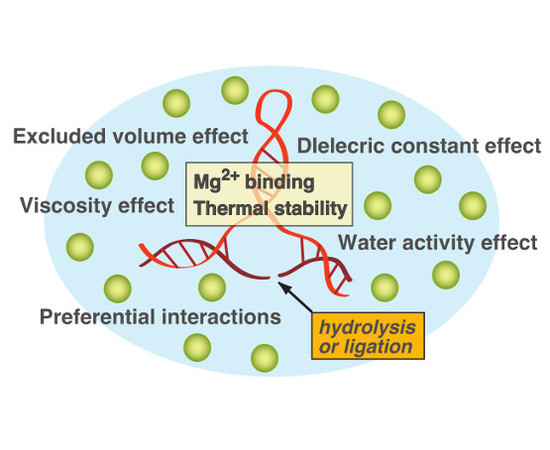
Catalytic Activities of Ribozymes and DNAzymes in Water and Mixed Aqueous Media
Abstract
:1. Introduction
2. Results
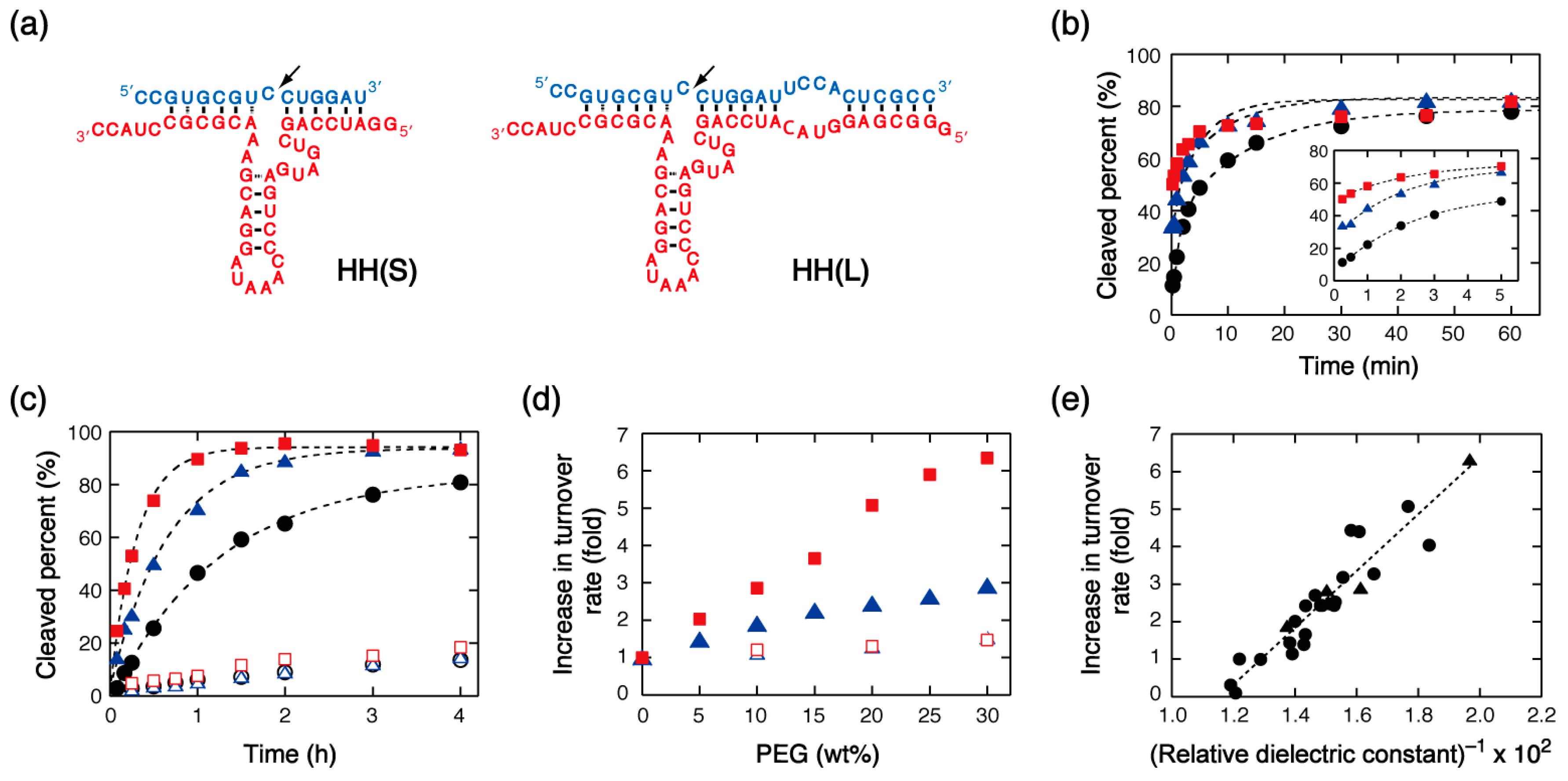
2.1. Substrate Cleavage by Ribozymes
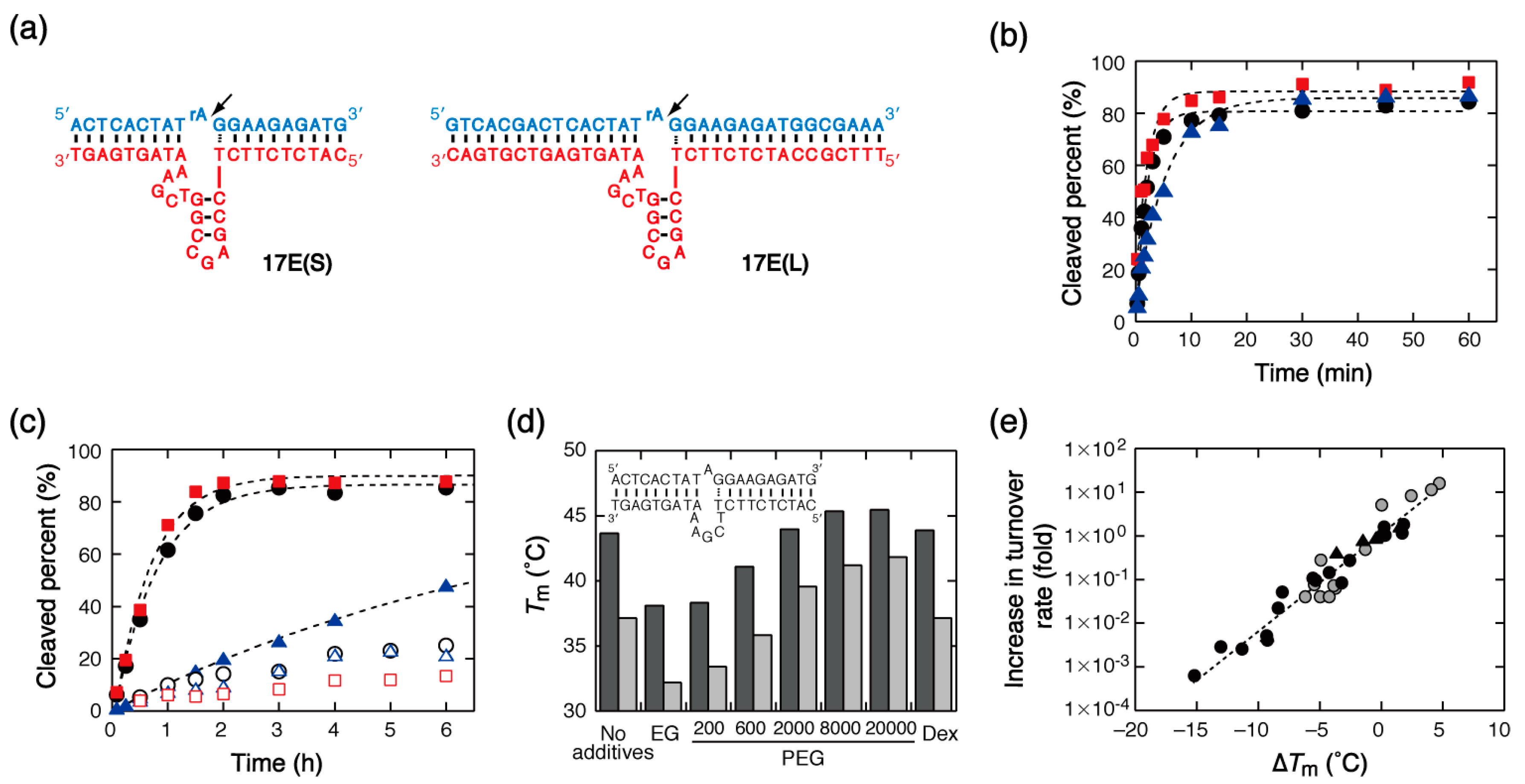
2.2. Substrate Cleavage by DNAzymes
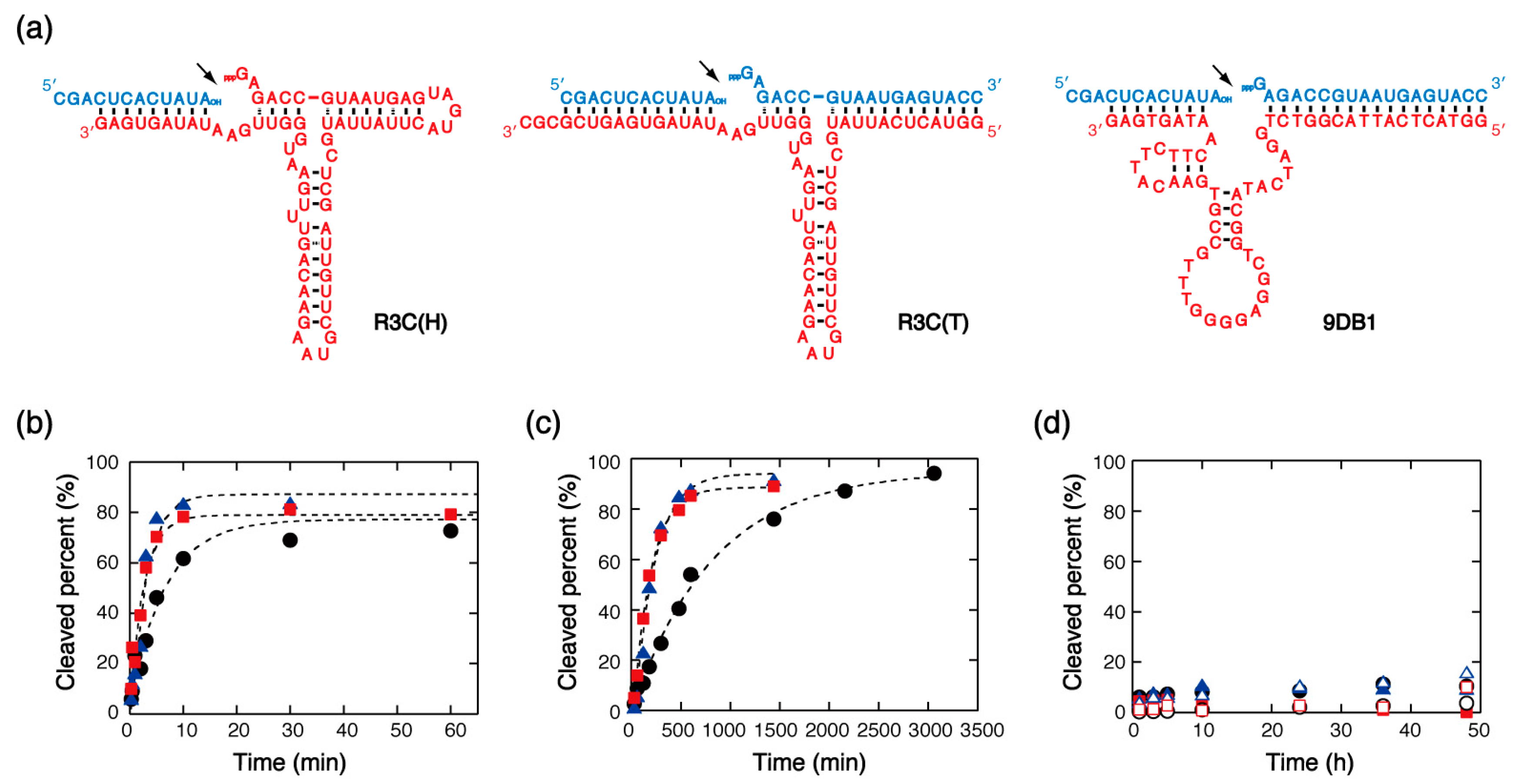
2.3. Substrate Ligation by Ribozyme and DNAzyme
3. Discussion
3.1. Classification of the Mixed Solutions Based on the Effects on Nucleic Acids
3.2. Comparison of the Effects of Mixed Solutions on the Nucleic Acid Enzymes
3.2.1. Effects on the Hammerhead Ribozyme
3.2.2. Effects on the 17E DNAzyme
3.2.3. Effects on the R3C Ribozyme and 9DB1 DNAzyme
4. Materials and Methods
4.1. Materials
4.2. Kinetic Studies on the Ribozymes and DNAzymes
4.3. Thermal Stability Measurements
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Jäschke, A.; Seelig, B. Evolution of DNA and RNA as catalysts for chemical reactions. Curr. Opin. Chem. Biol. 2000, 4, 257–262. [Google Scholar]
- Citti, L.; Rainaldi, G. Synthetic hammerhead ribozymes as therapeutic tools to control disease genes. Curr. Gene Ther. 2005, 5, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Fokina, A.A.; Stetsenko, D.A.; Francois, J.C. DNA enzymes as potential therapeutics: Towards clinical application of 10–23 DNAzymes. Expert Opin. Biol. Ther. 2015, 15, 689–711. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lu, Y. A highly sensitive and selective catalytic DNA biosensor for lead ions. J. Am. Chem. Soc. 2001, 122, 10466–10467. [Google Scholar] [CrossRef]
- Rueda, D.; Walter, N.G. Fluorescent energy transfer readout of an aptazyme-based biosensor. Methods Mol. Biol. 2006, 335, 289–310. [Google Scholar] [PubMed]
- Li, S.; Nosrati, M.; Kashani-Sabet, M. Knockdown of telomerase RNA using hammerhead ribozymes and RNA interference. Methods Mol. Biol. 2008, 405, 113–131. [Google Scholar]
- Liu, J.; Cao, Z.; Lu, Y. Functional nucleic acid sensors. Chem. Rev. 2009, 109, 1948–1998. [Google Scholar] [CrossRef] [PubMed]
