Are Directed Evolution Approaches Efficient in Exploring Nature’s Potential to Stabilize a Lipase in Organic Cosolvents?
Abstract
:1. Introduction
2. Results
2.1. Selection of Organic Cosolvent Concentrations for BSLA SSM Library Screening
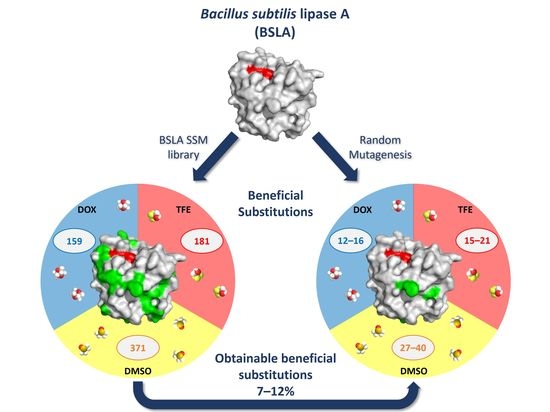
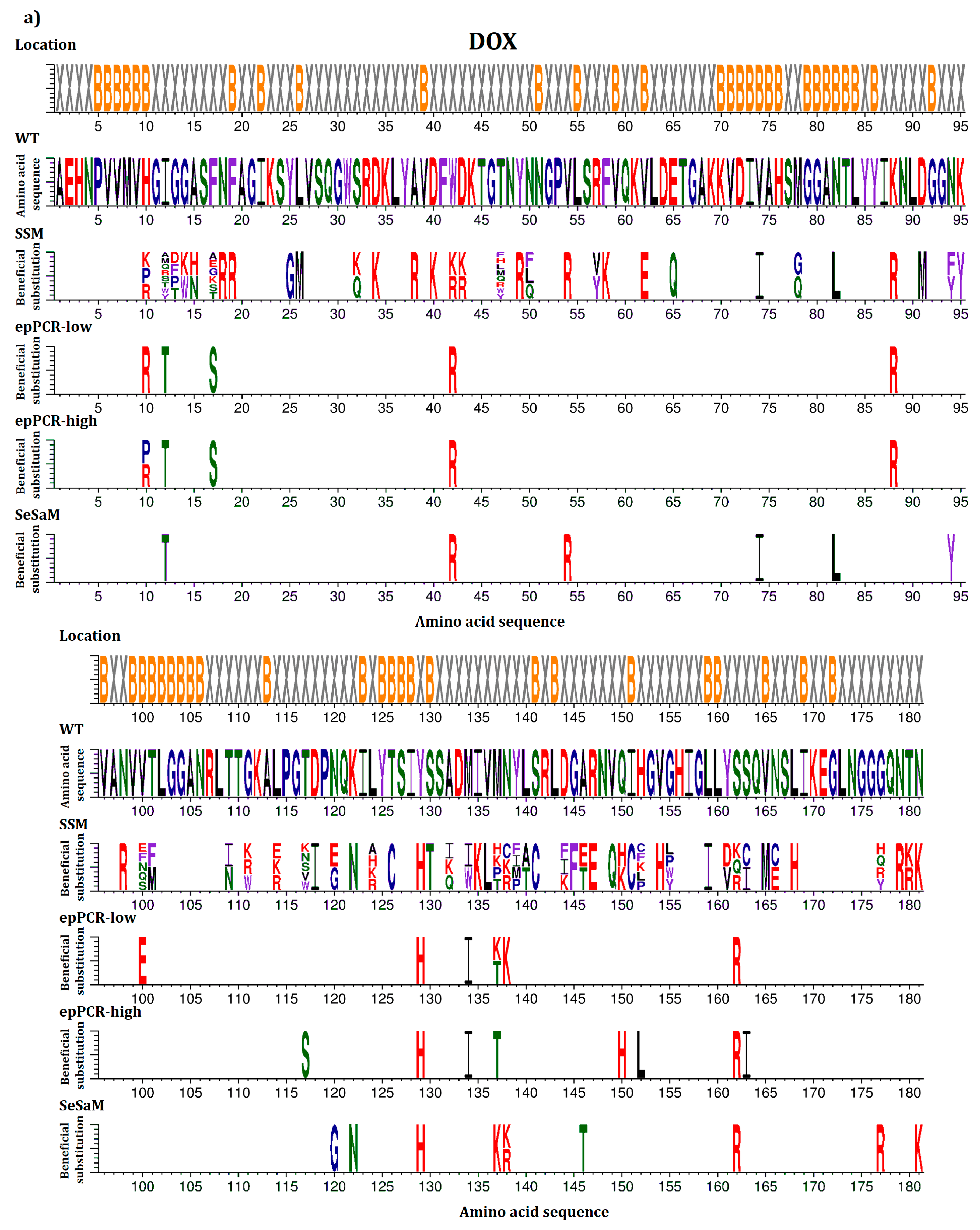
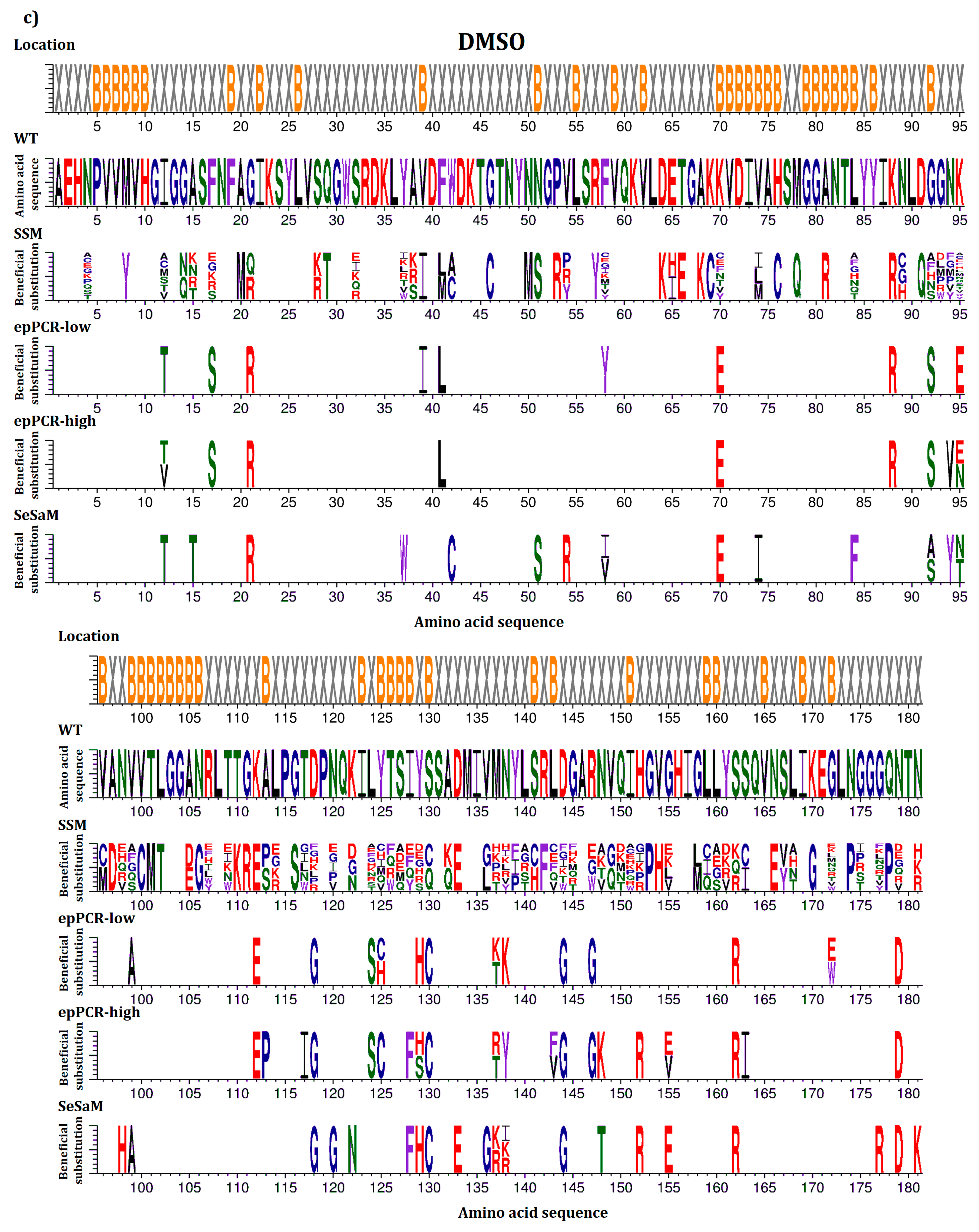
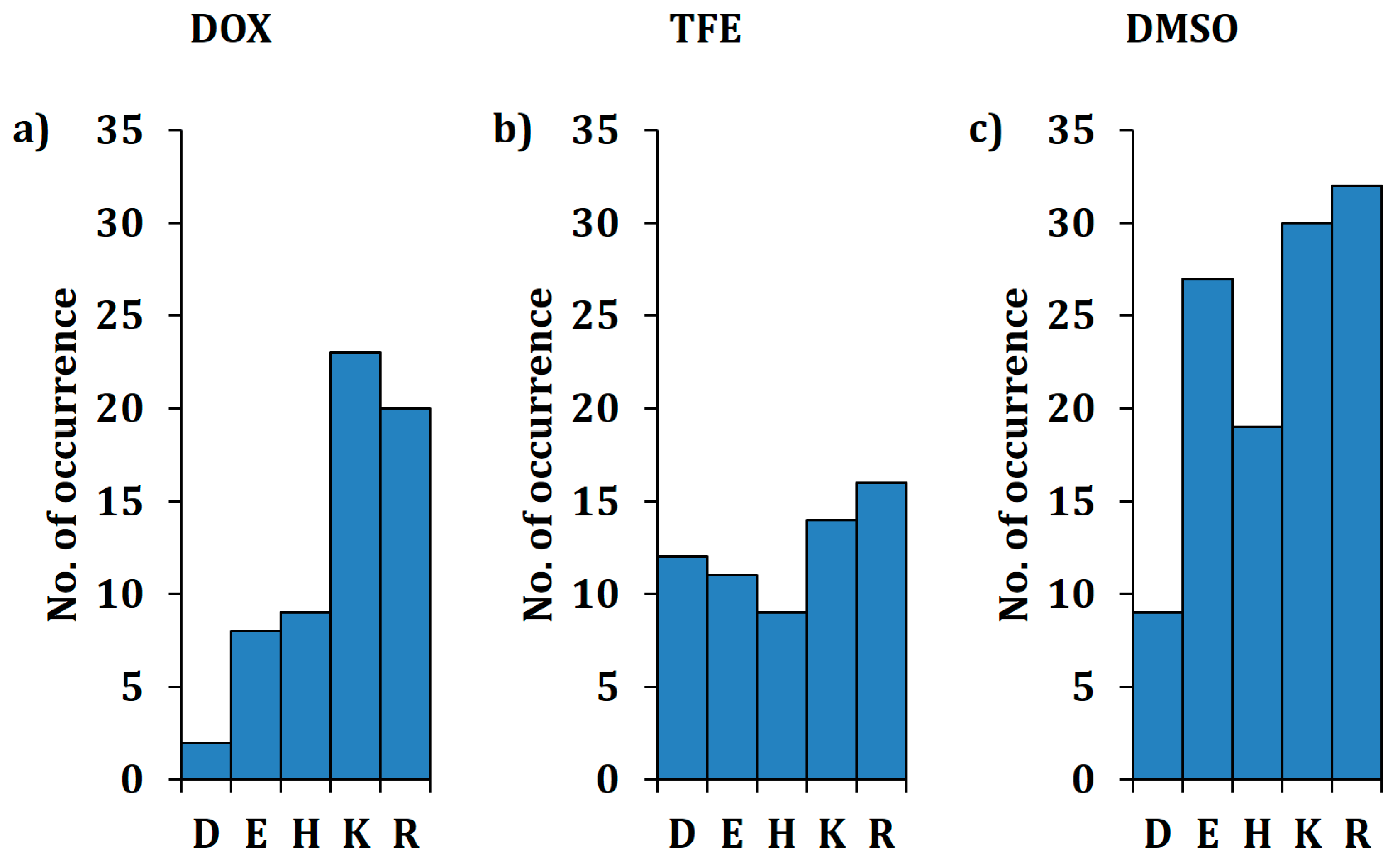
2.2. Overview of Beneficial Substitutions
2.3. Number of Beneficial Amino Acid Substitutions
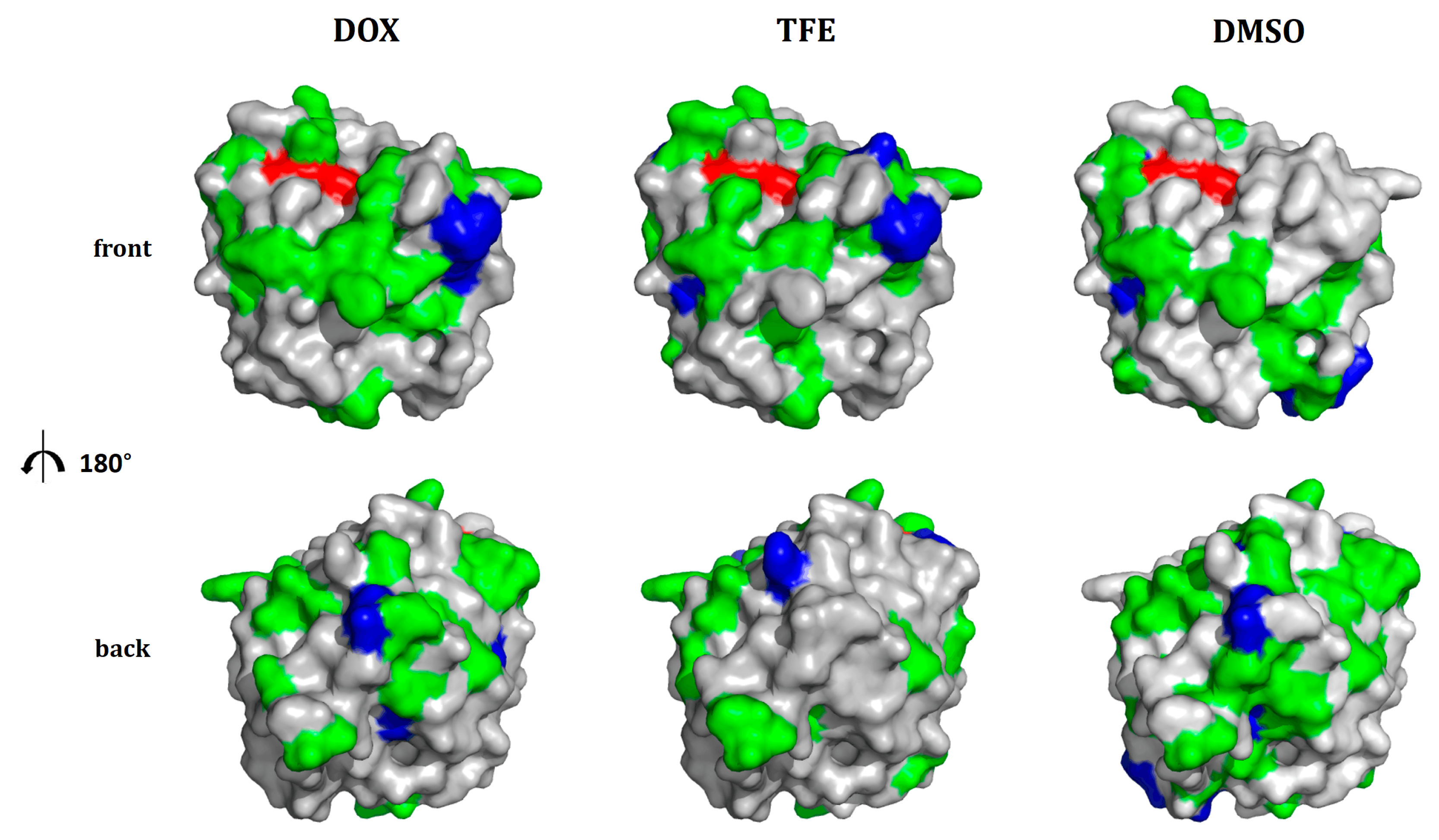
2.4. Location of Beneficial Amino Acid Substitutions in the BSLA 3D Structure
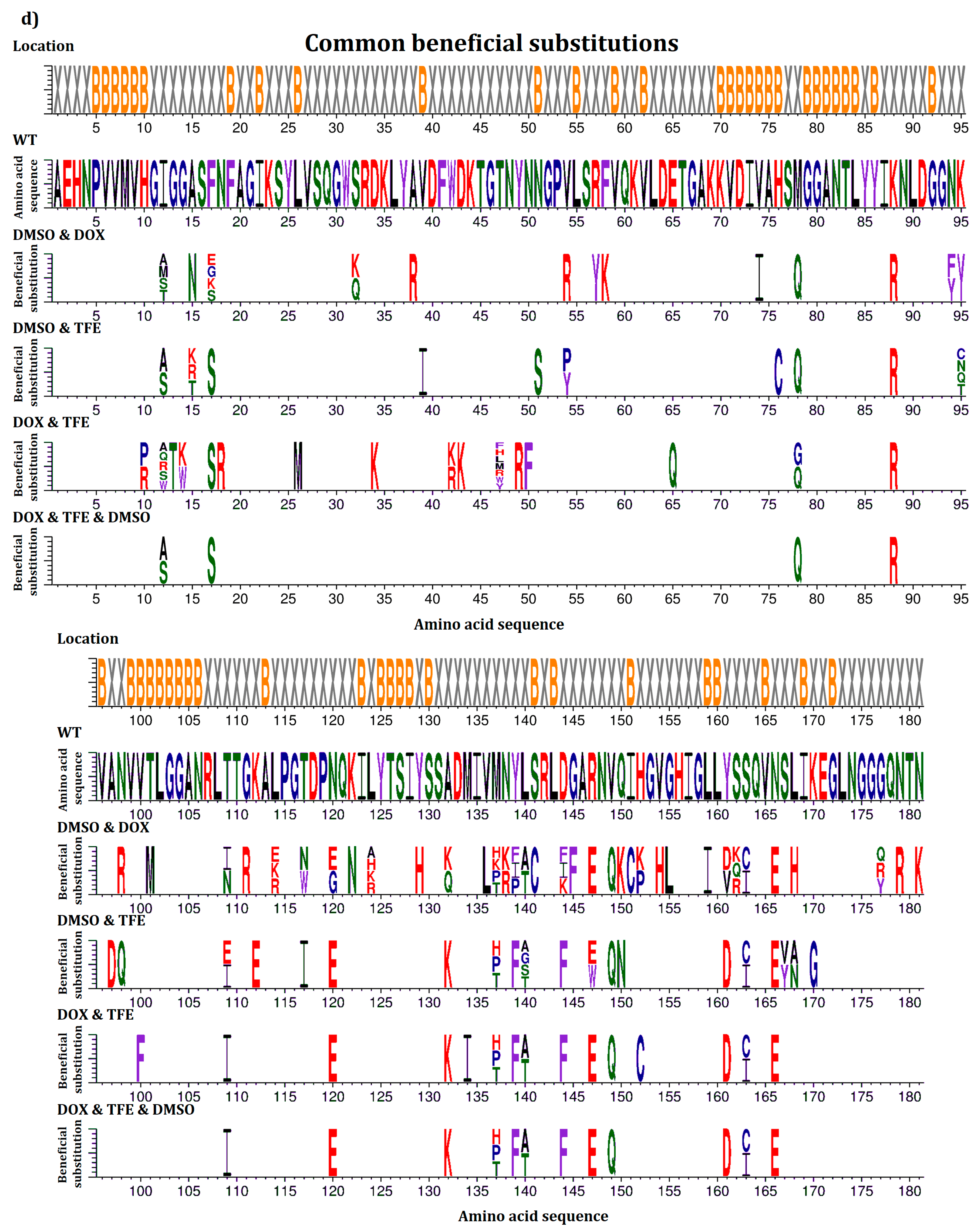
