Fluorescence Sensing Using DNA Aptamers in Cancer Research and Clinical Diagnostics
Abstract
:1. Introduction
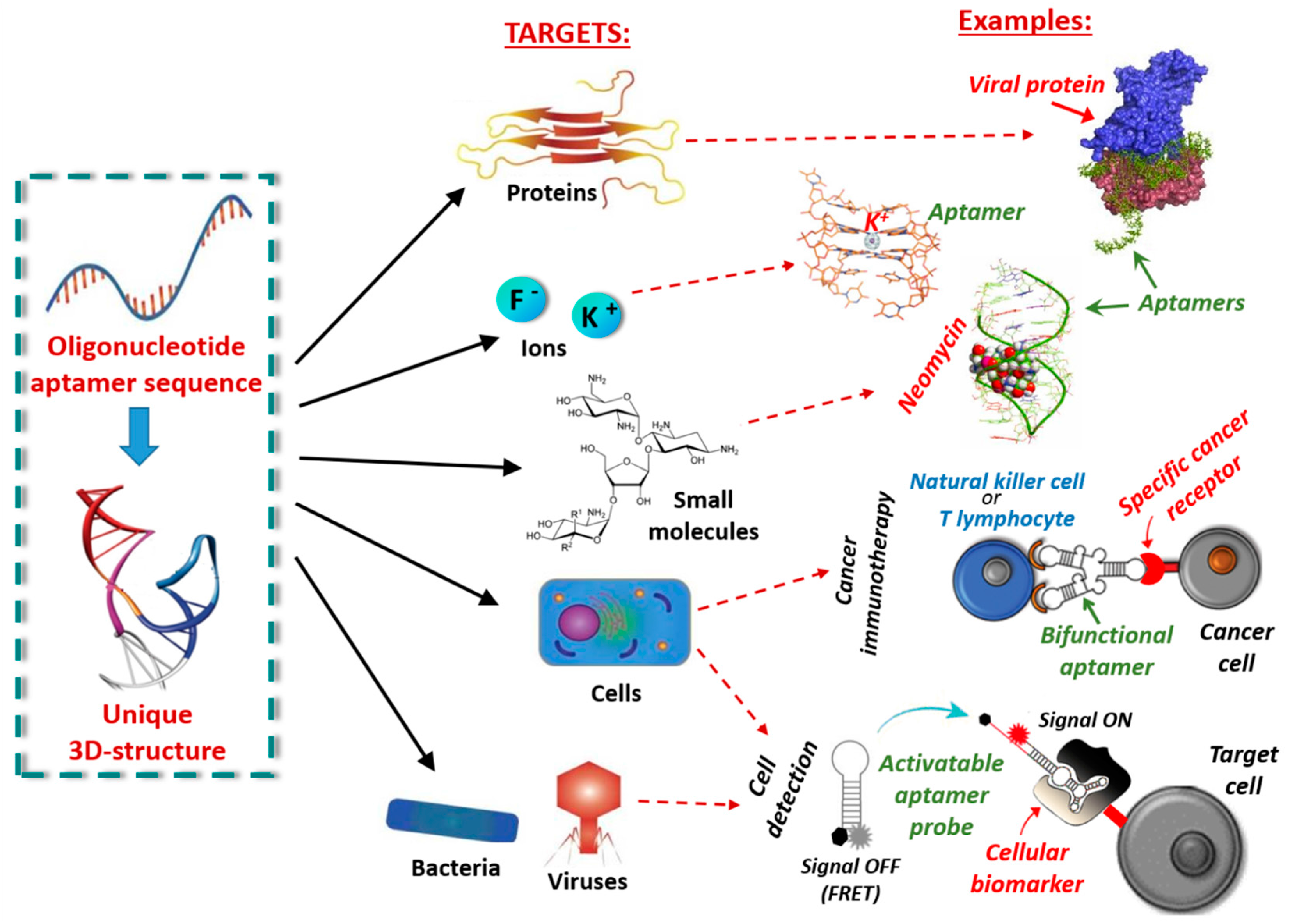
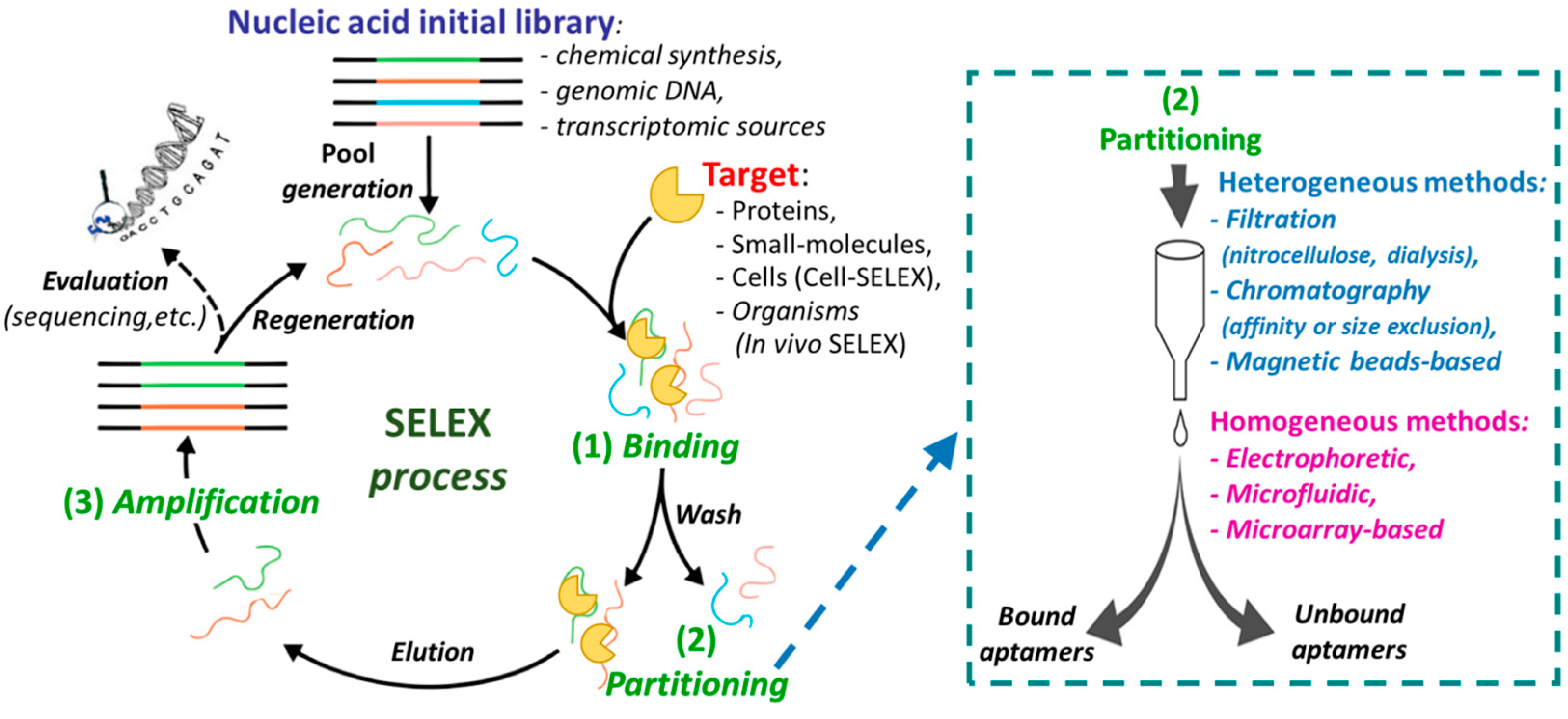
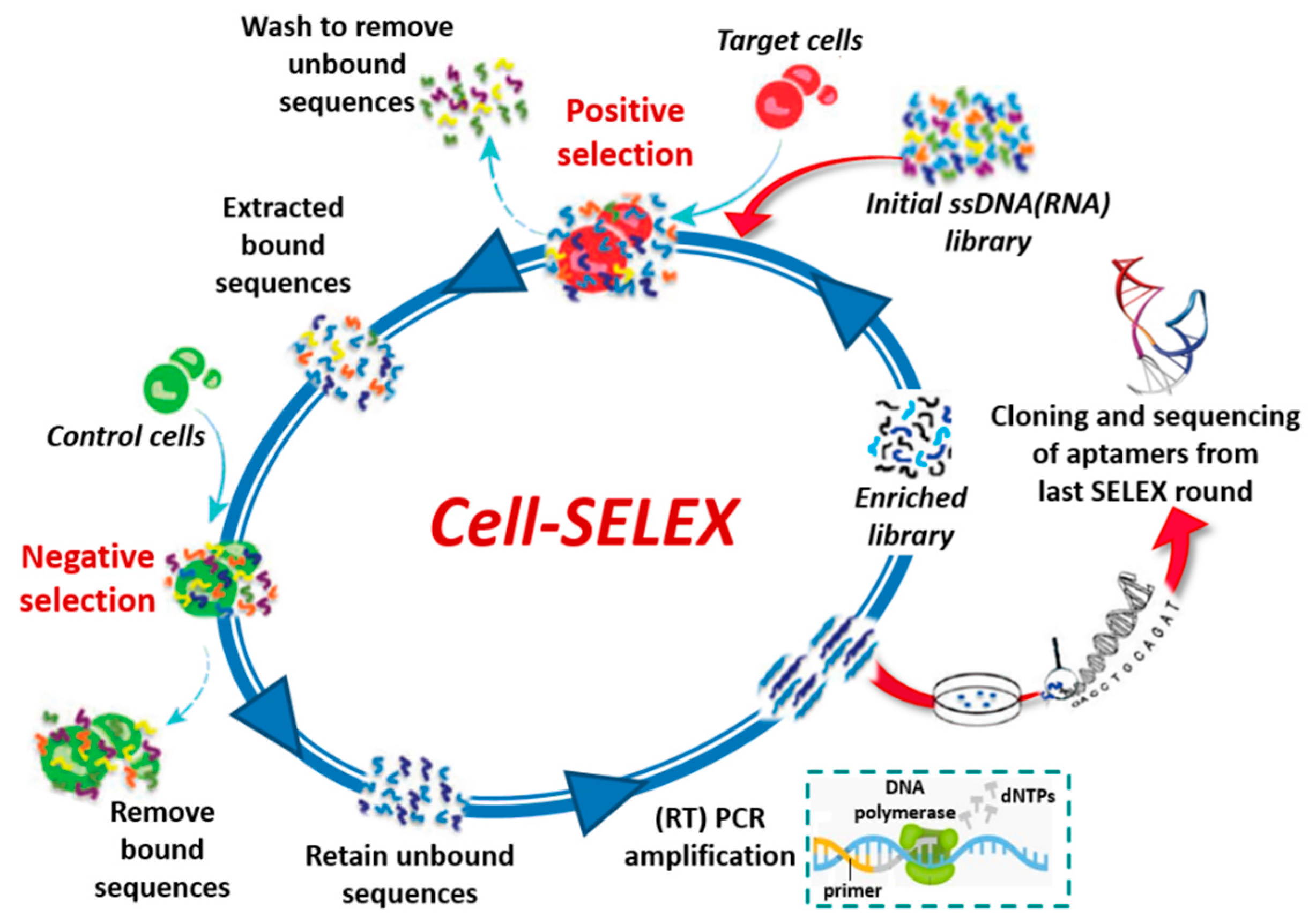
1.1. Nucleic Acid Aptamers and Their Selection Process
1.2. Therapeutic and Diagnostic Applications of Nucleic Acid Aptamers
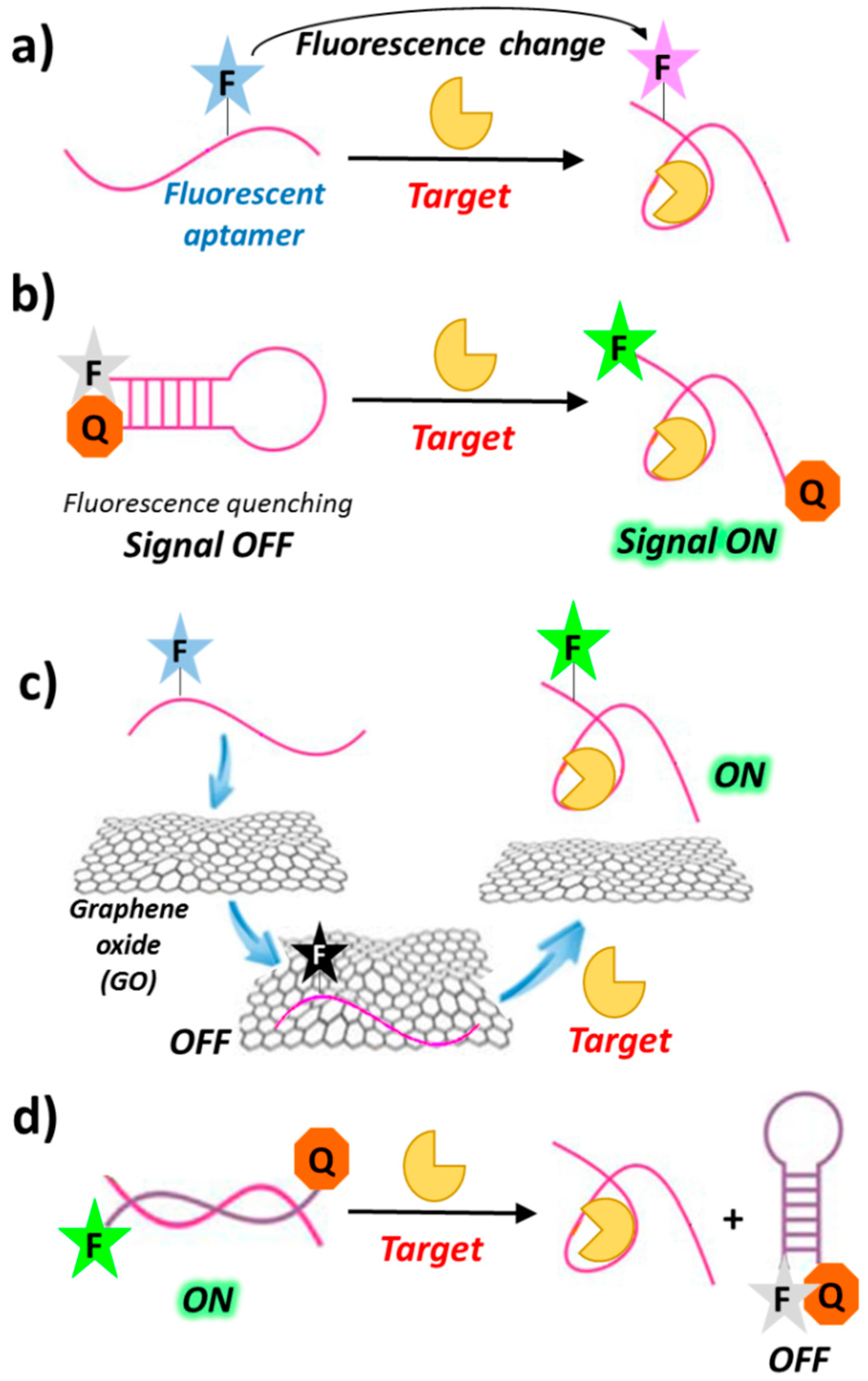
1.3. Aptamer-Sensing Systems Based on Fluorescence
1.4. Aim of This Review
2. Aptamer-Based Fluorescent Systems for the Specific Recognition of Cancer-Related Targets
2.1. Thrombin
2.2. PDGF
2.3. Angiogenin
2.4. Mucin 1
2.5. VEGF
2.6. Elastase
2.7. PTK7
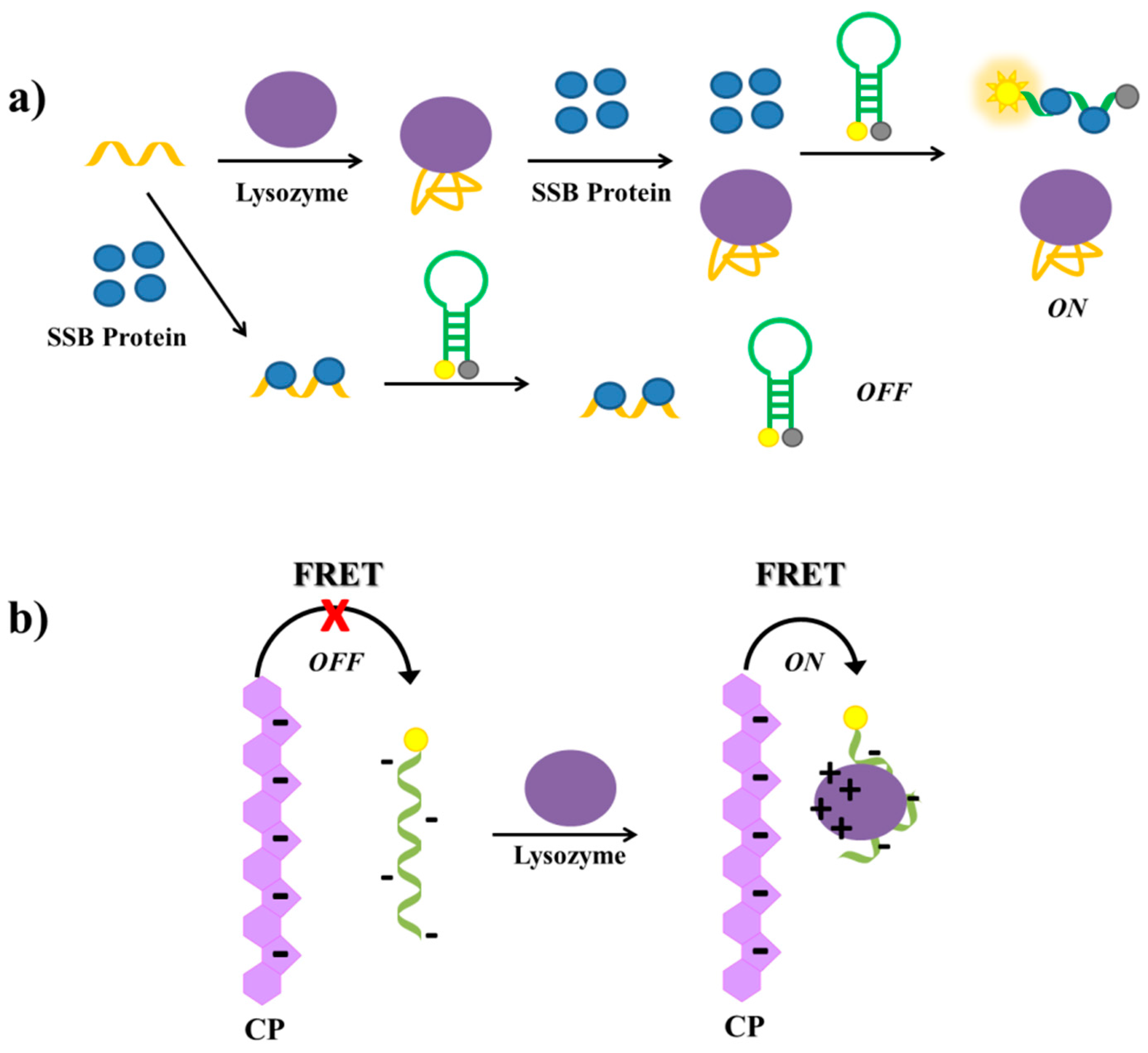
2.8. Lysozyme
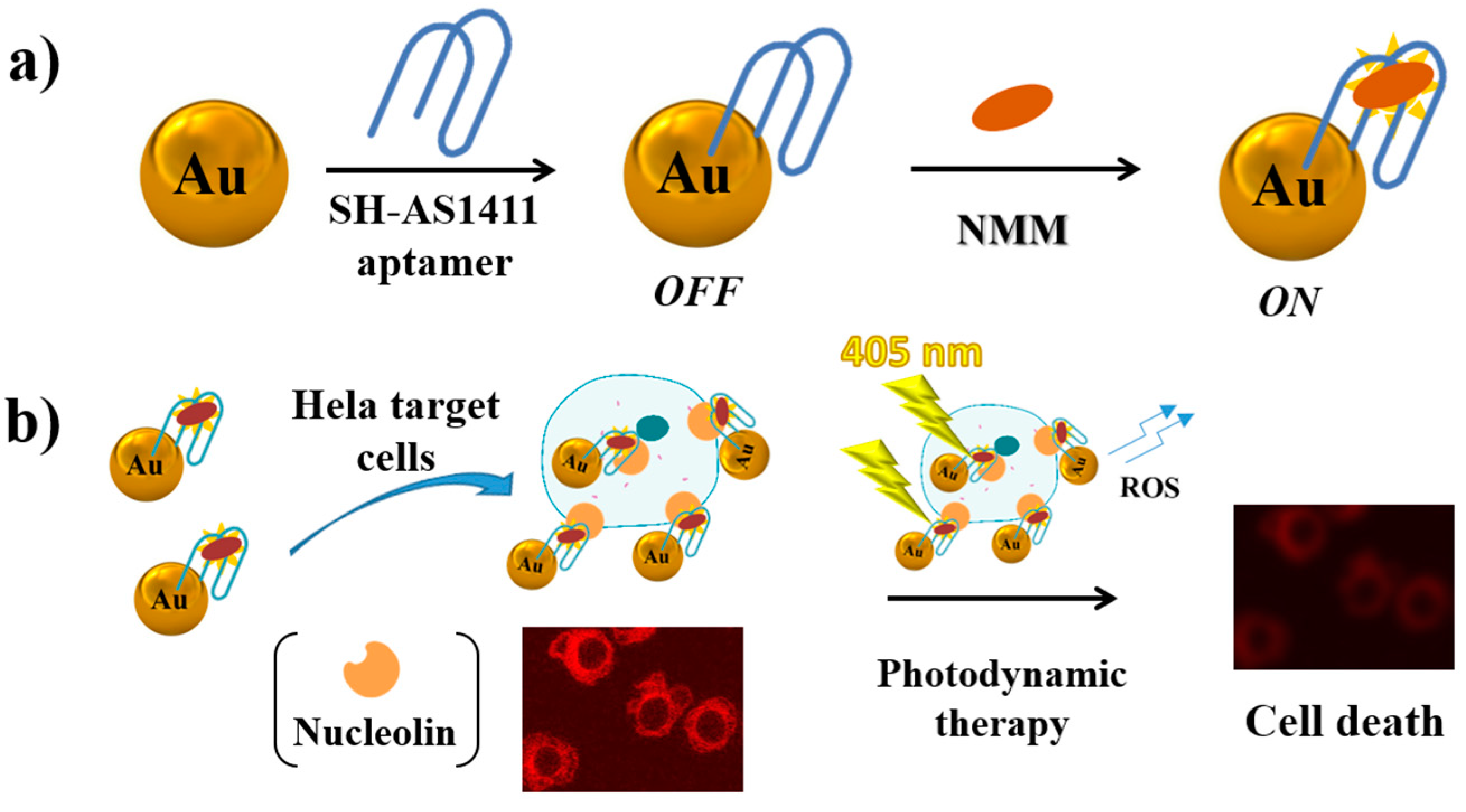
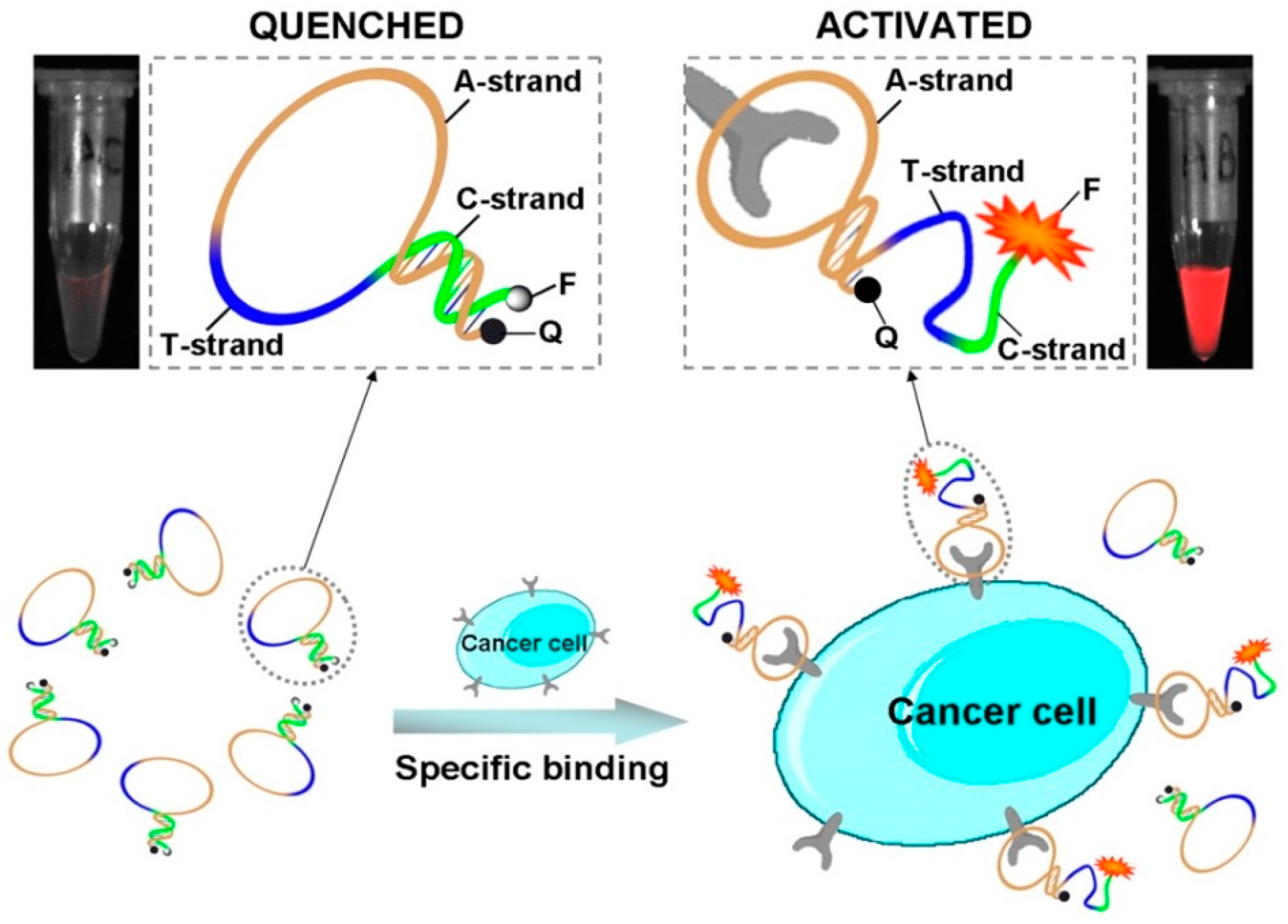
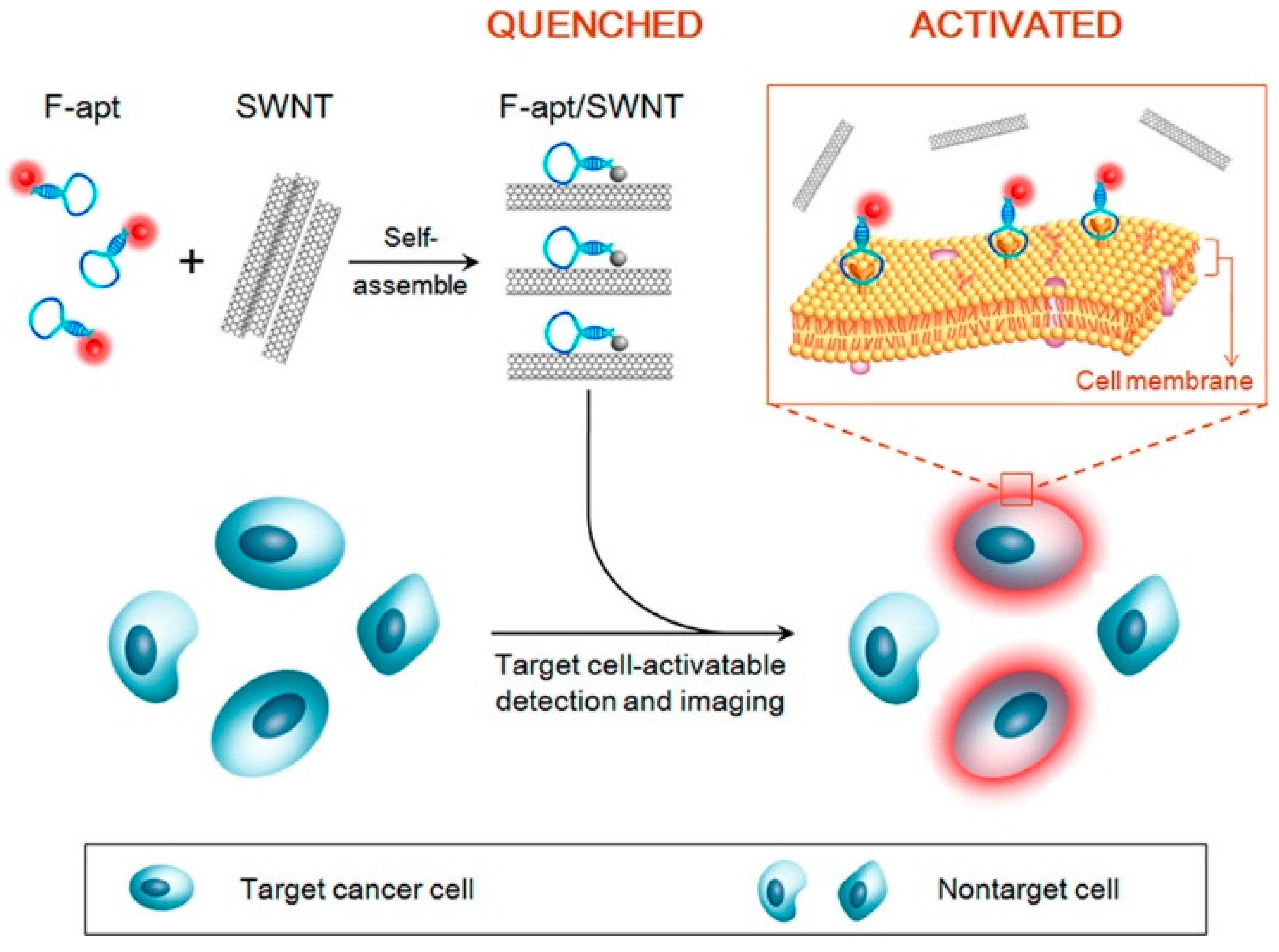
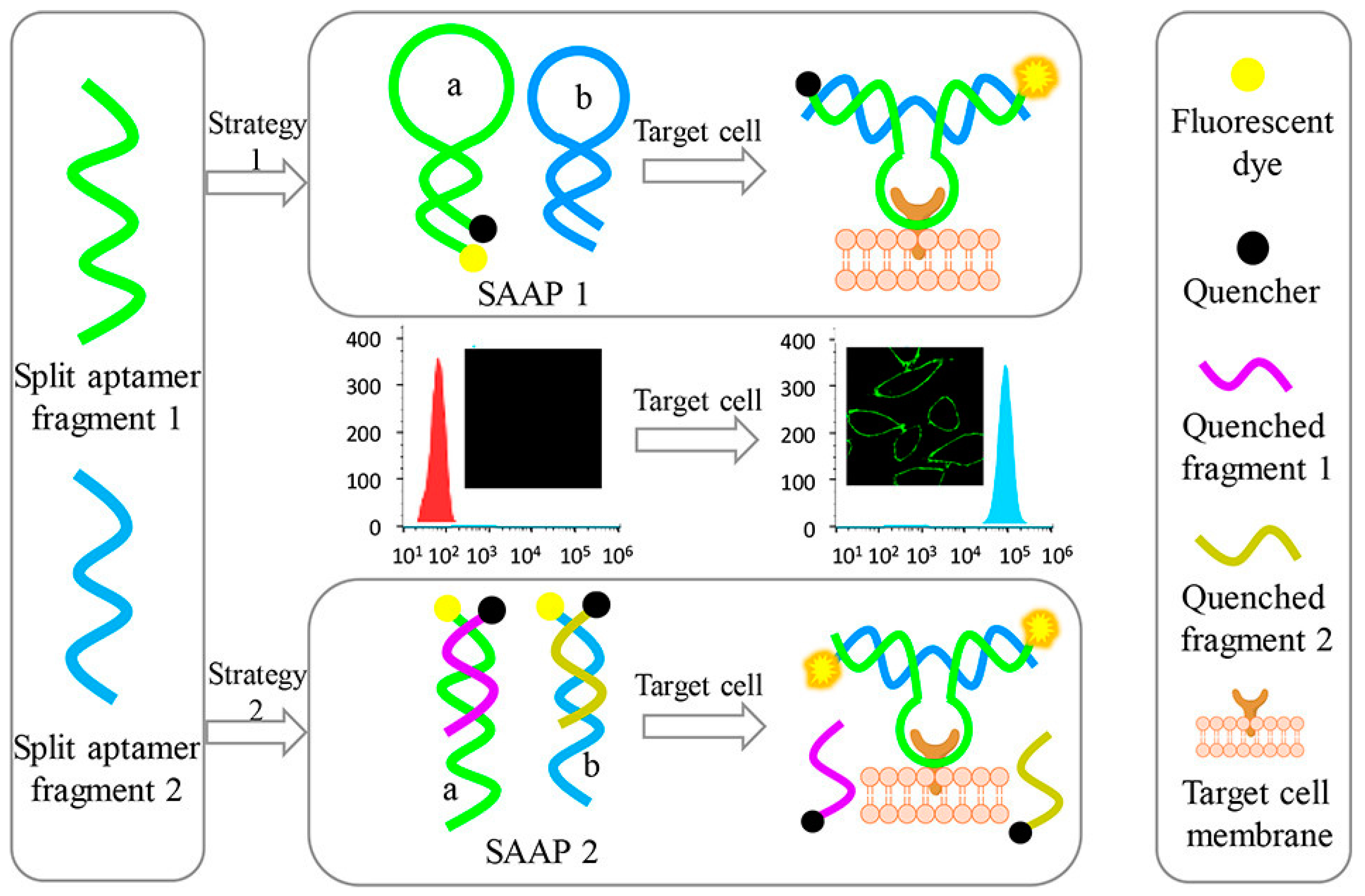
2.9. Cancer Cells
3. Conclusions
Acknowledgments
Conflicts of Interest
References
- Hernandez, L.; Machado, I.; Schafer, T.; Hernandez, F. Aptamers overview: Selection, features and applications. Curr. Top. Med. Chem. 2015, 15, 1066–1081. [Google Scholar] [CrossRef] [PubMed]
- Mayer, G. The chemical biology of aptamers. Angew. Chem. Int. Ed. Eng. 2009, 48, 2672–2689. [Google Scholar] [CrossRef] [PubMed]
- Darmostuk, M.; Rimpelova, S.; Gbelcova, H.; Ruml, T. Current approaches in SELEX: An update to aptamer selection technology. Biotechnol. Adv. 2015, 33, 1141–1161. [Google Scholar] [CrossRef] [PubMed]
- Vu, M.M.; Jameson, N.E.; Masuda, S.J.; Lin, D.; Larralde-Ridaura, R.; Lupták, A. Convergent evolution of adenosine aptamers spanning bacterial, human, and random sequences revealed by structure-based bioinformatics and genomic SELEX. Chem. Biol. 2012, 19, 1247–1254. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, Y.; Nakamura, Y.; Ohuchi, S. HEXIM1-binding elements on mRNAs identified through transcriptomic SELEX and computational screening. Biochimie 2012, 94, 1900–1909. [Google Scholar] [CrossRef] [PubMed]
- Keefe, A.D.; Cload, S.T. SELEX with modified nucleotides. Curr. Opin. Chem. Biol. 2008, 12, 448–456. [Google Scholar] [CrossRef] [PubMed]
- Gold, L.; Ayers, D.; Bertino, J.; Bock, C.; Bock, A.; Brody, E.N.; Carter, J.; Dalby, A.B.; Eaton, B.E.; Fitzwater, T.; et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE 2010, 5, e15004. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, V.B.; Taylor, A.I.; Cozens, C.; Abramov, M.; Renders, M.; Zhang, S.; Chaput, J.C.; Wengel, J.; Peak-Chew, S.Y.; McLaughlin, S.H.; et al. Synthetic genetic polymers capable of heredity and evolution. Science 2012, 336, 341–344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tolle, F.; Brändle, G.M.; Matzner, D.; Mayer, G. A versatile approach towards nucleobase-modified aptamers. Angew. Chem. Int. Ed. Eng. 2015, 54, 10971–10974. [Google Scholar] [CrossRef] [PubMed]
- Ishino, S.; Ishino, Y. DNA polymerases as useful reagents for biotechnology—The history of developmental research in the field. Front. Microbiol. 2014, 5, 465–473. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Romesberg, F.E. Directed polymerase evolution. FEBS Lett. 2014, 588, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Dellafiore, M.A.; Montserrat, J.M.; Iribarren, A.M. Modified nucleoside triphosphates for in-vitro selection techniques. Front. Chem. 2016, 4, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Berezovski, M.; Drabovich, A.; Krylova, S.M.; Musheev, M.; Okhonin, V.; Petrov, A.; Krylov, S.N. Nonequilibrium capillary electrophoresis of equilibrium mixtures: A universal tool for development of aptamers. J. Am. Chem. Soc. 2005, 127, 3165–3171. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, S.C.B. Methods developed for SELEX. Anal. Bioanal. Chem. 2007, 387, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Berezovski, M.V.; Musheev, M.U.; Drabovich, A.P.; Jitkova, J.V.; Krylov, S.N. Non-SELEX: Selection of aptamers without intermediate amplification of candidate oligonucleotides. Nat. Protoc. 2006, 1, 1359–1369. [Google Scholar] [CrossRef] [PubMed]
- Schütze, T.; Wilhelm, B.; Greiner, N.; Braun, H.; Peter, F.; Mörl, M.; Erdmann, V.A.; Lehrach, H.; Konthur, Z.; Menger, M.; Arndt, P.F.; Glökler, J. Probing the SELEX process with next-generation sequencing. PLoS ONE 2011, 6, e29604. [Google Scholar] [CrossRef] [PubMed]
- Quang, N.N.; Perret, G.; Ducongé, F. Applications of high-throughput sequencing for in vitro selection and characterization of aptamers. Pharmaceuticals 2016, 9, 76. [Google Scholar] [CrossRef] [PubMed]
- Chodosh, L.A. UV crosslinking of proteins to nucleic acids. Curr. Protoc. Mol. Biol. 2001, Chapter 12. Unit 12.5. [Google Scholar] [CrossRef]
- Wilkinson, K.A.; Merino, E.J.; Weeks, K.M. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): Quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006, 1, 1610–1616. [Google Scholar] [CrossRef] [PubMed]
- Papavassiliou, A.G. Determination of a transcription factor-binding site by nuclease protection footprinting onto southwestern blots. Methods Mol. Biol. 2009, 543, 201–218. [Google Scholar] [CrossRef] [PubMed]
- Shamah, S.M.; Healy, J.M.; Cload, S.T. Complex target SELEX. Acc. Chem. Res. 2008, 41, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Cerchia, L.; Giangrande, P.H.; Mcnamara, J.O.; De Franciscis, V.; Pansini, V. Cell-Specific aptamers for targeted therapies. Methods Mol. Biol. 2009, 535, 59–78. [Google Scholar] [CrossRef] [PubMed]
- Quang, N.N.; Miodek, A.; Cibiel, A.; Ducongé, F. Selection of aptamers against whole living cells: From cell-SELEX to identification of biomarkers. Methods Mol. Biol. 2017, 1575, 253–272. [Google Scholar] [CrossRef] [PubMed]
- Mercier, M.C.; Dontenwill, M.; Choulier, L. Selection of nucleic acid aptamers targeting tumor cell-surface protein biomarkers. Cancers 2017, 9, 69. [Google Scholar] [CrossRef] [PubMed]
- Khedri, M.; Rafatpanah, H.; Abnous, K.; Ramezani, P.; Ramezani, M. Cancer immunotherapy via nucleic acid aptamers. Int. Immunopharmacol. 2015, 29, 926–936. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.K.R. Monitoring intact viruses using aptamers. Biosensors 2016, 6, 40. [Google Scholar] [CrossRef] [PubMed]
- Ruigrok, V.J.B.; Levisson, M.; Eppink, M.H.M.; Smidt, H.; Van der Oost, J. Alternative affinity tools: More attractive than antibodies? Biochem. J. 2011, 436, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Keefe, A.D.; Pai, S.; Ellington, A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010, 9, 537–550. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.; Wang, H.; Chen, Y.; Zhang, X.; Zhu, H.; Yang, C.; Yang, R.; Liu, C. Molecular aptamers for drug delivery. Trends Biotechnol. 2011, 29, 634–640. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Ye, M.; Donovan, M.J.; Song, E.; Zhao, Z.; Tan, W. Nucleic acid aptamers: An emerging frontier in cancer therapy. Chem. Commun. 2012, 48, 10472–10480. [Google Scholar] [CrossRef] [PubMed]
- Maier, K.E.; Levy, M. From selection hits to clinical leads: Progress in aptamer discovery. Mol. Ther. Methods Clin. Dev. 2016, 5, 16014–16023. [Google Scholar] [CrossRef] [PubMed]
