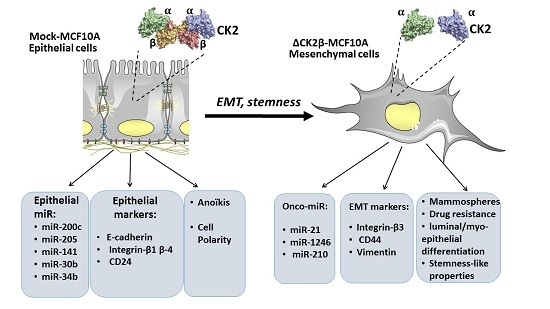
Stem Cell-Like Properties of CK2β-down Regulated Mammary Cells
Abstract
:1. Introduction
2. Results and Discussion
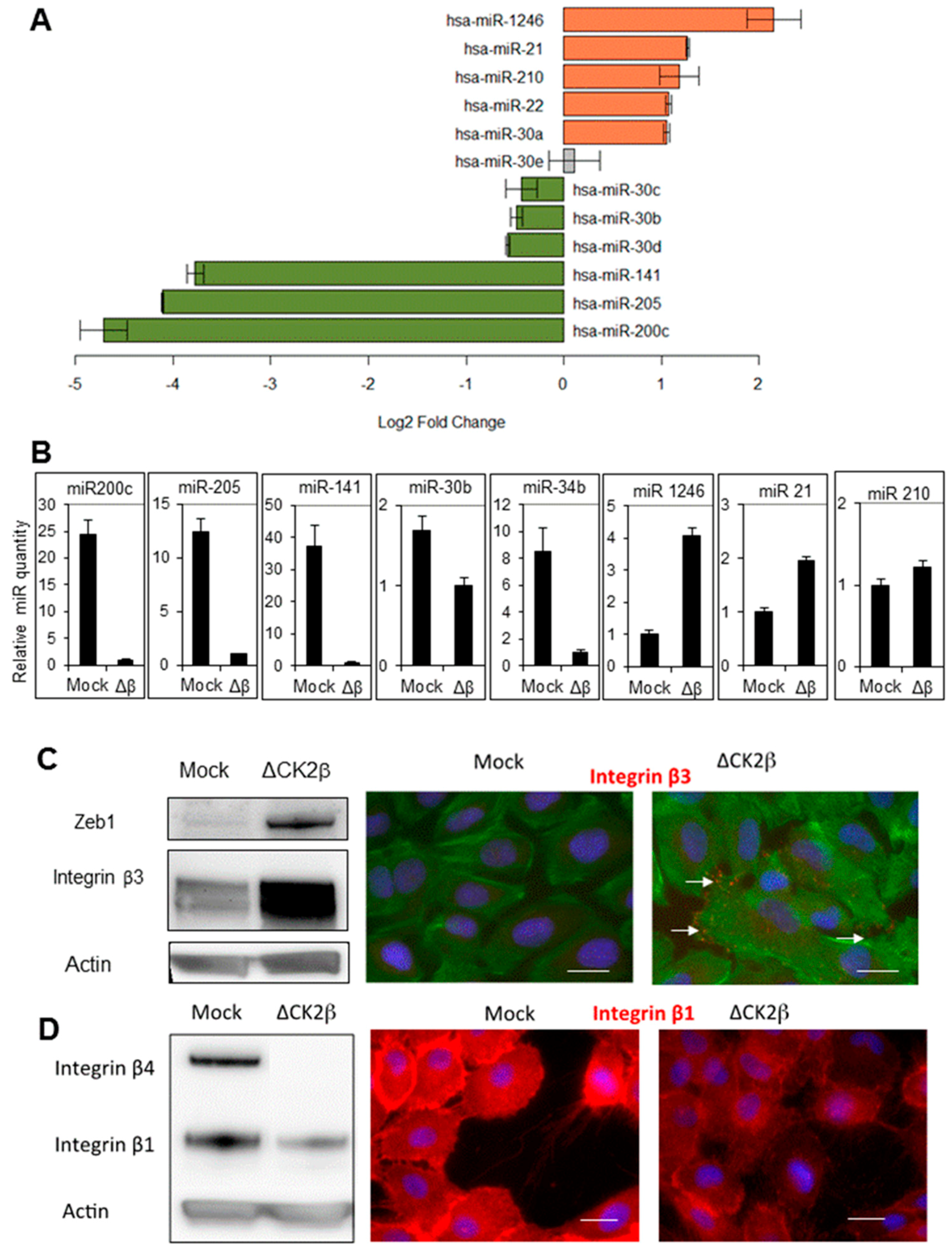
2.1. ∆CK2β-MCF10A Cells Have Reduced Expression of EMT miRNAs
2.2. ∆CK2β-MCF10A Cells Have Increased Expression of Specific miRNAs
2.3. Transcriptomic Analysis
2.4. ∆CK2β-MCF10A Cells Have Hallmarks of Stem Cells
2.4.1. CD44/CD24 Stem Cell Markers
2.4.2. Cell Proliferation and Viability
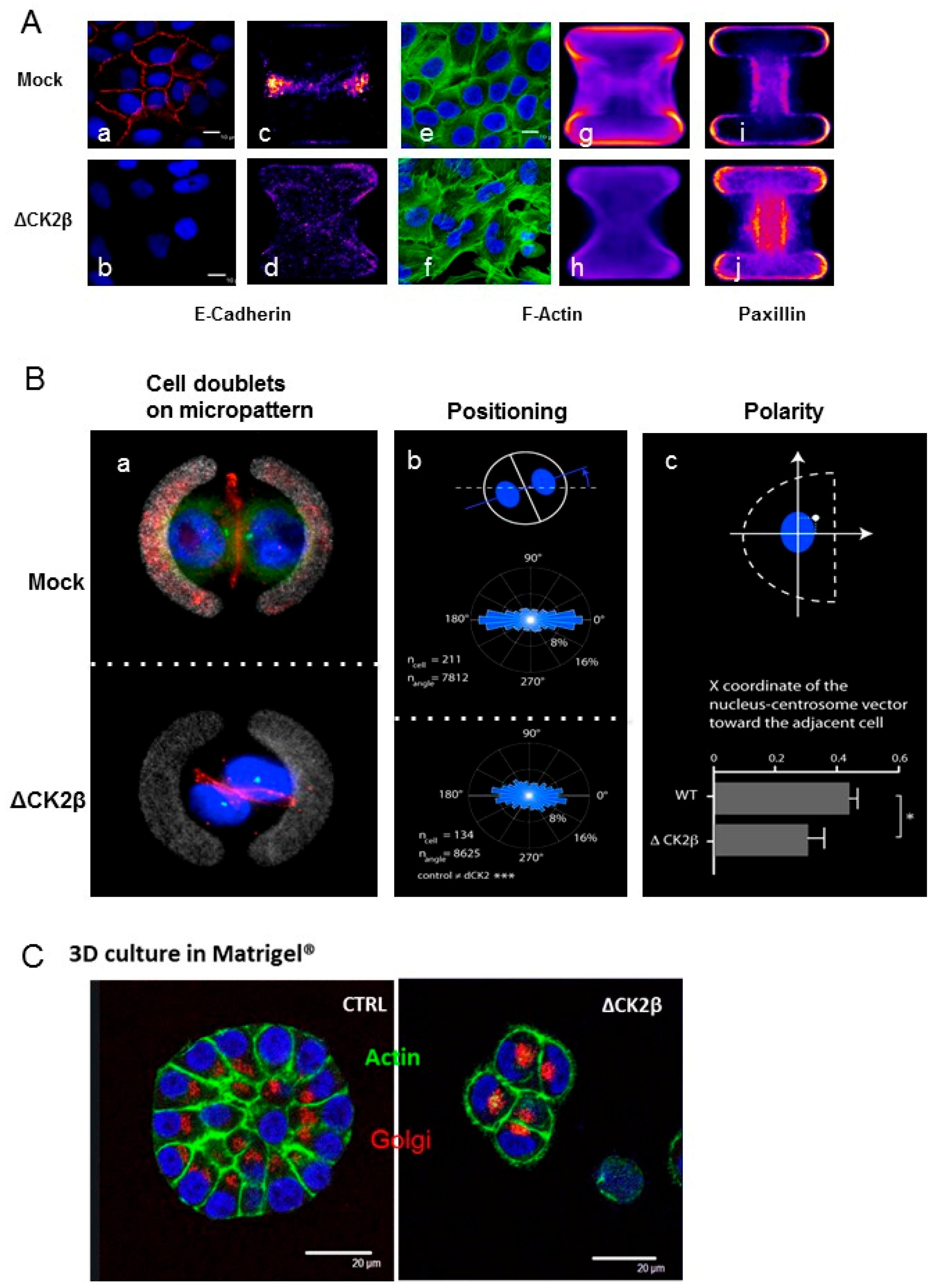
2.5. Cell Positioning and Polarity
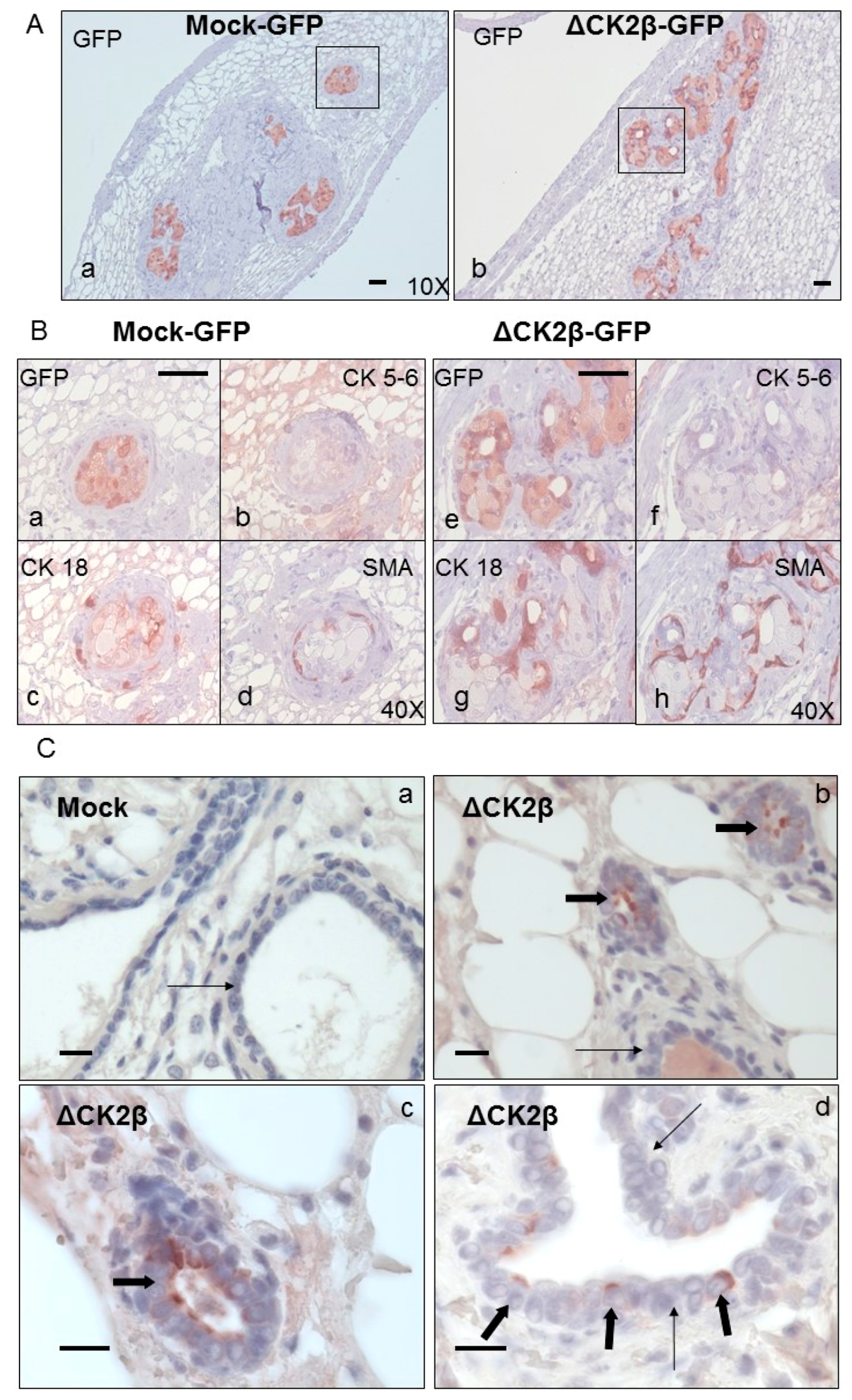
Analysis of Cell Phenotypes by Orthotopic Engraftment
3. Material & Methods
3.1. Cell Culture and Retroviral Infection
3.2. microRNA Profiling
3.3. Fluorescence-Activated Cell Sorting
3.4. Western Blot Analysis
3.5. Quantitative Real-Time PCR
3.6. Characterization of Resistance to Cytotoxic Agents
3.7. Colony Assay
3.8. Mouse Injection
3.9. Histological and Immunohistochemical Analysis
3.10. Gene Expression Microarray Analysis
3.11. Cell Micropatterning
3.12. Immunofluorescence
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Kreutzer, J.N.; Ruzzene, M.; Guerra, B. Enhancing chemosensitivity to gemcitabine via rna interference targeting the catalytic subunits of protein kinase ck2 in human pancreatic cancer cells. BMC Cancer 2010, 10, 440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deshiere, A.; Duchemin-Pelletier, E.; Spreux, E.; Ciais, D.; Forcet, C.; Cochet, C.; Filhol, O. Regulation of epithelial to mesenchymal transition: Ck2β on stage. Mol. Cell. Biochem. 2011, 356, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Filhol, O.; Nueda, A.; Martel, V.; Gerber-Scokaert, D.; Benitez, M.J.; Souchier, C.; Saoudi, Y.; Cochet, C. Live-cell fluorescence imaging reveals the dynamics of protein kinase ck2 individual subunits. Mol. Cell. Biol. 2003, 23, 975–987. [Google Scholar] [CrossRef] [PubMed]
- Filhol, O.; Cochet, C. Protein kinase ck2 in health and disease: Cellular functions of protein kinase ck2: A dynamic affair. Cell. Mol. Life Sci. 2009, 66, 1830–1839. [Google Scholar] [CrossRef] [PubMed]