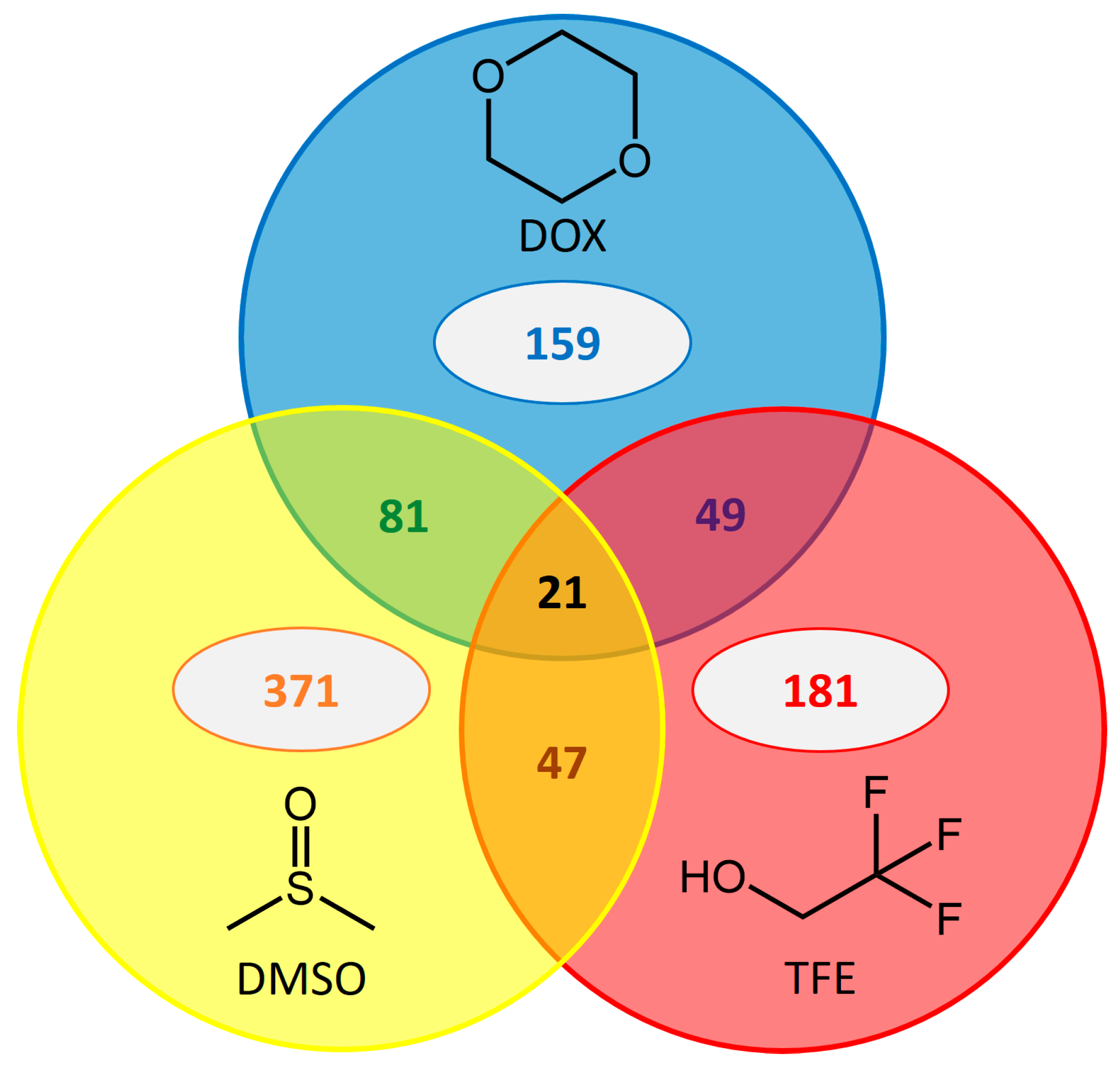
2.5. Common Beneficial Substitutions for Different Organic Cosolvents
2.6. Occurrence of Beneficial Substitutions
3. Discussion
3.1. Comparison of Multiple Mutagenesis Methods to Address Question (1) and (2)
3.2. General Design Principles Derived from the BSLA SSM Library to Address Question (3)
4. Materials and Methods
4.1. Generation of the BSLA SSM Library and Random Mutagenesis Libraries
4.2. Amino Acid Categorization
4.3. BSLA SSM Library Expression in 96-Well Plates
4.4. MTP-Based p-Nitrophenyl Butyrate (pNPB) Assay for BSLA Activity Measurement
4.5. Optimization of Screening Conditions in Organic Solvents