- Nimjee, S.M.; White, R.R.; Becker, R.C.; Sullenger, B.A. Aptamers as therapeutics. Annu. Rev. Pharmacol. Toxicol. 2017, 57, 61–79. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Rossi, J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2016, 16, 181–202. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.J.; Lee, J.W.; Ellington, A.D. Applications of aptamers as sensors. Annu. Rev. Anal. Chem. 2009, 2, 241–264. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Cao, Z.; Lu, Y. Functional nucleic acid sensors. Chem. Rev. 2009, 109, 1948–1998. [Google Scholar] [CrossRef] [PubMed]
- Santosh, B.; Yadava, P.K. Nucleic acid aptamers: Research tools in disease diagnostics and therapeutics. Biomed. Res. Int. 2014, 2014, 50451–50464. [Google Scholar] [CrossRef] [PubMed]
- Xiang, D.; Shigdar, S.; Qiao, G.; Wang, T.; Kouzani, A.Z.; Zhou, S.F.; Kong, L.; Li, Y.; Pu, C.; Duan, W. Nucleic acid aptamer-guided cancer therapeutics and diagnostics: The next generation of cancer medicine. Theranostics 2015, 5, 23–42. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Raston, N.H.A.; Gu, B.M. Aptamer-based nanobiosensors. Biosens. Bioelectron. 2016, 76, 2–19. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Tan, W.; Zu, Y. Aptamers: Versatile molecular recognition probes for cancer detection. Analyst 2016, 141, 403–415. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.E.; Wu, H.; Niu, Y.; Cai, J. Improving the stability of aptamers by chemical modification. Curr. Med. Chem. 2011, 18, 4126–4138. [Google Scholar] [CrossRef] [PubMed]
- Ng, E.W.M.; Shima, D.T.; Calias, P.; Cunningham, E.T.; Guyer, D.R.; Adamis, A.P. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006, 5, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Pratico, E.D.; Sullenger, B.A.; Nair, S.K. Identification and characterization of an agonistic aptamer against the T cell costimulatory receptor, OX40. Nucleic Acid Ther. 2013, 23, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Ramaswamy, V.; Monsalve, A.; Sautina, L.; Segal, M.S.; Dobson, J.; Allen, J.B. DNA aptamer assembly as a vascular endothelial growth factor receptor agonist. Nucleic Acid Ther. 2015, 25, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Yunn, N.O.; Koh, A.; Han, S.; Lim, J.H.; Park, S.; Lee, J.; Kim, E.; Jang, S.K.; Berggren, P.O.; Ryu, S.H. Agonistic aptamer to the insulin receptor leads to biased signaling and functional selectivity through allosteric modulation. Nucleic Acids Res. 2015, 43, 7688–7701. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Rossi, J.J. Cell-type-specific, aptamer-functionalized agents for targeted disease therapy. Mol. Ther. Nucleic Acids 2014, 3, e169. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Liang, C.; Lv, Q.; Li, D.; Xu, X.; Liu, B.; Lu, A.; Zhang, G. Molecular selection, modification and development of therapeutic oligonucleotide aptamers. Int. J. Mol. Sci. 2016, 17, 358. [Google Scholar] [CrossRef] [PubMed]
- Drolet, D.W.; Green, L.S.; Gold, L.; Janjic, N. Fit for the eye: Aptamers in ocular disorders. Nucleic Acid Ther. 2016, 26, 127–146. [Google Scholar] [CrossRef] [PubMed]
- Becker, R.C.; Povsic, T.; Mauricio, C.G.; Rusconi, C.P.; Sullenger, B. Nucleic acid aptamers as antithrombotic agents: Opportunities in extracellular therapeutics. Thromb. Haemost. 2010, 103, 586–595. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Yu, S.; Zheng, Y.; Zheng, Y.; Yang, H.; Zhang, J. Aptamer and its applications in neurodegenerative diseases. Cell. Mol. Life Sci. 2016, 74, 683–695. [Google Scholar] [CrossRef] [PubMed]
- Shum, K.T.; Zhou, J.; Rossi, J.J. Aptamer-based therapeutics: New approaches to combat human viral diseases. Pharmaceuticals 2013, 6, 1507–1542. [Google Scholar] [CrossRef] [PubMed]
- Musumeci, D.; Riccardi, C.; Montesarchio, D. G-Quadruplex forming oligonucleotides as anti-HIV agents. Molecules 2015, 20, 17511–17532. [Google Scholar] [CrossRef] [PubMed]
- Hermann, T.; Patel, D.J. Adaptive recognition by nucleic acid aptamers. Science 2000, 287, 820–825. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.K.H.; Sen, D.; Yu, H.Z. Design and testing of aptamer-based electrochemical biosensors for proteins and small molecules. Bioelectrochemistry 2009, 77, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.C.; Kouzani, A.Z.; Duan, W. Aptasensors: A review. J. Biomed. Nanotechnol. 2010, 6, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Iliuk, A.B.; Hu, L.; Tao, W.A. Aptamer in bioanalytical applications. Anal. Chem. 2011, 83, 4440–4452. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhou, L.; Zhu, Z.; Yang, C. Recent progress in aptamer-based functional probes for bioanalysis and biomedicine. Chem. A Eur. J. 2016, 22, 9886–9900. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Song, W.; Guo, X.; Wang, Z. Ultrasensitive detection of nucleic acids and proteins using quartz crystal microbalance and surface plasmon resonance sensors based on target-triggering multiple signal amplification strategy. Anal. Chim. Acta 2017, 978, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Zhang, X.; Yang, W.; Jia, H.; Li, Y. Fluorescence detection of adenosine triphosphate through an aptamer-molecular beacon multiple probe. Anal. Biochem. 2012, 424, 8–11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, F.; Dever, B.; Li, X.F.; Le, X.C. DNA-mediated homogeneous binding assays for nucleic acids and proteins. Chem. Rev. 2013, 113, 2812–2841. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Dai, S.; Wang, L. Optical aptasensors for quantitative detection of small biomolecules: A review. Biosens. Bioelectron. 2014, 59, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Lu, Y. DNA as sensors and imaging agents for metal Ions. Inorg. Chem. 2014, 53, 1125–1942. [Google Scholar] [CrossRef] [PubMed]
- Sassolas, A.; Blum, L.J.; Leca-Bouvier, B.D. Homogeneous assays using aptamers. Analyst 2011, 136, 257–274. [Google Scholar] [CrossRef] [PubMed]
- Jhaveri, S.D.; Kirby, R.; Conrad, R.; Maglott, E.J.; Bowser, M.; Kennedy, R.T.; Glick, G.; Ellington, A.D. Designed signaling aptamers that transduce molecular recognition to changes in fluorescence intensity. J. Am. Chem. Soc. 2000, 122, 2469–2473. [Google Scholar] [CrossRef]
- Katilius, E.; Katiliene, Z.; Woodbury, N.W. Signaling aptamers created using fluorescent nucleotide analogues. Anal. Chem. 2006, 78, 6484–6489. [Google Scholar] [CrossRef] [PubMed]
- Lerga, T.M.; O’Sullivan, C.K. Rapid determination of total hardness in water using fluorescent molecular aptamer beacon. Anal. Chim. Acta 2008, 610, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Swathi, R.S.; Sebastian, K.L. Resonance energy transfer from a dye molecule to graphene. J. Chem. Phys. 2008, 129, 54703. [Google Scholar] [CrossRef] [PubMed]