- Mani, S.A.; Guo, W.; Liao, M.J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef] [PubMed]
- Morel, A.P.; Lievre, M.; Thomas, C.; Hinkal, G.; Ansieau, S.; Puisieux, A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS ONE 2008, 3, e2888. [Google Scholar] [CrossRef] [PubMed]
- Deshiere, A.; Duchemin-Pelletier, E.; Spreux, E.; Ciais, D.; Combes, F.; Vandenbrouck, Y.; Coute, Y.; Mikaelian, I.; Giusiano, S.; Charpin, C.; et al. Unbalanced expression of ck2 kinase subunits is sufficient to drive epithelial-to-mesenchymal transition by snail1 induction. Oncogene 2013, 32, 1373–1383. [Google Scholar] [CrossRef] [PubMed]
- Golden, D.; Cantley, L.G. Casein kinase 2 prevents mesenchymal transformation by maintaining foxc2 in the cytoplasm. Oncogene 2015, 34, 4702–4712. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Hwan Kim, S. Ck2 inhibitor cx-4945 blocks tgf-beta1-induced epithelial-to-mesenchymal transition in a549 human lung adenocarcinoma cells. PLoS ONE 2013, 8, e74342. [Google Scholar] [CrossRef]
- Ferrer-Font, L.; Alcaraz, E.; Plana, M.; Candiota, A.P.; Itarte, E.; Arus, C. Protein kinase ck2 content in gl261 mouse glioblastoma. Pathol. Oncol. Res. 2016, 22, 633–637. [Google Scholar] [CrossRef] [PubMed]
- Roelants, C.; Giacosa, S.; Duchemin-Pelletier, E.; McLeer-Florin, A.; Tisseyre, C.; Aubert, C.; Champelovier, P.; Boutonnat, J.; Descotes, J.L.; Rambeaud, J.-J.; et al. Dysregulated expression of protein kinase ck2 in renal cancer. In Protein Kinase CK2 Cellular Function in Normal and Disease States; Ahmed, K., Issinger, O.-G., Szyszka, R., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 241–257. [Google Scholar]
- Blanpain, C.; Simons, B.D. Unravelling stem cell dynamics by lineage tracing. Nat. Rev. Mol. Cell Biol. 2013, 14, 489–502. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. Microrna expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The mir-200 family and mir-205 regulate epithelial to mesenchymal transition by targeting zeb1 and sip1. Nat. Cell Biol. 2008, 10, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Park, S.M.; Gaur, A.B.; Lengyel, E.; Peter, M.E. The mir-200 family determines the epithelial phenotype of cancer cells by targeting the e-cadherin repressors zeb1 and zeb2. Genes Dev. 2008, 22, 894–907. