.; Jang, S.M.; Marks, A.B.; Smith, O.K.; Zhang, Y.; Redon, C.E.; Shimizu, N.; Aladjem, M.I. Phosphorylated SIRT1 associates with replication origins to prevent excess replication initiation and preserve genomic stability. Nucleic Acids Res. 2017, 45, 7807–7824. [Google Scholar] [CrossRef] [PubMed]
- Mao, Z.; Hine, C.; Tina, X.; Van Meter, M.; Au, M.; Vaidya, A.; Seluanov, A.; Gorbunova, V. SIRT6 promotes DNA repair under stress by activating PARP1. Science 2011, 332, 1443–1446. [Google Scholar] [CrossRef] [PubMed]
- Toiber, D.; Erdel, F.; Bouazoune, K.; Silberman, D.M.; Zhong, L.; Mulligan, P.; Sebastian, C.; Cosentino, C.; Martinez-Pastor, B.; Giacosa, S.; et al. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol. Cell 2013, 51, 454–468. [Google Scholar] [CrossRef] [PubMed]
| MMR Status | Type | Mutation Rate | Genomic Alteration | Description |
|---|---|---|---|---|
| Proficient | CIN | Low | Gene amplification | |
| Chromosomal deletion | ||||
| Chromosomal rearrangement | Frequently occurs at common fragile sites | |||
| Tetraploidy/Aneuploidy | ||||
| Loss of heterozygousity | ||||
| Chromothripsis | Frequently occurs at common fragile sites | |||
| Chromoplexy | ||||
| Deficient | MSI | High | Alteration in the lengths of microsatellite fragments | Frequently occurs at common fragile sites |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Minakawa, Y.; Shimizu, A.; Matsuno, Y.; Yoshioka, K.-i. Genomic Destabilization Triggered by Replication Stress during Senescence. Cancers 2017, 9, 159. https://doi.org/10.3390/cancers9110159
Minakawa Y, Shimizu A, Matsuno Y, Yoshioka K-i. Genomic Destabilization Triggered by Replication Stress during Senescence. Cancers. 2017; 9(11):159. https://doi.org/10.3390/cancers9110159
Chicago/Turabian StyleMinakawa, Yusuke, Atsuhiro Shimizu, Yusuke Matsuno, and Ken-ichi Yoshioka. 2017. "Genomic Destabilization Triggered by Replication Stress during Senescence" Cancers 9, no. 11: 159. https://doi.org/10.3390/cancers9110159