4.6. Screening of the BSLA SSM Library in the Presence of the Three Organic Cosolvents
4.7. Computational Methods
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cheng, F.; Zhu, L.; Schwaneberg, U. Directed evolution 2.0: Improving and deciphering enzyme properties. Chem. Commun. 2015, 51, 9760–9772. [Google Scholar] [CrossRef] [PubMed]
- Arnold, F.H. Engineering enzymes for non-aqueous solvents. Trends Biotechnol. 1990, 8, 244–249. [Google Scholar] [CrossRef]
- Dordick, J.S. Enzymatic catalysis in monophasic organic solvents. Enzyme Microb. Technol. 1989, 11, 194–211. [Google Scholar] [CrossRef]
- Halling, P.J. Biocatalysis in multi-phase reaction mixtures containing organic liquids. Biotechnol. Adv. 1987, 5, 47–84. [Google Scholar] [CrossRef]
- Serdakowski, A.L.; Dordick, J.S. Enzyme activation for organic solvents made easy. Trends Biotechnol. 2008, 26, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Zaks, A.; Klibanov, A.M. Enzyme-catalyzed processes in organic solvents. Proc. Natl. Acad. Sci. USA 1985, 82, 3192–3196. [Google Scholar] [CrossRef] [PubMed]
- Dill, K.A. Dominant forces in protein folding. Biochemistry 1990, 29, 7133–7155. [Google Scholar] [CrossRef] [PubMed]