- Swathi, R.S.; Sebastian, K.L. Long range resonance energy transfer from a dye molecule to graphene has (distance)(-4) dependence. J. Chem. Phys. 2009, 130, 86101. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.; Tang, L.; Wang, Y.; Jiang, J.; Li, J. Graphene fluorescence resonance energy transfer aptasensor for the thrombin detection. Anal. Chem. 2010, 82, 2341–2346. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Lubin, A.A.; Baker, B.R.; Plaxco, K.W.; Heeger, A.J. Single-step electronic detection of femtomolar DNA by target-induced strand displacement in an electrode-bound duplex. Proc. Natl. Acad. Sci. USA 2006, 103, 16677–16680. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Ho, C.M. Aptamer-based optical probes with separated molecular recognition and signal transduction modules. J. Am. Chem. Soc. 2008, 130, 2380–2381. [Google Scholar] [CrossRef] [PubMed]
- Stojanovic, M.N.; de Prada, P.; Landry, D.W. Aptamer-based folding fluorescent sensor for cocaine. J. Am. Chem. Soc. 2001, 123, 4928–4931. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Huang, C.; Chang, H. Highly selective DNA-based sensor for lead(II) and mercury (II) ions. Anal. Chem. 2009, 81, 2383–2387. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Qin, C.; Li, T.; Wang, L.; Dong, S. Flourescent switch constructed based on hemin-sensitive anionic conjugated polymer and its applications in DNA-related sensors. Anal. Chem. 2009, 81, 3544–3550. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Yang, C.; Zhou, X.; Qin, J. Label-free aptamer-based sensors for L-argininamide by using nucleic acid minor groove binding dyes. Chem. Commun. 2011, 47, 3192–3194. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Lu, Y. Label-free fluorescent aptamer sensor based on regulation of malachite green fluorescence. Anal. Chem. 2010, 82, 574–578. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.C.; Chang, H.T. Aptamer-based fluorescence sensor for rapid detection of potassium ions in urine. Chem. Commun. 2008, 1461–1463. [Google Scholar] [CrossRef] [PubMed]
- Huntington, J.A.; Baglin, T.P. Targeting thrombin—Rational drug design from natural mechanisms. Trends Pharmacol. Sci. 2003, 24, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Huntington, J.A. Molecular recognition mechanisms of thrombin. J. Thromb. Haemost. 2005, 3, 1861–1872. [Google Scholar] [CrossRef] [PubMed]
- Crawley, J.T.B.; Zanardelli, S.; Chion, C.K.N.K.; Lane, D.A. The central role of thrombin in hemostasis. J. Thromb. Haemost. 2007, 5, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Wolberg, A.S. Thrombin generation and fibrin clot structure. Blood Rev. 2007, 21, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Di Cera, E. Thrombin. Mol. Aspects Med. 2008, 29, 203–254. [Google Scholar] [CrossRef] [PubMed]
- Licari, L.G.; Kovacic, J.P. Thrombin physiology and pathophysiology. J. Vet. Emerg. Crit. Care 2009, 19, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Sokolova, E.; Reiser, G. Prothrombin/thrombin and the thrombin receptors PAR-1 and PAR-4 in the brain: Localization, expression and participation in neurodegenerative diseases. Thromb. Haemost. 2008, 100, 576–581. [Google Scholar] [CrossRef] [PubMed]
- Franchini, M.; Mannucci, P.M. Thrombin and cancer: From molecular basis to therapeutic implications. Semin. Thromb. Hemost. 2012, 38, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Danckwardt, S.; Hentze, M.W.; Kulozik, A.E. Pathologies at the nexus of blood coagulation and inflammation: Thrombin in hemostasis, cancer, and beyond. J. Mol. Med. 2013, 91, 1257–1271. [Google Scholar] [CrossRef] [PubMed]
- Shuman, M.A.; Majerus, P.W. The measurement of thrombin in clotting blood by radioimmunoassay. J. Clin. Invest. 1976, 58, 1249–1258. [Google Scholar] [CrossRef] [PubMed]
- Brummel-Ziedins, K.E.; Vossen, C.Y.; Butenas, S.; Mann, K.G.; Rosendaal, F.R. Thrombin generation profiles in deep venous thrombosis. J. Thromb. Haemost. 2005, 3, 2497–2505. [Google Scholar] [CrossRef] [PubMed]
- Bock, L.C.; Griffin, L.C.; Latham, J.A.; Vermaas, E.H.; Toole, J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 1992, 355, 564–566. [Google Scholar] [CrossRef] [PubMed]
- Macaya, R.F.; Schultze, P.; Smith, F.W.; Roet, J.A.; Feigon, J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl. Acad. Sci. USA 1993, 90, 3745–3749. [Google Scholar] [CrossRef] [PubMed]
- Musumeci, D.; Montesarchio, D. Polyvalent nucleic acid aptamers and modulation of their activity: A focus on the thrombin binding aptamer. Pharmacol. Ther. 2012, 136, 202–215. [Google Scholar] [CrossRef] [PubMed]
- Tasset, D.M.; Kubik, M.F.; Steiner, W. Oligonucleotide inhibitors of human thrombin that bind distinct epitopes. J. Mol. Biol. 1997, 272, 688–698. [Google Scholar] [CrossRef] [PubMed]
- Deng, B.; Lin, Y.; Wang, C.; Li, F.; Wang, Z.; Zhang, H.; Li, X.F.; Le, X.C. Aptamer binding assays for proteins: The thrombin example-a review. Anal. Chim. Acta 2014, 837, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Leung, C.H.; Chan, D.S.H.; He, H.Z.; Cheng, Z.; Yang, H.; Ma, D.L. Luminescent detection of DNA-binding proteins. Nucleic Acids Res. 2012, 40, 941–955. [Google Scholar] [CrossRef] [PubMed]
- Platella, C.; Riccardi, C.; Montesarchio, D.; Roviello, G.N.; Musumeci, D. G-quadruplex-based aptamers against protein targets in therapy and diagnostics. BBA Gen. Subj. 2017, 1861, 1429–1447. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Huang, R.; Zhou, Y.; Zhang, M.; Deng, M.; Wang, X.; Weng, X.; Zhou, X. Aptamer-based turn-on fluorescent four-branched quaternary ammonium pyrazine probe for selective thrombin detection. Chem. Commun. 2011, 47, 1273–1275. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Hu, R.; Shao, N. Label-free sensing of thrombin based on quantum dots and thrombin binding aptamer. Talanta 2013, 107, 140–145. [Google Scholar] [CrossRef] [PubMed]
- Du, C.; Hu, Y.; Zhang, Q.; Guo, Z.; Ge, G.; Wang, S.; Zhai, C.; Zhu, M. Competition-derived FRET-switching cationic conjugated polymer-Ir(III) complex probe for thrombin detection. Biosens. Bioelectron. 2017, 100, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Fang, X.; Tan, W. Molecular aptamer beacons for real-time protein recognition. Biochem. Biophys. Res. Commun. 2002, 292, 31–40. [Google Scholar] [CrossRef] [PubMed]
- De Tito, S.; Morvan, F.; Meyer, A.; Vasseur, J.J.; Cummaro, A.; Petraccone, L.; Pagano, B.; Novellino, E.; Randazzo, A.; Giancola, C.; et al. Fluorescence enhancement upon G-quadruplex folding: Synthesis, structure, and biophysical characterization of a dansyl/cyclodextrin-tagged thrombin binding aptamer. Bioconjugate Chem. 2013, 24, 1917–1927. [Google Scholar] [CrossRef] [PubMed]
- Coppola, C.; Paciello, A.; Mangiapia, G.; Licen, S.; Boccalon, M.; De Napoli, L.; Paduano, L.; Tecilla, P.; Montesarchio, D. Design, synthesis and characterisation of a fluorescently labelled CyPLOS ionophore. Chemistry 2010, 16, 13757–13772. [Google Scholar] [CrossRef] [PubMed]
- Riccardi, C.; Russo Krauss, I.; Musumeci, D.; Morvan, F.; Meyer, A.; Vasseur, J.J.; Paduano, L.; Montesarchio, D. Fluorescent thrombin binding aptamer-tagged nanoparticles for an efficient and reversible control of thrombin activity. ACS Appl. Mater. Interfaces 2017, 9, 35574–35587. [Google Scholar] [CrossRef] [PubMed]