- Lan, T.; Lu, Y. Metal ion-dependent DNAzymes and their applications as biosensors. Met. Ions Life Sci. 2012, 10, 217–248. [Google Scholar] [PubMed]
- Gong, L.; Zhao, Z.; Lv, Y.F.; Huan, S.Y.; Fu, T.; Zhang, X.B.; Shen, G.L.; Yu, R.Q. DNAzyme-based biosensors and nanodevices. Chem. Commun. 2015, 51, 979–995. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lu, Y. Adenosine-dependent assembly of aptazyme-functionalized gold nanoparticles and its application as a colorimetric biosensor. Anal. Chem. 2004, 76, 1627–1632. [Google Scholar] [CrossRef] [PubMed]
- Win, M.N.; Smolke, C.D. A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proc. Natl. Acad. Sci. USA 2007, 104, 14283–14288. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Liu, X.; Feng, K.; Jiang, J.; Shen, G.; Yu, R. An aptazyme-based electrochemical biosensor for the detection of adenosine. Anal. Chim. Acta 2010, 669, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Auslander, S.; Ketzer, P.; Hartig, J.S. A ligand-dependent hammerhead ribozyme switch for controlling mammalian gene expression. Mol. Biosyst. 2010, 6, 807–814. [Google Scholar] [CrossRef] [PubMed]
- Bevilacqua, P.C.; Brown, T.S.; Nakano, S.; Yajima, R. Catalytic roles for proton transfer and protonation in ribozymes. Biopolymers 2004, 73, 90–109. [Google Scholar] [CrossRef] [PubMed]
- Doudna, J.A.; Lorsch, J.R. Ribozyme catalysis: Not different, just worse. Nat. Struct. Mol. Biol. 2005, 12, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Lilley, D.M. Structure, folding and mechanisms of ribozymes. Curr. Opin. Struct. Biol. 2005, 15, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Wedekind, J.E. Metal ion binding and function in natural and artificial small RNA enzymes from a structural perspective. Met. Ions Life Sci. 2011, 9, 299–345. [Google Scholar] [PubMed]
- Auffinger, P.; Grover, N.; Westhof, E. Metal ion binding to RNA. Met. Ions Life Sci. 2011, 9, 1–35. [Google Scholar] [PubMed]
- Ward, W.L.; Plakos, K.; DeRose, V.J. Nucleic acid catalysis: Metals, nucleobases, and other cofactors. Chem. Rev. 2014, 114, 4318–4342. [Google Scholar] [CrossRef] [PubMed]