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Deng, H.; Yao, H.; Liu, Q.; Su, F.; Song, E. Mir-30 reduction maintains self-renewal and inhibits apoptosis in breast tumor-initiating cells. Oncogene 2010, 29, 4194–4204. [Google Scholar] [CrossRef] [PubMed]
- Li, X.J.; Ren, Z.J.; Tang, J.H. Microrna-34a: A potential therapeutic target in human cancer. Cell Death Dis. 2014, 5, e1327. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.Z.; Li, C.; Chen, Q.; Jing, Y.; Carpenter, R.; Jiang, Y.; Kung, H.F.; Lai, L.; Jiang, B.H. Mir-21 induced angiogenesis through akt and erk activation and hif-1alpha expression. PLoS ONE 2011, 6, e19139. [Google Scholar] [CrossRef]
- Yan, L.X.; Huang, X.F.; Shao, Q.; Huang, M.Y.; Deng, L.; Wu, Q.L.; Zeng, Y.X.; Shao, J.Y. Microrna mir-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA 2008, 14, 2348–2360. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.; Luo, W.; Sun, X.; Lin, J.; Wang, M.; Zhang, Y.; Luo, W.; Zhang, Y. Microrna21 promotes migration and invasion of glioma cells via activation of sox2 and betacatenin signaling. Mol. Med. Rep. 2017, 15, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Nguyen, H.T.; Zhuang, Y.; Lin, Y.; Flemington, E.K.; Guo, W.; Guenther, J.; Burow, M.E.; Morris, G.F.; Sullivan, D.; et al. Post-transcriptional up-regulation of mir-21 by type i collagen. Mol. Carcinog. 2011, 50, 563–570. [Google Scholar] [CrossRef] [PubMed]
- Ferraro, A.; Kontos, C.K.; Boni, T.; Bantounas, I.; Siakouli, D.; Kosmidou, V.; Vlassi, M.; Spyridakis, Y.; Tsipras, I.; Zografos, G.; et al. Epigenetic regulation of mir-21 in colorectal cancer: Itgb4 as a novel mir-21 target and a three-gene network (mir-21-itgbeta4-pdcd4) as predictor of metastatic tumor potential. Epigenetics 2014, 9, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Selcuklu, S.D.; Donoghue, M.T.; Kerin, M.J.; Spillane, C. Regulatory interplay between mir-21, jag1 and 17beta-estradiol (e2) in breast cancer cells. Biochem. Biophys. Res. Commun. 