- Hamaguchi, N.; Ellington, A.; Stanton, M. Aptamer beacons for the direct detection of proteins. Anal. Biochem. 2001, 294, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Nutiu, R.; Li, Y. Structure-switching signaling aptamers. J. Am. Chem. Soc. 2003, 125, 4771–4778. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Chen, C.; Qian, M.; Zhao, X.S. Aptamer biosensor for protein detection using gold nanoparticles. Anal. Biochem. 2008, 373, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Nambi Krishnan, J.; Park, S.-H.; Kim, S. Aptamer-based single-step assay by the fluorescence enhancement on electroless plated nano Au substrate. Sensors 2017, 17, 2044. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Tang, Z.; Yan, J.; Kang, H.; Kim, Y.; Zhu, Z.; Tan, W. Noncovalent assembly of carbon nanotubes and single-stranded DNA: An effective sensing platform for probing biomolecular interactions. Anal. Chem. 2008, 80, 7408–7413. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhang, Y.; Luo, Y.; Sun, X. Nano-C60: A novel, effective, fluorescent sensing platform for biomolecular detection. Small 2011, 7, 1562–1568. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.M.; Yang, L.R.; Liang, X.F.; Dong, T.T.; Liu, H.Z. Bare magnetic nanoparticles as fluorescence quenchers for detection of thrombin. Analyst 2015, 140, 4114–4120. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, B.; Jin, Y. Label-free fluorescent detection of thrombin using G-quadruplex-based DNAzyme as sensing platform. Analyst 2011, 136, 3268–3273. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Zou, R.; Lou, X. Label-free fluorescent detection of ions, proteins, and small molecules using structure-switching aptamers, SYBR Gold, and exonuclease I. Anal. Chem. 2012, 84, 3554–3560. [Google Scholar] [CrossRef] [PubMed]
- Levy, M.; Cater, S.F.; Ellington, A.D. Quantum-dot aptamer beacons for the detection of proteins. ChemBioChem 2005, 6, 2163–2166. [Google Scholar] [CrossRef] [PubMed]
- Heyduk, E.; Heyduk, T. Nucleic acid-based fluorescence sensors for detecting proteins. Anal. Chem. 2005, 77, 1147–1156. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; Zhou, X.; Xing, D. Sensitive and homogeneous protein detection based on target-triggered aptamer hairpin switch and nicking enzyme assisted fluorescence signal amplification. Anal. Chem. 2012, 84, 3507–3513. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Zhou, W.; Liu, J.; Chai, Y.; Xiang, Y.; Yuan, R. Label-free and homogeneous aptamer proximity binding assay for fluorescent detection of protein biomarkers in human serum. Talanta 2015, 141, 230–234. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Jiang, Y.; Hou, S.; Ma, B.; Fang, X.; Li, M. Detection of oncoprotein platelet-derived growth factor using a fluorescent signaling complex of an aptamer and TOTO. Anal. Bioanal. Chem. 2006, 384, 1175–1180. [Google Scholar] [CrossRef] [PubMed]
- Penmatsa, V.; Ruslinda, A.R.; Beidaghi, M.; Kawarada, H.; Wang, C. Platelet-derived growth factor oncoprotein detection using three-dimensional carbon microarrays. Biosens. Bioelectron. 2013, 39, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Jiang, A.; Hou, T.; Li, H.; Li, F. Enzyme-free and label-free fluorescence aptasensing strategy for highly sensitive detection of protein based on target-triggered hy- bridization chain reaction ampli fi cation. Biosens. Bioelectron. 2015, 70, 324–329. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Sen, A.; Vicens, M.; Tan, W. Synthetic DNA aptamers to detect protein molecular variants in a high-throughput fluorescence quenching assay. ChemBioChem 2003, 4, 829–834. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Wei, R.; He, S.; Liu, Y.; Guo, L.; Li, L. A highly sensitive and selective aptasensor based on graphene oxide fluorescence resonance energy transfer for the rapid determination of oncoprotein PDGF-BB. Analyst 2013, 138, 1726–1732. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.J.; Jockusch, S.; Vicens, M.; Turro, N.J.; Tan, W. Light-switching excimer probes for rapid protein monitoring in complex biological fluids. Proc. Natl. Acad. Sci. USA 2005, 102, 17278–17283. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Cao, Z.; Beck, T.; Tan, W.; Biotechnologies, T.; Drive, N.R.; Diego, S. Molecular aptamer for real-time oncoprotein platelet-derived growth factor monitoring by fluorescence anisotropy. Anal. Biochem. 2001, 73, 5752–5757. [Google Scholar] [CrossRef]
- Zhang, D.; Lu, M.; Wang, H. Fluorescence anisotropy analysis for mapping aptamer-protein interaction at the single nucleotide level. J. Am. Chem. Soc. 2011, 133, 9188–9191. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Schafer, S.; Choi, J.; Yamanaka, Y.J.; Lombardi, M.L.; Bose, S.; Carlson, A.L.; Phillips, J.A.; Teo, W.; Droujinine, I.A.; et al. Cell-surface sensors for real-time probing of cellular environments. Nat. Nanotechnol. 2011, 6, 524–531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, J.-W.; Jo, B.-G.; deMello, A.J.; Choo, J.; Kim, H.Y. Streptavidin-triggered signal amplified fluorescence polarization for analysis of DNA–protein interactions. Analyst 2016, 141, 6499–6502. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, K.; Tan, W.; Ma, C.; Yang, X. Aptamer-based analysis of angiogenin by fluorescence anisotropy. Analyst 2007, 132, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yang, X.; Wang, K.; Tan, W.; Li, H.; Ma, C. FRET-based aptamer probe for rapid angiogenin detection. Talanta 2008, 75, 770–774. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yang, X.; Wang, K.; Tan, W.; He, Y.; Guo, Q.; Tang, H.; Liu, J. Real-time imaging of protein internalization using aptamer conjugates. Anal. Chem. 2008, 80, 5002–5008. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Liu, H.; Tian, T.; Song, X.; Yu, J.; Yan, M. A simple and rapid detection assay for peptides based on the specific recognition of aptamer and signal amplification of hybridization chain reaction. Biosens. Bioelectron. 2016, 83, 15–18. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Lin, Y.; Tang, H.; Pang, D. A graphene oxide-based fluorescent aptasensor for the turn-on detection of epithelial tumor marker mucin 1. Nanoscale 2012, 4, 2054–2059. [Google Scholar] [CrossRef] [PubMed]
- Martínez, O.; Bellard, E.; Golzio, M.; Mechiche-Alami, S.; Rols, M.-P.; Teissié, J.; Ecochard, V.; Paquereau, L. Direct validation of aptamers as powerful tools to image solid tumor. Nucleic Acid Ther. 2014, 24, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhao, J.; Zhang, M.; Yang, H.; Ma, Y.; Gu, Y. MUC1 aptamer-based near-infrared fluorescence probes for tumor imaging. Mol. Imaging Biol. 2015, 17, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Sun, K.; Chen, Z.; Shi, J.; Zhou, D.; Xie, G. A fluorescence biosensor for VEGF detection based on DNA assembly structure switching and isothermal amplification. Biosens. Bioelectron. 2017, 89, 964–969. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ding, X.; Fan, J. Nicking endonuclease-assisted signal amplification of a split molecular aptamer beacon for biomolecule detection using graphene oxide as a sensing platform. Analyst 2015, 140, 7918–7925. [Google Scholar] [CrossRef] [PubMed]