- Nakano, S.; Miyoshi, D.; Sugimoto, N. Effects of molecular crowding on the structures, interactions, and functions of nucleic acids. Chem. Rev. 2014, 114, 2733–2758. [Google Scholar] [CrossRef] [PubMed]
- Winship, P.R. An improved method for directly sequencing PCR amplified material using dimethyl sulphoxide. Nucl. Acids Res. 1989, 17, 1266. [Google Scholar] [CrossRef] [PubMed]
- Jensen, M.A.; Fukushima, M.; Davis, R.W. DMSO and betaine greatly improve amplification of GC-rich constructs in de novo synthesis. PLoS ONE 2010, 5, e11024. [Google Scholar] [CrossRef] [PubMed]
- Dave, N.; Liu, J. Fast molecular beacon hybridization in organic solvents with improved target specificity. J. Phys. Chem. B 2010, 114, 15694–15699. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Shang, C.; Duan, R.; Hakeem, A.; Zhang, Z.; Lou, X.; Xia, F. Polar organic solvents accelerate the rate of DNA strand replacement reaction. Analyst 2015, 140, 2023–2028. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Abe, N.; Shibata, A.; Ito, K.; Tanaka, Y.; Ito, M.; Saneyoshi, H.; Shuto, S.; Ito, Y. Structure formation and catalytic activity of DNA dissolved in organic solvents. Angew. Chem. Int. Ed. Engl. 2012, 51, 6475–6479. [Google Scholar] [CrossRef] [PubMed]
- Behera, A.K.; Schlund, K.J.; Mason, A.J.; Alila, K.O.; Han, M.; Grout, R.L.; Baum, D.A. Enhanced deoxyribozyme-catalyzed RNA ligation in the presence of organic cosolvents. Biopolymers 2013, 99, 382–391. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Saran, R.; Chen, Q.; Ding, J.; Liu, J. A new Na+-dependent RNA-cleaving DNAzyme with over 1000-fold rate acceleration by ethanol. ChemBioChem 2016, 17, 159–163. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Zhou, W.; Liu, J. Ultrasensitive DNAzyme-based Ca2+ detection boosted by ethanol and a solvent compatible scaffold for aptazyme design. ChemBioChem 2017. [Google Scholar] [CrossRef] [PubMed]
- Leamy, K.A.; Assmann, S.M.; Mathews, D.H.; Bevilacqua, P.C. Bridging the gap between in vitro and in vivo RNA folding. Q. Rev. Biophys. 2016, 49, e10. [Google Scholar] [CrossRef] [PubMed]