2012, 423, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.C.; Chin, T.M.; Yang, H.; Nga, M.E.; Lunny, D.P.; Lim, E.K.; Sun, L.L.; Pang, Y.H.; Leow, Y.N.; Malusay, S.R.; et al. Tumour-initiating cell-specific mir-1246 and mir-1290 expression converge to promote non-small cell lung cancer progression. Nat. Commun. 2016, 7, 11702. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.C.; Banerjee, J.; Choi, S.Y.; Sen, C.K. Mir-210: The master hypoxamir. Microcirculation 2012, 19, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Takeshita, F.; Hino, Y.; Fukunaga, S.; Kudo, Y.; Tamaki, A.; Matsunaga, J.; Takahashi, R.U.; Takata, T.; Shimamoto, A.; et al. Mir-22 represses cancer progression by inducing cellular senescence. J. Cell Biol. 2011, 193, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Polyak, K.; Weinberg, R.A. Transitions between epithelial and mesenchymal states: Acquisition of malignant and stem cell traits. Nat. Rev. Cancer 2009, 9, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Charafe-Jauffret, E.; Ginestier, C.; Monville, F.; Finetti, P.; Adelaide, J.; Cervera, N.; Fekairi, S.; Xerri, L.; Jacquemier, J.; Birnbaum, D.; et al. Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene 2006, 25, 2273–2284. [Google Scholar] [CrossRef] [PubMed]
- Sarrio, D.; Rodriguez-Pinilla, S.M.; Hardisson, D.; Cano, A.; Moreno-Bueno, G.; Palacios, J. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008, 68, 989–997. [Google Scholar] [CrossRef] [PubMed]
- Gotzmann, J.; Fischer, A.N.; Zojer, M.; Mikula, M.; Proell, V.; Huber, H.; Jechlinger, M.; Waerner, T.; Weith, A.; Beug, H.; et al. A crucial function of pdgf in tgf-beta-mediated cancer progression of hepatocytes. Oncogene 2006, 25, 3170–3185. [Google Scholar] [CrossRef] [PubMed]
- Jechlinger, M.; Sommer, A.; Moriggl, R.; Seither, P.; Kraut, N.; Capodiecci, P.; Donovan, M.; Cordon-Cardo, C.; Beug, H.; Grunert, S. Autocrine pdgfr signaling promotes mammary cancer metastasis. J. Clin. Investig. 2006, 116, 1561–1570. [Google Scholar] [CrossRef] [PubMed]