- Mita, C.; Abe, K.; Fukaya, T.; Ikebukuro, K. Vascular endothelial growth factor (VEGF) detection using an aptamer and PNA-based bound/free separation system. Materials 2014, 7, 1046–1054. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.E.; Huang, Y.; Hu, K.; Tian, J.; Zhao, S. A highly sensitive and selective aptasensor based on fluorescence polarization for the rapid determination of oncoprotein vascular endothelial growth factor (VEGF). Anal. Methods 2014, 6, 62–66. [Google Scholar] [CrossRef]
- Wang, S.E.; Si, S. A fluorescent nanoprobe based on graphene oxide fluorescence resonance energy transfer for the rapid determination of oncoprotein vascular endothelial growth factor (VEGF). Appl. Spectrosc. 2013, 67, 1270–1274. [Google Scholar] [CrossRef] [PubMed]
- Charlton, J.; Sennello, J.; Smith, D. In vivo imaging of inflammation using an aptamer inhibitor of human neutrophil elastase. Chem. Biol. 1997, 4, 809–816. [Google Scholar] [CrossRef]
- He, J.L.; Wu, Z.S.; Zhang, S.B.; Shen, G.L.; Yu, R.Q. Fluorescence aptasensor based on competitive-binding for human neutrophil elastase detection. Talanta 2010, 80, 1264–1268. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.M.; Wu, Z.; Liu, S.J.; Chu, X. Structure-switching aptamer triggering hybridization chain reaction on the cell surface for activatable theranostics. Anal. Chem. 2015, 87, 6470–6474. [Google Scholar] [CrossRef] [PubMed]
- Calzada, V.; Moreno, M.; Newton, J.; González, J.; Fernández, M.; Gambini, J.P.; Ibarra, M.; Chabalgoity, A.; Deutscher, S.; Quinn, T.; Cabral, P.; Cerecetto, H. Development of new PTK7-targeting aptamer-fluorescent and -radiolabelled probes for evaluation as molecular imaging agents: Lymphoma and melanoma in vivo proof of concept. Biorgan. Med. Chem. 2017, 25, 1163–1171. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Liao, D.; Zhu, Q.; Wang, F.; Jiao, H.; Zhang, Y.; Yu, C. Fuorescence turn-on detection of a protein through the displaced single-stranded DNA binding protein binding to a molecular beacon. Chem. Commun. 2011, 47, 5485–5487. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhao, J.; Jiang, J.; Yu, R. A novel exonuclease III-aided amplification assay for lysozyme based on graphene oxide platform. Talanta 2012, 101, 357–361. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhu, Z.; Bamrungsap, S.; Zhu, G.Z.; You, M.X.; He, X.X.; Wang, K.M.; Tan, W.H. Competition-mediated pyrene-switching aptasensor: Probing lysozyme in human serum with a monomer-excimer fluorescence switch. Anal. Chem. 2010, 82, 10158–10163. [Google Scholar] [CrossRef] [PubMed]
- Zou, M.; Chen, Y.; Xu, X.; Huang, H.; Liu, F.; Li, N. The homogeneous fluorescence anisotropic sensing of salivary lysozyme using the 6-carboxyfluorescein-labeled DNA aptamer. Biosens. Bioelectron. 2012, 32, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, B. Fluorescence resonance energy transfer between an anionic conjugated polymer and a dye-labeled lysozyme aptamer for specific lysozyme detection. Chem. Commun. 2009, 2284–2286. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Ke, G.; Wang, C.; Yang, C.J. A cyclic enzymatic amplification method for sensitive and selective detection of nucleic acids. Analyst 2010, 135, 2069–2073. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; Zhou, X.; Xing, D. Highly sensitive protein detection based on aptamer probe and isothermal nicking enzyme assisted fluorescence signal amplification. Chem. Commun. 2010, 46, 7373–7375. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Li, G.; Hu, Y. Aptamer-involved fluorescence amplification strategy facilitated by directional enzymatic hydrolysis for bioassays based on a metal-organic framework platform: Highly selective and sensitive determination of thrombin and oxytetracycline. Microchim. Acta 2017, 184, 2365–2373. [Google Scholar] [CrossRef]
- Fredriksson, L.; Li, H.; Eriksson, U. The PDGF family: Four gene products form five dimeric isoforms. Cytokine Growth Factor Rev. 2004, 15, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Ross, R.; Glomset, J.; Kariya, B.; Harker, L. A platelet-dependent serum factor that stimulates the proliferation of arterial smooth muscle cells in vitro. Proc. Natl. Acad. Sci. USA 1974, 71, 1207–1210. [Google Scholar] [CrossRef] [PubMed]
- Kohler, N.; Lipton, A. Platelets as a source of fibroblast growth-promoting activity. Exp. Cell Res. 1974, 87, 297–301. [Google Scholar] [CrossRef]
- Westermark, B.; Wasteson, A. A platelet factor stimulating human normal glial cells. Exp. Cell Res. 1976, 98, 170–174. [Google Scholar] [CrossRef]
- Kazlauskas, A. PDGFs and their receptors. Gene 2017, 614, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Tallquist, M.; Kazlauskas, A. PDGF signaling in cells and mice. Cytokine Growth Factor Rev. 2004, 15, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.H.H.; Chen, X.; He, X. Platelet-derived growth factors and their receptors: Structural and functional perspectives. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2013, 1834, 2176–2186. [Google Scholar] [CrossRef] [PubMed]
- Östman, A.; Heldin, C.H. PDGF receptors as targets in tumor treatment. Adv. Cancer Res. 2007, 97, 247–274. [Google Scholar] [CrossRef] [PubMed]
- Andrae, J.; Gallini, R.; Betsholtz, C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 2008, 22, 1276–1312. [Google Scholar] [CrossRef] [PubMed]
- Heldin, C.H. Targeting the PDGF signaling pathway in tumor treatment. Cell Commun. Signal. 2013, 11, 97. [Google Scholar] [CrossRef] [PubMed]
- Farooqi, A.A.; Siddik, Z.H. Platelet-derived growth factor (PDGF) signalling in cancer: Rapidly emerging signalling landscape. Cell Biochem. Funct. 2015, 33, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Appiah-Kubi, K.; Wang, Y.; Qian, H.; Wu, M.; Yao, X.; Wu, Y.; Chen, Y. Platelet-derived growth factor receptor/platelet-derived growth factor (PDGFR/PDGF) system is a prognostic and treatment response biomarker with multifarious therapeutic targets in cancers. Tumor Biol. 2016, 37, 10053–10066. [Google Scholar] [CrossRef] [PubMed]
- Green, L.S.; Jellinek, D.; Jenison, R.; Arne, O.; Heldin, C.H.; Janjic, N. Inhibitory DNA ligands to platelet-derived growth factor B-chain. Biochemistry 1996, 35, 14413–14424. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhao, Q.; Zhao, B.; Wang, H. Fluorescence anisotropy reduction of allosteric aptamer for sensitive and specific protein signaling. Anal. Chem. 2012, 84, 3070–3074. [Google Scholar] [CrossRef] [PubMed]
- Fredriksson, S.; Gullberg, M.; Jarvius, J.; Olsson, C.; Pietras, K.; Gústafsdóttir, S.M.; Östman, A.; Landegren, U. Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 2002, 20, 473–477. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Cao, Z.; Lau, C.; Lu, J. Label-free technology for the amplified detection of microRNA based on the allosteric hairpin DNA switch and hybridization chain reaction. Analyst 2014, 139, 6022–6027. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Chai, Y.; Xie, S.; Yuan, Y.; Zhang, J.; Yuan, R. Cleavage-based hybridization chain reaction for electrochemical detection of thrombin. Analyst 2014, 139, 4264–4269. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Hu, S.; Cao, Y.; Zhang, B.; Li, G. Electrochemical detection of protein based on hybridization chain reaction-assisted formation of copper nanoparticles. Biosens. Bioelectron. 2015, 66, 327–331. [Google Scholar] [CrossRef] [PubMed]