- Nakano, S.; Sugimoto, N. Model studies of the effects of intracellular crowding on nucleic acid interactions. Mol. Biosyst. 2016, 13, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Nakano, S.; Sugimoto, N. The structural stability and catalytic activity of DNA and RNA oligonucleotides in the presence of organic solvents. Biophys. Rev. 2016, 8, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Osborne, E.M.; Schaak, J.E.; DeRose, V.J. Characterization of a native hammerhead ribozyme derived from Schistosomes. RNA 2005, 11, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Nelson, J.A.; Uhlenbeck, O.C. Minimal and extended hammerheads utilize a similar dynamic reaction mechanism for catalysis. RNA 2008, 14, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Nakano, S.; Karimata, H.T.; Kitagawa, Y.; Sugimoto, N. Facilitation of RNA enzyme activity in the molecular crowding media of cosolutes. J. Am. Chem. Soc. 2009, 131, 16881–16888. [Google Scholar] [CrossRef] [PubMed]
- Nakano, S.; Kitagawa, Y.; Yamashita, H.; Miyoshi, D.; Sugimoto, N. Effects of cosolvents on the folding and catalytic activities of the hammerhead ribozyme. ChemBioChem 2015, 16, 1803–1810. [Google Scholar] [CrossRef] [PubMed]
- Santoro, S.W.; Joyce, G.F. A general purpose RNA-cleaving DNA enzyme. Proc. Natl. Acad. Sci. USA 1997, 94, 4262–4266. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zheng, W.; Kwon, A.H.; Lu, Y. In vitro selection and characterization of a highly efficient Zn(II)-dependent RNA-cleaving deoxyribozyme. Nucl. Acids Res. 2000, 28, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Hwang, K.; Wu, P.; Kim, T.; Lei, L.; Tian, S.; Wang, Y.; Lu, Y. Photocaged DNAzymes as a general method for sensing metal ions in living cells. Angew. Chem. Int. Ed. Engl. 2014, 53, 13798–13802. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Chen, Q.; Huang, P.J.; Ding, J.; Liu, J. DNAzyme hybridization, cleavage, degradation, and sensing in undiluted human blood serum. Anal. Chem. 2015, 87, 4001–4007. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Y.; Niu, C.G.; Guo, L.J.; Hu, L.Y.; Wu, S.Q.; Zeng, G.M.; Li, F. A fluorescence sensor for lead(II) ions determination based on label-free gold nanoparticles (GNPs)-DNAzyme using time-gated mode in aqueous solution. J. Fluoresc. 2017, 27, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Nakano, S.; Karimata, H.; Ohmichi, T.; Kawakami, J.; Sugimoto, N. The effect of molecular crowding with nucleotide length and cosolute structure on DNA duplex stability. J. Am. Chem. Soc. 2004, 126, 14330–14331. [Google Scholar] [CrossRef] [PubMed]
- Knowles, D.B.; LaCroix, A.S.; Deines, N.F.; Shkel, I.; Record, M.T., Jr. Separation of preferential interaction and excluded volume effects on DNA duplex and hairpin stability. Proc. Natl. Acad. Sci. USA 2011, 108, 12699–12704. [Google Scholar] [CrossRef] [PubMed]
- Nakano, S.; Yamaguchi, D.; Tateishi-Karimata, H.; Miyoshi, D.; Sugimoto, N. Hydration changes upon DNA folding studied by osmotic stress experiments. Biophys. J. 2012, 102, 2808–2817. [Google Scholar] [CrossRef] [PubMed]
- Nakano, S.; Kitagawa, Y.; Miyoshi, D.; Sugimoto, N. Hammerhead ribozyme activity and oligonucleotide duplex stability in mixed solutions of water and organic compounds. FEBS Open Bio 2014, 4, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Rogers, J.; Joyce, G.F. The effect of cytidine on the structure and function of an RNA ligase ribozyme. RNA 2001, 7, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Kurihara, E.; Uchida, S.; Umehara, T.; Tamura, K. Development of a functionally minimized mutant of the R3C ligase ribozyme offers insight into the plausibility of the RNA world hypothesis. Biology 2014, 3, 452–465. [Google Scholar] [CrossRef] [PubMed]
- Purtha, W.E.; Coppins, R.L.; Smalley, M.K.; Silverman, S.K. General deoxyribozyme-catalyzed synthesis of native 3′-5′ RNA linkages. J. Am. Chem. Soc. 2005, 127, 13124–13125. [Google Scholar] [CrossRef] [PubMed]