- Boquest, A.C.; Shahdadfar, A.; Fronsdal, K.; Sigurjonsson, O.; Tunheim, S.H.; Collas, P.; Brinchmann, J.E. Isolation and transcription profiling of purified uncultured human stromal stem cells: Alteration of gene expression after in vitro cell culture. Mol. Biol. Cell 2005, 16, 1131–1141. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.; Wu, D.; Pal, B.; Bouras, T.; Asselin-Labat, M.L.; Vaillant, F.; Yagita, H.; Lindeman, G.J.; Smyth, G.K.; Visvader, J.E. Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast Cancer Res. 2010, 12, R21. [Google Scholar] [CrossRef] [PubMed]
- Pece, S.; Tosoni, D.; Confalonieri, S.; Mazzarol, G.; Vecchi, M.; Ronzoni, S.; Bernard, L.; Viale, G.; Pelicci, P.G.; Di Fiore, P.P. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell 2010, 140, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Schuetz, C.S.; Bonin, M.; Clare, S.E.; Nieselt, K.; Sotlar, K.; Walter, M.; Fehm, T.; Solomayer, E.; Riess, O.; Wallwiener, D.; et al. Progression-specific genes identified by expression profiling of matched ductal carcinomas in situ and invasive breast tumors, combining laser capture microdissection and oligonucleotide microarray analysis. Cancer Res. 2006, 66, 5278–5286. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wyckoff, J.B.; Goswami, S.; Wang, Y.; Sidani, M.; Segall, J.E.; Condeelis, J.S. Coordinated regulation of pathways for enhanced cell motility and chemotaxis is conserved in rat and mouse mammary tumors. Cancer Res. 2007, 67, 3505–3511. [Google Scholar] [CrossRef] [PubMed]
- Poola, I.; DeWitty, R.L.; Marshalleck, J.J.; Bhatnagar, R.; Abraham, J.; Leffall, L.D. Identification of mmp-1 as a putative breast cancer predictive marker by global gene expression analysis. Nat. Med. 2005, 11, 481–483. [Google Scholar] [CrossRef] [PubMed]
- Dontu, G.; Al-Hajj, M.; Abdallah, W.M.; Clarke, M.F.; Wicha, M.S. Stem cells in normal breast development and breast cancer. Cell Prolif. 2003, 36 (Suppl. 1), 59–72. [Google Scholar] [CrossRef] [PubMed]