- He, B.S.; Song, B.; Li, D.; Zhu, C.; Qi, W.; Wen, Y.; Wang, L.; Song, S.; Fang, H.; Fan, C. A graphene nanoprobe for rapid, sensitive, and multicolor fluorescent DNA analysis. Adv. Funct. Mater. 2010, 20, 453–459. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Springer: Heidelberg, Germany, 2006. [Google Scholar]
- Karp, J.M.; Sock, G.; Teo, L. Mesenchymal stem cell homing: The devil is in the details. Cell Stem Cell 2009, 4, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Ankrum, J.; Karp, J.M. Mesenchymal stem cell therapy: Two steps forward, one step back. Trends Mol. Med. 2010, 16, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, D.; Vemula, P.K.; Teo, G.S.L.; Spelke, D.; Karnik, R.; Wee, L.Y.; Karp, J.M. Chemical engineering of mesenchymal stem cells to induce a cell rolling response. Bioconj. Chem. 2008, 19, 2105–2109. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, D.; Vemula, P.K.; Zhao, W.; Gupta, A.; Karnik, R.; Karp, J.M. Engineered mesenchymal stem cells with self-assembled vesicles for systemic cell targeting. Biomaterials 2010, 31, 5266–5274. [Google Scholar] [CrossRef] [PubMed]
- Tello-Montoliu, A.; Patel, J.V.; Lip, G.Y.H. Angiogenin: A review of the pathophysiology and potential clinical applications. J. Thromb. Haemost. 2006, 4, 1864–1874. [Google Scholar] [CrossRef] [PubMed]
- Harper, J.W.; Fox, E.A.; Shapiro, R.; Vallee, B.L. Mutagenesis of residues flanking Lys-40 enhances the enzymatic activity and reduces the angiogenic potency of angiogenin. Biochemistry 1990, 29, 7297–7302. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, N.; Wang, L.; Kishimoto, K.; Tsuji, T.; Hu, G. A therapeutic target for prostate cancer based on angiogenin-stimulated angiogenesis and cancer cell proliferation. Proc. Natl. Acad. Sci. USA 2006, 103, 14519–14524. [Google Scholar] [CrossRef] [PubMed]
- Sheng, J.; Xu, Z. Three decades of research on angiogenin: A review and perspective. Acta Biochim. Biophys. 2016, 48, 399–410. [Google Scholar] [CrossRef] [PubMed]
- Landt, S.; Mordelt, K.; Schwidde, I.; Barinoff, J.; Korlach, S.; Stöblen, F.; Lichtenegger, W.; Sehouli, J.; Kümmel, S. Prognostic significance of the angiogenic factors angiogenin, endoglin and endostatin in cervical cancer. Anticancer Res. 2011, 31, 2651–2656. [Google Scholar] [PubMed]
- Fang, S.; Repo, H.; Joensuu, H.; Orpana, A.; Salven, P. High serum angiogenin at diagnosis predicts for failure on long-term treatment response and for poor overall survival in non-Hodgkin lymphoma. Eur. J. Cancer 2011, 47, 1708–1716. [Google Scholar] [CrossRef] [PubMed]
- Li, D.D.; Bell, J.; Brown, A.; Berry, C.L. The observation of angiogenin and basic fibroblast growth factor gene expression in human colonic adenocarcinomas, gastric adenocarcinomas, and hepatocellular carcinomas. J. Phatol. 1994, 172, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Musolino, C.; Alonci, A.; Bellomo, G.; Loteta, B.; Quartarone, E.; Gangemi, D.; Massara, E.; Calabro, L. Levels of soluble angiogenin in chronic myeloid malignancies: Clinical implications. Eur. J. Haematol. 2004, 72, 416–419. [Google Scholar] [CrossRef] [PubMed]
- Nobile, V.; Russo, N.; Hu, G.F.; Riordan, J.F. Inhibition of human angiogenin by DNA aptamers: Nuclear colocalization of an angiogenin-inhibitor complex. Biochemistry 1998, 37, 6857–6863. [Google Scholar] [CrossRef] [PubMed]
- Kufe, D.W. Mucins in cancer: Function, prognosis and therapy. Nat. Rev. Cancer 2009, 9, 874–885. [Google Scholar] [CrossRef] [PubMed]
- Rahn, J.J.; Dabbagh, L.; Pasdar, M.; Hugh, J.C. The importance of MUC1 cellular localization in patients with breast carcinoma. Cancer 2001, 91, 1973–1982. [Google Scholar] [CrossRef]
- Wreesmann, V.B.; Sieczka, E.M.; Socci, N.D.; Hezel, M.; Belbin, T.J.; Childs, G.; Patel, S.G.; Patel, K.N.; Tallini, G.; Prystowsky, M.; et al. Genome-wide profiling of papillary thyroid cancer identifies MUC1 as an independent prognostic marker. Cancer Res. 2004, 64, 3780–3789. [Google Scholar] [CrossRef] [PubMed]
- Khodarev, N.N.; Pitroda, S.P.; Beckett, M.A.; MacDermed, D.M.; Huang, L.; Kufe, D.W.; Weichselbaum, R.R. MUC1-induced transcriptional programs associated with tumorigenesis predict outcome in breast and lung cancer. Cancer Res. 2009, 69, 2833–2837. [Google Scholar] [CrossRef] [PubMed]
- Karsten, U.; Von Mensdorff-Pouilly, S.; Goletz, S. What makes MUC1 a tumor antigen? Tumor Biol. 2005, 26, 217–220. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, C.S.M.; Matthews, C.S.; Missailidis, S. DNA aptamers that bind to MUC1 tumour marker: Design and characterization of MUC1-binding single-stranded DNA aptamers. Tumor Biol. 2006, 27, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Nabavinia, M.S.; Gholoobi, A.; Charbgoo, F.; Nabavinia, M.; Ramezani, M.; Abnous, K. Anti-MUC1 aptamer: A potential opportunity for cancer treatment. Med. Res. Rev. 2017, 37, 1518–1539. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, C.S.M.; Cheung, M.C.; Missailidis, S.; Bisland, S.; Gariépy, J. Phototoxic aptamers selectively enter and kill epithelial cancer cells. Nucleic Acids Res. 2009, 37, 866–876. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Neoh, K.G.; Kang, E.T.; Choe, W.S.; Su, X. PEGylated anti-MUC1 aptamer-doxorubicin complex for targeted drug delivery to MCF7 breast cancer cells. Macromol. Biosci. 2011, 11, 1331–1335. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Hu, Y.; Duan, J.; Yuan, W.; Wang, C.; Xu, H.; Yang, X. Da Novel aptamer-nanoparticle bioconjugates enhances delivery of anticancer drug to MUC1-positive cancer cells in vitro. PLoS ONE 2011, 6, e24077. [Google Scholar] [CrossRef]
- Hu, Y.; Duan, J.; Zhan, Q.; Wang, F.; Lu, X.; Yang, X. Da Novel MUC1 aptamer selectively delivers cytotoxic agent to cancer cells in vitro. PLoS ONE 2012, 7, e31970. [Google Scholar] [CrossRef]
- Ferrara, N.; Davis-Smyth, T. The biology of vascular endothelial growth factor. Endocr. Rev. 1997, 18, 4–25. [Google Scholar] [CrossRef] [PubMed]
- Niu, G.; Chen, X. Vascular endothelial growth factor as an anti-angiogenic target for cancer therapy. Curr. Drug Targets 2010, 11, 1000–1017. [Google Scholar] [CrossRef] [PubMed]
- Nonaka, Y.; Yoshida, W.; Abe, K.; Ferri, S.; Schulze, H.; Bachmann, T.T.; Ikebukuro, K. Affinity improvement of a VEGF aptamer by in silico maturation for a sensitive VEGF-detection system. Anal. Chem. 2013, 85, 1132–1137. [Google Scholar] [CrossRef] [PubMed]
- Nathan, C. Neutrophils and immunity: Challenges and opportunities. Nat. Rev. Immunol. 2006, 6, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Burg, N.D.; Pillinger, M.H. The Neutrophil: Function and regulation in innate and humoral immunity. Clin. Immunol. 2001, 99, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Takahashi, S.; Mizumoto, T.; Harao, M.; Akizuki, M.; Takasugi, M.; Fukutomi, T.; Yamashita, J. Neutrophil elastase and cancer. Surg. Oncol. 2006, 15, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Ho, A.S.; Chen, C.H.; Cheng, C.C.; Wang, C.C.; Lin, H.C.; Luo, T.Y.; Lien, G.S.; Chang, J. Neutrophil elastase as a diagnostic marker and therapeutic target in colorectal cancers. Oncotarget 2014, 5, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Charlton, J.; Kirschenheuter, G.P.; Smith, D. Highly potent irreversible inhibitors of neutrophil elastase generated by selection from a randomized DNA-valine phosphonate library. Biochemistry 1997, 36, 3018–3026. [Google Scholar] [CrossRef] [PubMed]