- Bevers, S.; Xiang, G.; McLaughlin, L.W. Importance of specific adenosine N3-nitrogens for efficient cleavage by a hammerhead ribozyme. Biochemistry 1996, 35, 6483–6490. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.S.; Lopez, C.S.; Giambasu, G.M.; Martick, M.; Scott, W.G.; York, D.M. Role of Mg2+ in hammerhead ribozyme catalysis from molecular simulation. J. Am. Chem. Soc. 2008, 130, 3053–3064. [Google Scholar] [CrossRef] [PubMed]
- Asami, K.; Hanai, T.; Koizumi, N. Dielectric properties of yeast cells. J. Membr. Biol. 1976, 28, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Cuervo, A.; Dans, P.D.; Carrascosa, J.L.; Orozco, M.; Gomila, G.; Fumagalli, L. Direct measurement of the dielectric polarization properties of DNA. Proc. Natl. Acad. Sci. USA 2014, 111, E3624–E3630. [Google Scholar] [CrossRef] [PubMed]
- Grubbs, R.D. Intracellular magnesium and magnesium buffering. Biometals 2002, 15, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Raz, M.H.; Hollenstein, M. Probing the effect of minor groove interactions on the catalytic efficiency of DNAzymes 8–17 and 10–23. Mol. Biosyst. 2015, 11, 1454–1461. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Rasnik, I.; Liu, J.; Ha, T.; Lu, Y. Dissecting metal ion-dependent folding and catalysis of a single DNAzyme. Nat. Chem. Biol. 2007, 3, 763–768. [Google Scholar] [CrossRef] [PubMed]
- Nakano, S.; Watabe, T.; Sugimoto, N. Modulation of the ribozyme and deoxyribozyme activities using tetraalkylammonium ions. ChemPhysChem 2017. [Google Scholar] [CrossRef] [PubMed]
- Ponce-Salvatierra, A.; Wawrzyniak-Turek, K.; Steuerwald, U.; Hobartner, C.; Pena, V. Crystal structure of a DNA catalyst. Nature 2016, 529, 231–234. [Google Scholar] [CrossRef] [PubMed]
| Solution 1 | 10 mM MgCl2 | 1 mM MgCl2 | Solution 1 | 10 mM MgCl2 | 1 mM MgCl2 | Solution 1 | 10 mM MgCl2 | 1 mM MgCl2 |
|---|---|---|---|---|---|---|---|---|
| PEG20000 | 3.4 | 13 | PDO | 1.7 | 2.8 | FA | 0.32 | <0.02 |
| PEG8000 | 5.1 | 17 | MME | 2.4 | 3.9 | DMF | 1.3 | <0.02 |
| PEG2000 | 3.3 | 10 | DME | 4.4 | 11 | AcAm | 1.0 | <0.02 |
| PEG600 | 2.5 | 6.8 | MeOH | 2.4 | 4.9 | AcCN | 2.7 | 6.5 |