- Sleeman, K.E.; Kendrick, H.; Ashworth, A.; Isacke, C.M.; Smalley, M.J. Cd24 staining of mouse mammary gland cells defines luminal epithelial, myoepithelial/basal and non-epithelial cells. Breast Cancer Res. 2006, 8, R7. [Google Scholar] [CrossRef] [PubMed]
- Zoller, M. Cd44: Can a cancer-initiating cell profit from an abundantly expressed molecule? Nat. Rev. Cancer 2011, 11, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Zeilstra, J.; Joosten, S.P.; Dokter, M.; Verwiel, E.; Spaargaren, M.; Pals, S.T. Deletion of the wnt target and cancer stem cell marker cd44 in apc(min/+) mice attenuates intestinal tumorigenesis. Cancer Res. 2008, 68, 3655–3661. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Weinberg, R.A. Epithelial-mesenchymal transition: At the crossroads of development and tumor metastasis. Dev. Cell 2008, 14, 818–829. [Google Scholar] [CrossRef] [PubMed]
- Bhat-Nakshatri, P.; Appaiah, H.; Ballas, C.; Pick-Franke, P.; Goulet, R., Jr.; Badve, S.; Srour, E.F.; Nakshatri, H. Slug/snai2 and tumor necrosis factor generate breast cells with cd44+/cd24- phenotype. BMC Cancer 2010, 10, 411. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Cong, Y.; Wang, D.; Sun, Y.; Deng, L.; Liu, Y.; Martin-Trevino, R.; Shang, L.; McDermott, S.P.; Landis, M.D.; et al. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Rep. 2014, 2, 78–91. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.B.; Onder, T.T.; Jiang, G.; Tao, K.; Kuperwasser, C.; Weinberg, R.A.; Lander, E.S. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell 2009, 138, 645–659. [Google Scholar] [CrossRef] [PubMed]
- Yauch, R.L.; Januario, T.; Eberhard, D.A.; Cavet, G.; Zhu, W.; Fu, L.; Pham, T.Q.; Soriano, R.; Stinson, J.; Seshagiri, S.; et al. Epithelial versus mesenchymal phenotype determines in vitro sensitivity and predicts clinical activity of erlotinib in lung cancer patients. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2005, 11, 8686–8698. [Google Scholar] [CrossRef] [PubMed]