- Shin, W.S.; Maeng, Y.S.; Jung, J.W.; Min, J.K.; Kwon, Y.G.; Lee, S.T. Soluble PTK7 inhibits tube formation, migration, and invasion of endothelial cells and angiogenesis. Biochem. Biophys. Res. Commun. 2008, 371, 793–798. [Google Scholar] [CrossRef] [PubMed]
- Prebet, T.; Lhoumeau, A.C.; Arnoulet, C.; Aulas, A.; Marchetto, S.; Audebert, S.; Puppo, F.; Chabannon, C.; Sainty, D.; Santoni, M.J.; et al. The cell polarity PTK7 receptor acts as a modulator of the chemotherapeutic response in acute myeloid leukemia and impairs clinical outcome. Blood 2010, 116, 2315–2323. [Google Scholar] [CrossRef] [PubMed]
- Na, H.W.; Shin, W.S.; Ludwig, A.; Lee, S.T. The cytosolic domain of protein-tyrosine kinase 7 (PTK7), generated from sequential cleavage by a disintegrin and metalloprotease 17 (ADAM17) and γ-secretase, enhances cell proliferation and migration in colon cancer cells. J. Biol. Chem. 2012, 287, 25001–25009. [Google Scholar] [CrossRef] [PubMed]
- Golubkov, V.S.; Prigozhina, N.L.; Zhang, Y.; Stoletov, K.; Lewis, J.D.; Schwartz, P.E.; Hoffman, R.M.; Strongin, A.Y. Protein-tyrosine pseudokinase 7 (PTK7) directs cancer cell motility and metastasis. J. Biol. Chem. 2014, 289, 24238–24239. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Qi, S.; Yang, X.; Chen, W. Prognostic significance of PTK7 in human malignancies. Histol. Histopathol. 2017, 11933. [Google Scholar] [CrossRef]
- Shangguan, D.; Tang, Z.; Mallikaratchy, P.; Xiao, Z.; Tan, W. Aptamers Selected by Cell-SELEX for Theranostics; Tan, W., Fang, X., Eds.; Springer: Berlin, Germany, 2007; Volume 8, pp. 603–606. [Google Scholar]
- Shangguan, D.; Cao, Z.; Meng, L.; Mallikaratchy, P.; Sefah, K.; Wang, H.; Li, Y.; Tan, W. Cell-specific aptamer probes for membrane protein elucidation in cancer cells. J. Proteome Res. 2008, 7, 2133–2139. [Google Scholar] [CrossRef] [PubMed]
- Dhar, S.; Daniel, W.L.; Giljohann, D.A.; Mirkin, C.A.; Lippard, S.J. Polyvalent oligonucleotide gold nanoparticle conjugates as delivery vehicles for platinum(IV) warheads. J. Am. Chem. Soc. 2009, 131, 14652–14653. [Google Scholar] [CrossRef] [PubMed]
- Perera, S.; Uddin, M.; Hayes, J.A. Salivary lysozyme: A noninvasive marker for the study of the effects of stress of natural immunity. Int. J. Behav. Med. 1997, 4, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Sava, G.; Benetti, A.; Ceschia, V.; Pacor, S. Lysozyme and cancer: Role of exogenous lysozyme as anticancer agent (review). Anticancer Res. 1989, 9, 583–591. [Google Scholar] [PubMed]
- Vizoso, F.; Plaza, E.; Vazquez, J.; Serra, C.; Lamelas, M.L.; Gonzalez, L.O.; Merino, A.M.; Mendez, J. Lysozyme expression by breast carcinomas, correlation with clinicopathologic parameters, and prognostic significance. Ann. Surg. Oncol. 2001, 8, 667–674. [Google Scholar] [CrossRef] [PubMed]
- Serra, C.; Vizoso, F.; Alonso, L.; Rodriguez, J.; Gonzalez, L.; Fernandez, M.; Lamelas, M.; Sanchez, L.; Garcia-Muniz, J.; Baltasar, A.; et al. Expression and prognostic significance of lysozyme in male breast cancer. Breast Cancer Res. 2002, 4, R16. [Google Scholar] [CrossRef] [PubMed]
- Levinson, S.S.; Elin, R.J.; Yam, L. Light chain proteinuria and lysozymuria in a patient with acute monocytic leukemia. Clin. Chem. 2002, 48, 1131–1132. [Google Scholar] [PubMed]
- Tran, D.T.; Janssen, K.P.F.; Pollet, J.; Lammertyn, E.; Anné, J.; Van Schepdael, A.; Lammertyn, J. Selection and characterization of DNA aptamers for egg white lysozyme. Molecules 2010, 15, 1127–1140. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.C.; Ellington, A.D. Automated selection of anti-protein aptamers. Biorgan. Med. Chem. 2001, 9, 2525–2531. [Google Scholar] [CrossRef]
- Teller, C.; Shimron, S.; Willner, I. Aptamer-DNAzyme hairpins for amplified biosensing. Anal. Chem. 2009, 81, 9114–9119. [Google Scholar] [CrossRef] [PubMed]
- Garland, M.; Yim, J.J.; Bogyo, M. A bright future for precision medicine: Advances in fluorescent chemical probe design and their clinical application. Cell Chem. Biol. 2016, 23, 122–136. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Yu, F.; Lv, C.; Choo, J.; Chen, L. Fluorescent chemical probes for accurate tumor diagnosis and targeting therapy. Chem. Soc. Rev. 2017, 46, 2237–2271. [Google Scholar] [CrossRef] [PubMed]
- Cibiel, A.; Pestourie, C.; Ducongé, F. In vivo uses of aptamers selected against cell surface biomarkers for therapy and molecular imaging. Biochimie 2012, 94, 1595–1606. [Google Scholar] [CrossRef] [PubMed]
- Dougherty, C.A.; Cai, W.; Hong, H. Applications of aptamers in targeted imaging: State of the art. Curr. Top. Med. Chem. 2015, 15, 1138–1152. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Estevez, M.C.; Zhu, Z.; Huang, Y.F.; Chen, Y.; Wang, L.; Tan, W. Using aptamer-conjugated fluorescence resonance energy transfer nanoparticles for multiplexed cancer cell monitoring. Anal. Chem. 2009, 81, 7009–7014. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Rajendran, R.; Jeong, M.-S.; Ko, H.Y.; Joo, J.Y.; Cho, S.; Chang, Y.W.; Kim, S. Bioimaging of targeting cancers using aptamer-conjugated carbon nanodots. Chem. Commun. 2013, 49, 6543–6545. [Google Scholar] [CrossRef] [PubMed]
- Jie, G.; Zhao, Y.; Qin, Y. A fluorescent polymeric quantum dot/aptamer superstructure and its application for imaging cancer cells. Chem. Asian J. 2014, 9, 1261–1264. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Ye, X.; He, X.; Wang, K.; Cui, W.; He, D.; Li, D.; Jia, X. Au@Ag/Au nanoparticles assembled with activatable aptamer probes as smart “nano-doctors” for image-guided cancer thermotherapy. Nanoscale 2014, 6, 8754–8761. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Tang, Z.; Kim, Y.; Nie, H.; Huang, Y.F.; He, X.; Deng, K.; Wang, K.; Tan, W. In vivo fluorescence imaging of tumors using molecular aptamers generated by cell-SELEX. Chem. Asian J. 2010, 5, 2209–2213. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Cui, W.; He, X.; Guo, Q.; Wang, K.; Ye, X.; Tang, J. Whole Cell-SELEX aptamers for highly specific fluorescence molecular imaging of carcinomas in vivo. PLoS ONE 2013, 8, e70476. [Google Scholar] [CrossRef] [PubMed]
- Bates, P.J.; Reyes-reyes, E.M.; Malik, M.T.; Murphy, E.M.; Toole, M.G.O.; Trent, J.O. G-quadruplex oligonucleotide AS1411 as a cancer-targeting agent : Uses and mechanisms. BBA Gen. Subj. 2017, 1861, 1414–1428. [Google Scholar] [CrossRef] [PubMed]