| PEG200 | 2.4 | 5.5 | EtOH | 2.5 | 4.8 | DMSO | 1.4 | 1.2 |
| EG | 2.0 | 1.3 | PrOH | 3.2 | 6.0 | DOX | 1.9 | 4.6 |
| Glyc | 1.1 | 1.3 | Urea | 0.10 | <0.02 | Dex | 1.4 | 2.5 |
| Solution | 10 mM MgCl2 | 1 mM MgCl2 | Solution | 10 mM MgCl2 | 1 mM MgCl2 | Solution | 10 mM MgCl2 | 1 mM MgCl2 |
|---|---|---|---|---|---|---|---|---|
| PEG20000 | 1.8 | 16 | PDO | 0.14 | 0.073 | FA | <0.001 | <0.03 |
| PEG8000 | 1.2 | 11 | MME | 0.0041 | <0.03 | DMF | <0.001 | <0.03 |
| PEG2000 | 1.0 | 8.2 | DME | 0.052 | 0.28 | AcAm | <0.001 | <0.03 |
| PEG600 | 0.27 | 0.49 | MeOH | 0.022 | 0.076 | AcCN | <0.001 | <0.03 |
| PEG200 | 0.095 | 0.064 | EtOH | 0.0026 | 0.041 | DMSO | 0.0051 | <0.03 |
| EG | 0.11 | 0.040 | PrOH | <0.001 | <0.03 | DOX | 0.0029 | <0.03 |
| Glyc | 0.083 | 0.040 | Urea | <0.001 | <0.03 | Dex | 1.6 | 5.1 |
| Solution | R3C(H) | 9DB1 | Solution | R3C(H) | 9DB1 | Solution | R3C(H) | 9DB1 |
|---|---|---|---|---|---|---|---|---|
| PEG20000 | 1.6 | 4.2 | PDO | 1.2 | 2.6 | FA | <0.001 | <0.001 |
| PEG8000 | 2.0 | 4.6 | MME | 1.9 | 3.3 | DMF | 0.26 | <0.001 |
| PEG2000 | 1.6 | 6.8 | DME | 1.2 | 1.9 | AcAm | 0.30 | <0.001 |
| PEG600 | 1.3 | 4.2 | MeOH | 1.5 | 4.5 | AcCN | 1.8 | 4.5 |
| PEG200 | 2.0 | 4.1 | EtOH | 2.2 | 2.4 | DMSO | 1.7 | 2.5 |
| EG | 1.0 | 2.8 | PrOH | 1.8 | 3.9 | DOX | 1.3 | 3.8 |
| Glyc | 1.2 | 1.1 | Urea | <0.001 | <0.001 | Dex | 1.3 | 0.89 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakano, S.-i.; Horita, M.; Kobayashi, M.; Sugimoto, N. Catalytic Activities of Ribozymes and DNAzymes in Water and Mixed Aqueous Media. Catalysts 2017, 7, 355. https://doi.org/10.3390/catal7120355
Nakano S-i, Horita M, Kobayashi M, Sugimoto N. Catalytic Activities of Ribozymes and DNAzymes in Water and Mixed Aqueous Media. Catalysts. 2017; 7(12):355. https://doi.org/10.3390/catal7120355
Chicago/Turabian StyleNakano, Shu-ichi, Masao Horita, Miku Kobayashi, and Naoki Sugimoto. 2017. "Catalytic Activities of Ribozymes and DNAzymes in Water and Mixed Aqueous Media" Catalysts 7, no. 12: 355. https://doi.org/10.3390/catal7120355