- Tseng, Q.; Duchemin-Pelletier, E.; Deshiere, A.; Balland, M.; Guillou, H.; Filhol, O.; Thery, M. Spatial organization of the extracellular matrix regulates cell-cell junction positioning. Proc. Natl. Acad. Sci. USA 2012, 109, 1506–1511. [Google Scholar] [CrossRef] [PubMed]
- Burute, M.; Prioux, M.; Blin, G.; Truchet, S.; Letort, G.; Tseng, Q.; Bessy, T.; Lowell, S.; Young, J.; Filhol, O.; et al. Polarity reversal by centrosome repositioning primes cell scattering during epithelial-to-mesenchymal transition. Dev. Cell 2017, 40, 168–184. [Google Scholar] [CrossRef] [PubMed]
- Deshiere, A.; Theis-Febvre, N.; Martel, V.; Cochet, C.; Filhol, O. Protein kinase ck2 and cell polarity. Mol. Cell. Biochem. 2008, 316, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Debnath, J.; Muthuswamy, S.K.; Brugge, J.S. Morphogenesis and oncogenesis of mcf-10a mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 2003, 30, 256–268. [Google Scholar] [CrossRef]
- Reich, M.; Liefeld, T.; Gould, J.; Lerner, J.; Tamayo, P.; Mesirov, J.P. GenePattern 2.0. Nat. Genet. 2006, 38, 500–501. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Tam, W.L.; Shibue, T.; Kaygusuz, Y.; Reinhardt, F.; Ng Eaton, E.; Weinberg, R.A. Distinct emt programs control normal mammary stem cells and tumour-initiating cells. Nature 2015, 525, 256–260. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duchemin-Pelletier, E.; Baulard, M.; Spreux, E.; Prioux, M.; Burute, M.; Mograbi, B.; Guyon, L.; Théry, M.; Cochet, C.; Filhol, O. Stem Cell-Like Properties of CK2β-down Regulated Mammary Cells. Cancers 2017, 9, 114. https://doi.org/10.3390/cancers9090114
Duchemin-Pelletier E, Baulard M, Spreux E, Prioux M, Burute M, Mograbi B, Guyon L, Théry M, Cochet C, Filhol O. Stem Cell-Like Properties of CK2β-down Regulated Mammary Cells. Cancers. 2017; 9(9):114. https://doi.org/10.3390/cancers9090114
Chicago/Turabian StyleDuchemin-Pelletier, Eve, Megghane Baulard, Elodie Spreux, Magali Prioux, Mithila Burute, Baharia Mograbi, Laurent Guyon, Manuel Théry, Claude Cochet, and Odile Filhol. 2017. "Stem Cell-Like Properties of CK2β-down Regulated Mammary Cells" Cancers 9, no. 9: 114. https://doi.org/10.3390/cancers9090114