- Ai, J.; Li, T.; Li, B.; Xu, Y.; Li, D.; Liu, Z.; Wang, E. In situ labeling and imaging of cellular protein via a bi-functional anticancer aptamer and its fluorescent ligand. Anal. Chim. Acta 2012, 741, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Ai, J.; Xu, Y.; Lou, B.; Li, D.; Wang, E. Multifunctional AS1411-functionalized fluorescent gold nanoparticles for targeted cancer cell imaging and efficient photodynamic therapy. Talanta 2014, 118, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Chen, Z.Z.; Chen, M.Y.; Tang, H.W.; Pang, D.W. MUC-1 aptamer-conjugated dye-doped silica nanoparticles for MCF-7 cells detection. Biomaterials 2013, 34, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; He, X.; Wang, K.; Wu, X.; Ye, X.; Guo, Q.; Tan, W.; Qing, Z.; Yang, X.; Zhou, B. Activatable aptamer probe for contrast-enhanced in vivo cancer imaging based on cell membrane protein-triggered conformation alteration. Proc. Natl. Acad. Sci. USA 2011, 108, 3900–3905. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Shi, H.; He, X.; Wang, K.; Tang, J.; Chen, M.; Ye, X.; Xu, F.; Lei, Y. A versatile activatable fluorescence probing platform for cancer cells in vitro and in vivo based on self-assembled aptamer/carbon nanotube ensembles. Anal. Chem. 2014, 86, 9271–9277. [Google Scholar] [CrossRef] [PubMed]
- Motaghi, H.; Mehrgardi, M.A.; Bouvet, P. Carbon dots-AS1411 aptamer nanoconjugate for ultrasensitive spectrofluorometric detection of cancer cells. Sci. Rep. 2017, 7, 10513. [Google Scholar] [CrossRef] [PubMed]
- Yuan, B.; Sun, Y.; Guo, Q.; Huang, J.; Yang, X.; Chen, Y.; Wen, X.; Meng, X.; Liu, J.; Wang, K. High signal-to-background ratio detection of cancer cells with activatable strategy based on target-induced self-assembly of split aptamers. Anal. Chem. 2017, 89, 9347–9353. [Google Scholar] [CrossRef] [PubMed]
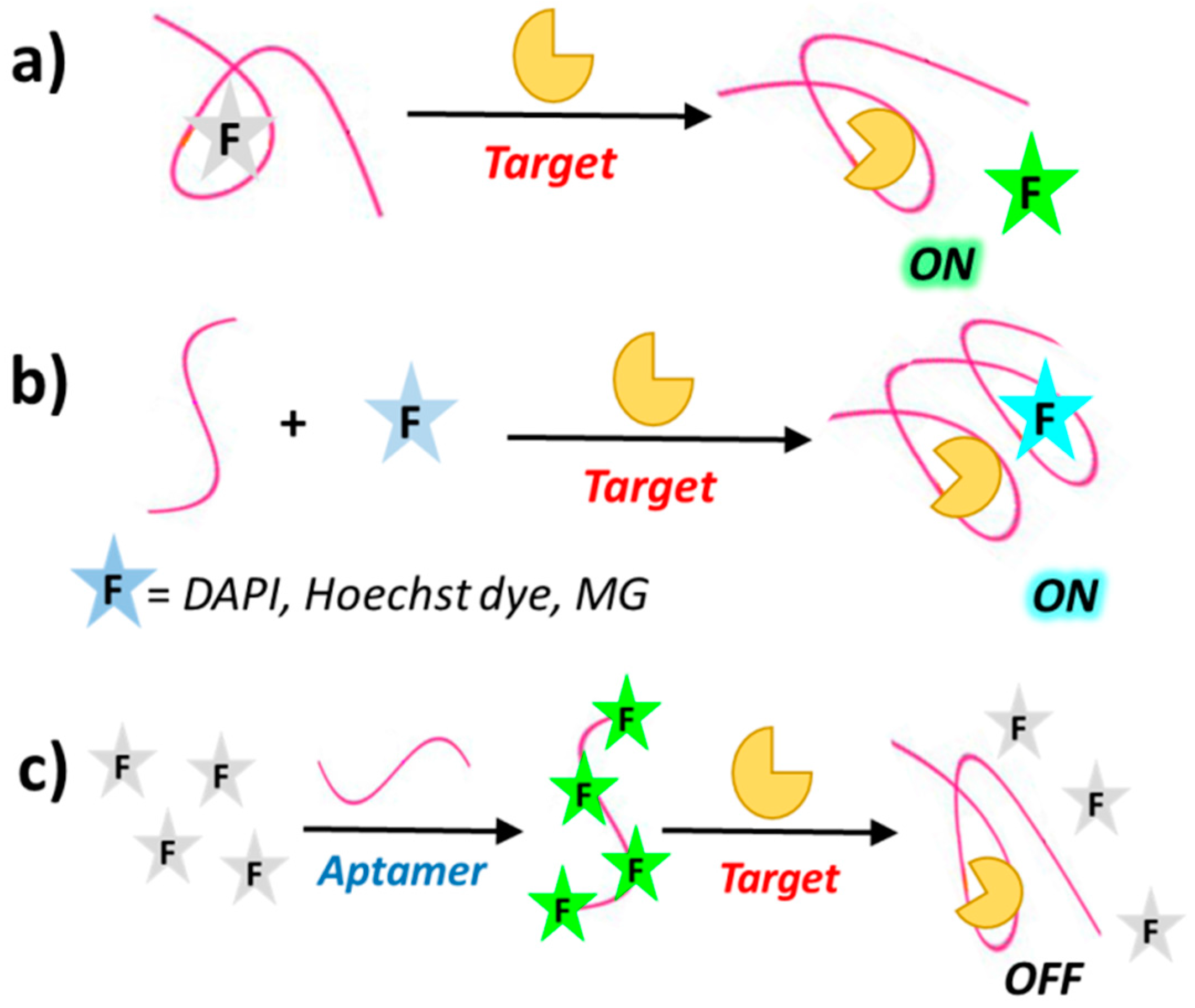
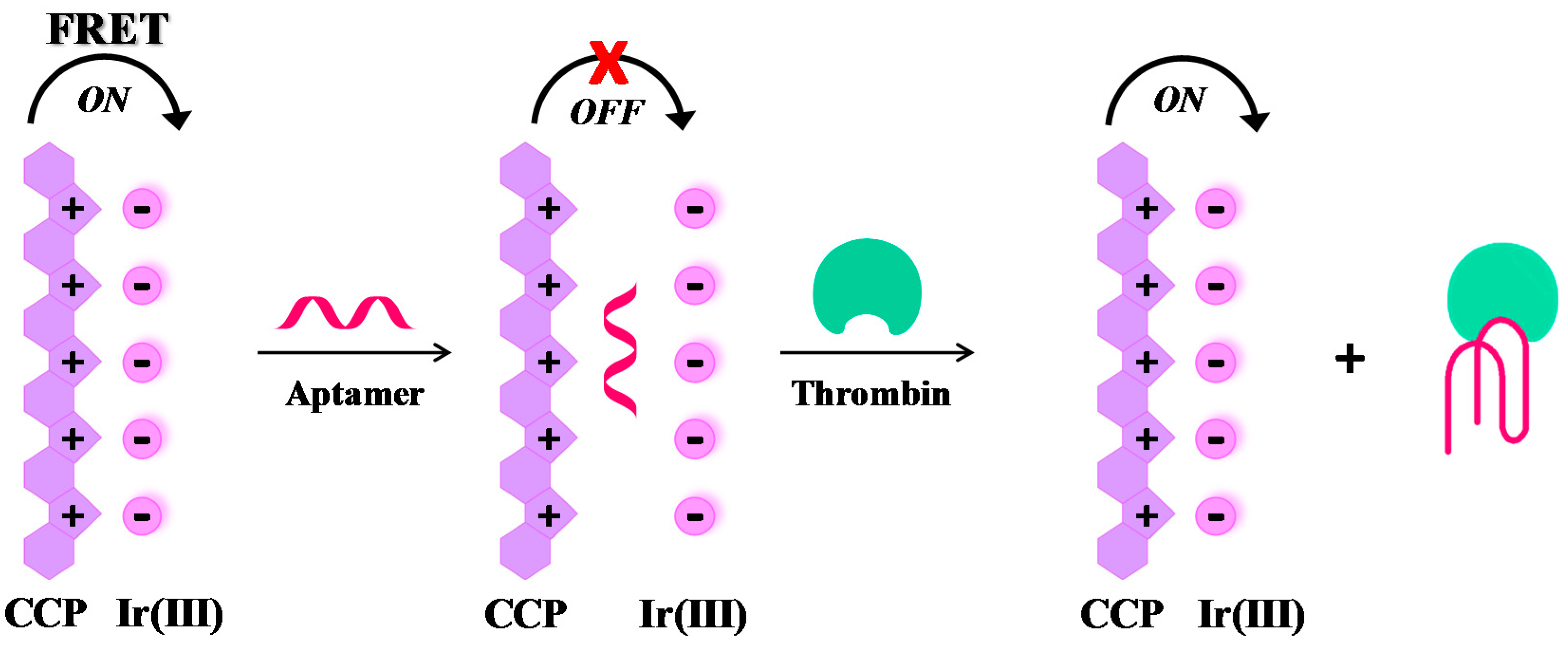
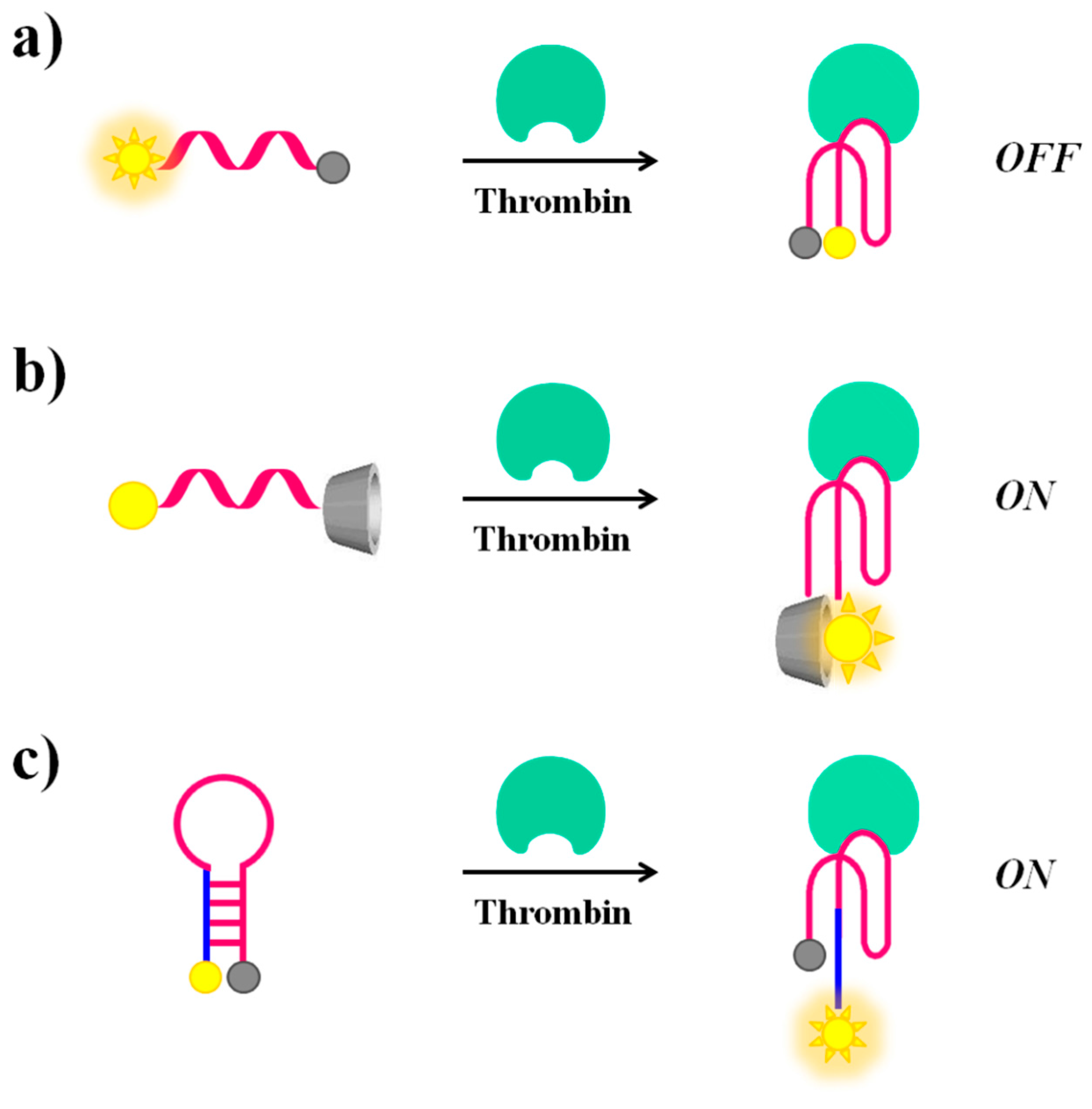

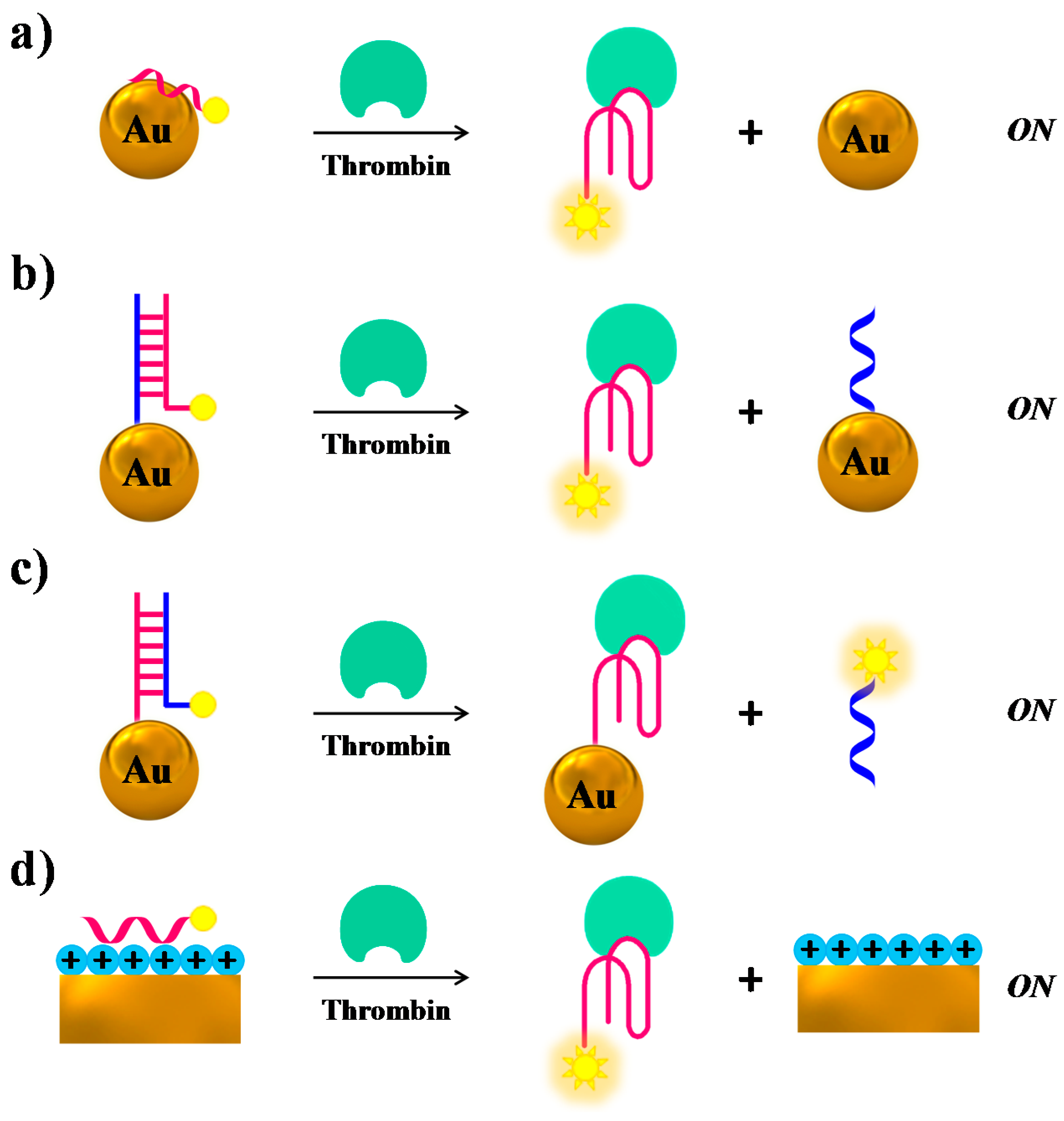
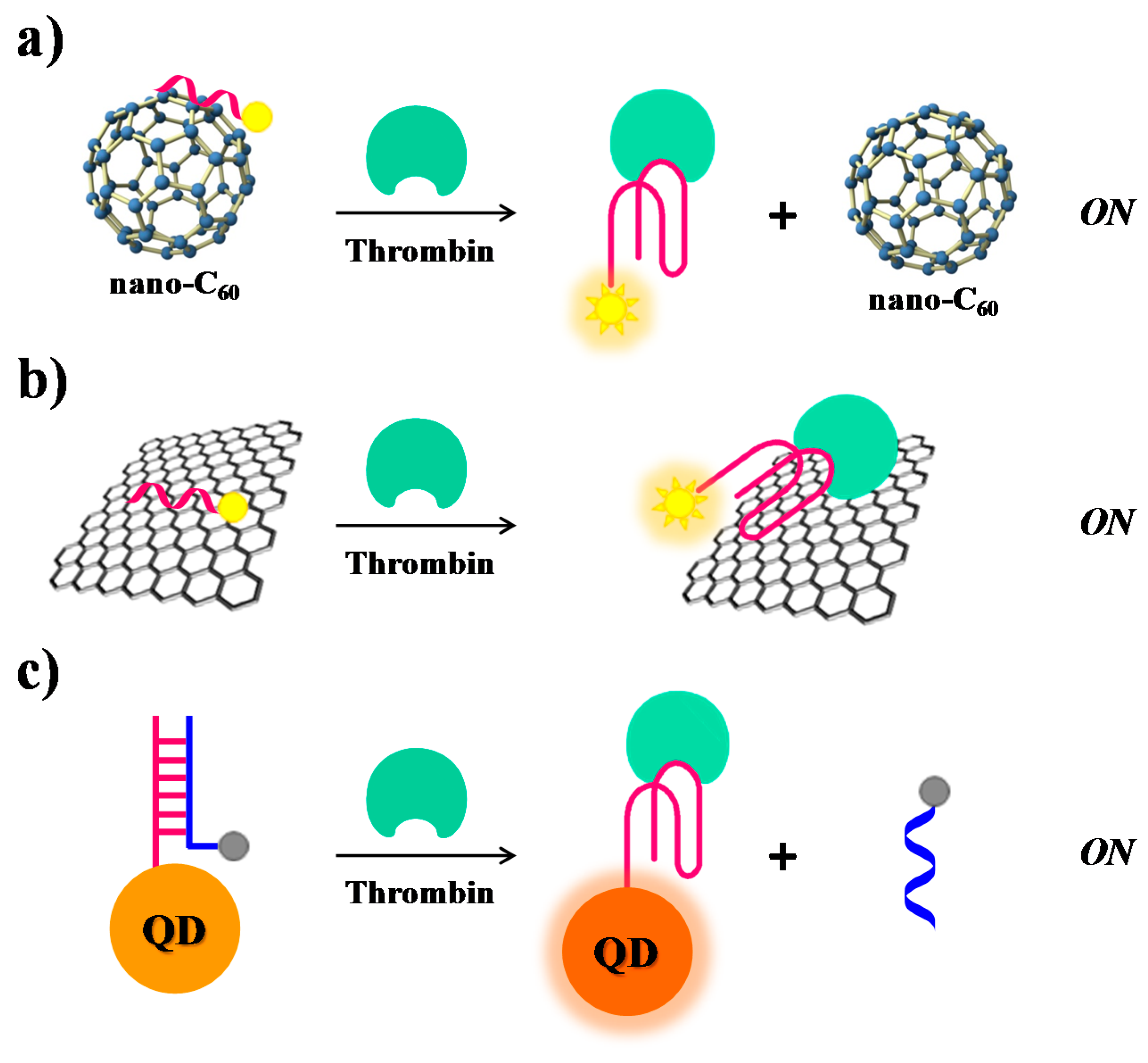

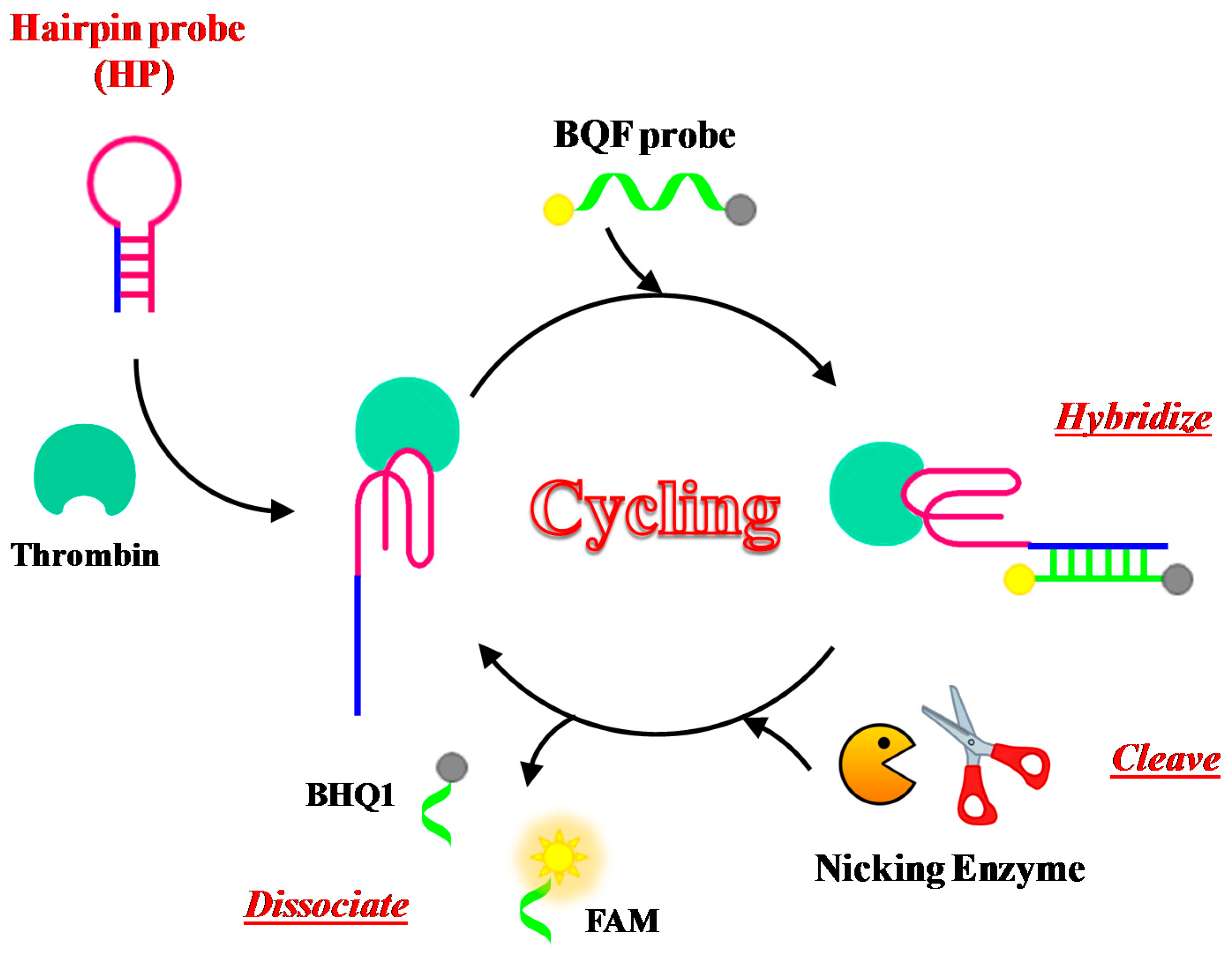



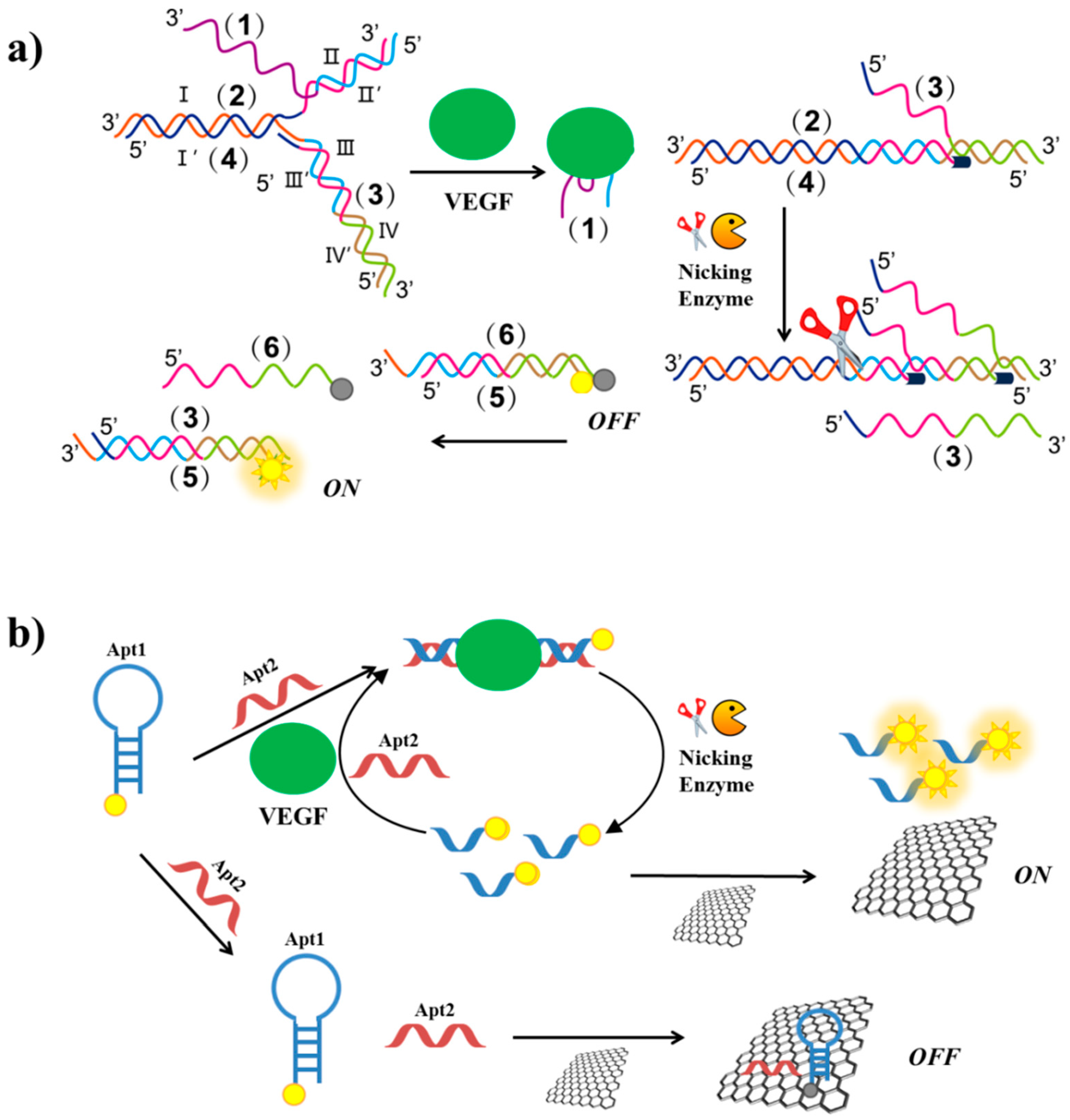
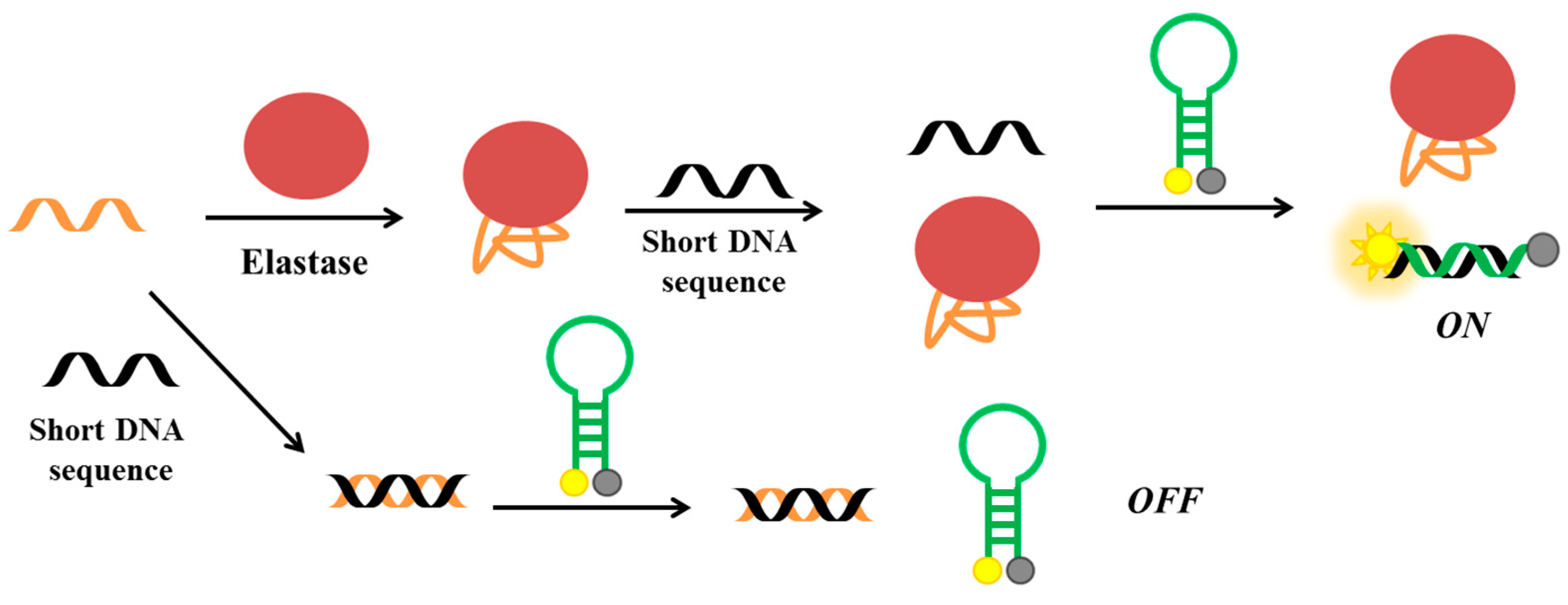
| Target | Labelling Method | Fluorophore a/Quencher b | Fluorescent Dye c | Mode | Sample | Limit of Detection | Reference |
|---|---|---|---|---|---|---|---|
| Thrombin | Label-free | -- | Ir(III) complex | Signal-ON | serum, urine, saliva | 0.05 pM | [97] |
| Label-free | -- | hemin | Signal-ON | buffer solution | 1 pM | [109] | |
| Label-free | -- | SYBR Gold | Signal-ON | serum | 680 nM | [110] | |
| Covalent | 6-carboxyfluorescein/dabcyl | -- | Signal-OFF | buffer solution | 112 pM | [98] | |
| Covalent | dansyl/β-cyclodextrin | -- | Signal-ON | buffer solution | n.d. | [99] | |
| Covalent | fluorescein/dabcyl | -- | Signal-ON | buffer solution | 0.2 μM | [102] | |
| Covalent | fluorescein/dabcyl | -- | Signal-ON | buffer solution | 3 μM | [103] | |
| Covalent | pyrrolo dC | --- | Signal-ON | buffer solution | 10 μM | [70] | |
| Covalent | 6-carboxyfluorescein/Iowa Black FQ | -- | Signal-OFF | buffer solution | 10 μM | [70] | |
| Covalent | 6-carboxytetramethylrhodamine/AuNP | -- | Signal-ON | buffer solution | 0.14 nM | [104] | |
| Covalent | Cy3/AuNP | Signal-ON | buffer solution | 10 pM | [105] | ||
| Covalent | 6-carboxyfluorescein/SWCT | -- | Signal-ON | buffer solution | 1.8 nM | [106] | |
| Covalent | 6-carboxyfluorescein/nano-C60 | -- | Signal-ON | serum | 1 nM | [107] | |
| Covalent | 6-carboxyfluorescein/graphene | -- | Signal-ON | serum | 31.3 pM | [68] | |
| Covalent | Cy3/MNP | -- | Signal-ON | serum | 0.5 nM | [108] | |
| Covalent | Quantum dots/dabcyl | -- | Signal-ON | buffer solution | 1 μM | [111] | |
| Covalent | fluorescein/dabcyl | -- | Signal-OFF | cell extracts, plasma | 1 nM | [112] | |
| Covalent | 6-carboxyfluorescein/ Black Hole Quencher 1 | -- | Signal-ON | serum | 100 pM | [113] | |
| PDGF | Label-free | -- | NMM | Signal-ON | serum | 3.2 nM | [114] |
| Label-free | -- | TOTO | Signal-OFF | buffer solution | 100 pM | [115] | |
| Label-free | -- | TOTO | Signal-OFF | buffer solution | 5 pM | [116] | |
| Label-free | -- | SYBR Green I | Signal-ON | serum | 1.25 pM | [117] | |
| Covalent | 6-amino fluorescein/dabcyl | -- | Signal-OFF | serum, cell culture media | 110 pM | [118] | |
| Covalent | fluorescein/graphene oxide | -- | Signal-ON | serum | 167 pM | [119] | |
| Covalent | pyrene | -- | Fluorescence emission shift | cell culture media | pM range | [120] | |
| Covalent | fluorescein | -- | Fluorescence anisotropy increase | buffer solution | 220 pM | [121] | |
| Covalent | tetramethylrhodamine | -- | Fluorescence anisotropy decrease | buffer solution | pM range | [122] | |
| Covalent | fluorescein/dabcyl | -- | Signal-OFF | cell culture media | pM range | [123] | |
| Angiogenin | Label free | -- | Alexa Fluor 488 | Fluorescence anisotropy increase | buffer solution | 6.3 nM | [124] |
| Covalent | fluorescein | -- | Fluorescence anisotropy increase | serum | 1 nM | [125] | |
| Covalent | 6-carboxyfluorescein/6-carboxytetramethyl-rhodamine | -- | Signal-ON | serum | 200 pM | [126] | |
| Covalent | Cy5 | -- | Signal-ON | cell culture media | n.d. | [127] | |
| Mucin | Label-free | -- | fluorescein | Signal-ON | serum | 3.33 pM | [128] |
| Covalent | Cy5/graphene oxide | -- | Signal-ON | serum | 28 nM | [129] | |
| Covalent | Cy3 | -- | Signal-ON | cell culture media | n.d. | [130] | |
| Covalent | MPA | -- | Signal-ON | cell culture media, nude mice | n.d. | [131] | |
| VEGF | Label-free | -- | 6-carboxyfluorescein | Signal-ON | serum | 3.5 pg/mL | [132] |
| Covalent | fluorescein | -- | Signal-ON | buffer solution | 1 pM | [133] | |
| Covalent | fluorescein | -- | Signal-ON | buffer solution | 25 nM | [134] | |
| Covalent | 6-carboxyfluorescein | -- | Fluorescence anisotropy increase | buffer solution | 320 pM | [135] | |
| Covalent | 6-carboxyfluorescein | -- | Signal-ON | serum | 250 pM | [136] | |
| Elastase | Covalent | fluorescein | -- | Signal-ON | cell culture media, rats | n.d. | [137] |
| Covalent | -- | fluorescein | Signal-ON | buffer solution | 47 pM | [138] | |
| PTK7 | Label-free | -- | Cy5 | Signal-ON | cell culture media | 1 pM | [139] |
| Covalent | Alexa Fluor 647 | -- | Signal-ON | cell culture media, nude mice | n.d. | [140] | |
| Lysozyme | Label-free | -- | 6-carboxyfluorescein | Signal-ON | saliva | 200 pM | [141] |
| Label-free | -- | 6-carboxyfluorescein | Signal-ON | buffer solution | 0.125 µg/mL | [142] | |
| Label-free | -- | Pyrene | Fluorescence emission shift | serum | 200 pM | [143] | |
| Covalent | 6-carboxyfluorescein | -- | Fluorescence anisotropy increase | saliva | 4.9 nM | [144] | |
| Covalent | 6-carboxyfluorescein | -- | Signal-ON | buffer solution | 0.80 µg/mL | [145] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Musumeci, D.; Platella, C.; Riccardi, C.; Moccia, F.; Montesarchio, D. Fluorescence Sensing Using DNA Aptamers in Cancer Research and Clinical Diagnostics. Cancers 2017, 9, 174. https://doi.org/10.3390/cancers9120174
Musumeci D, Platella C, Riccardi C, Moccia F, Montesarchio D. Fluorescence Sensing Using DNA Aptamers in Cancer Research and Clinical Diagnostics. Cancers. 2017; 9(12):174. https://doi.org/10.3390/cancers9120174
Chicago/Turabian StyleMusumeci, Domenica, Chiara Platella, Claudia Riccardi, Federica Moccia, and Daniela Montesarchio. 2017. "Fluorescence Sensing Using DNA Aptamers in Cancer Research and Clinical Diagnostics" Cancers 9, no. 12: 174. https://doi.org/10.3390/cancers9120174