- Pace, C.N.; Shirley, B.A.; McNutt, M.; Gajiwala, K. Forces contributing to the conformational stability of proteins. FASEB J. 1996, 10, 75–83. [Google Scholar] [PubMed]
- Prabhu, N.; Sharp, K. Protein-solvent interactions. Chem. Rev. 2006, 106, 1616–1623. [Google Scholar] [CrossRef] [PubMed]
- Wang, W. Instability, stabilization, and formulation of liquid protein pharmaceuticals. Int. J. Pharm. 1999, 185, 129–188. [Google Scholar] [CrossRef]
- Bone, S.; Pethig, R. Dielectric studies of protein hydration and hydration-induced flexibility. J. Mol. Biol. 1985, 181, 323–326. [Google Scholar] [CrossRef]
- Daniel, R.M.; Dunn, R.V.; Finney, J.L.; Smith, J.C. The role of dynamics in enzyme activity. Annu. Rev. Biophys. Biomol. Struct. 2003, 32, 69–92. [Google Scholar] [CrossRef] [PubMed]
- Gorman, L.A.S.; Dordick, J.S. Organic solvents strip water off enzymes. Biotechnol. Bioeng. 1992, 39, 392–397. [Google Scholar] [CrossRef] [PubMed]
- Poole, P.L.; Finney, J.L. Hydration-induced conformational and flexibility changes in lysozyme at low water content. Int. J. Biol. Macromol. 1983, 5, 308–310. [Google Scholar] [CrossRef]
- Rupley, J.A.; Careri, G. Protein hydration and function. Adv. Protein Chem. 1991, 41, 37–172. [Google Scholar] [PubMed]
- Gao, X.G.; Maldonado, E.; Pérez-Montfort, R.; Garza-Ramos, G.; de Gómez-Puyou, M.T.; Gómez-Puyou, A.; Rodríguez-Romero, A. Crystal structure of triosephosphate isomerase from Trypanosoma cruzi in hexane. Proc. Natl. Acad. Sci. USA 1999, 96, 10062–10067. [Google Scholar] [CrossRef] [PubMed]
- Zaks, A.; Klibanov, A.M. The effect of water on enzyme action in organic media. J. Biol. Chem. 1988, 263, 8017–8021. [Google Scholar] [PubMed]
- Micaêlo, N.M.; Soares, C.M. Modeling hydration mechanisms of enzymes in nonpolar and polar organic solvents. FEBS J. 2007, 274, 2424–2436. [Google Scholar] [CrossRef] [PubMed]
- Wedberg, R.; Abildskov, J.; Peters, G.H. Protein dynamics in organic media at varying water activity studied by molecular dynamics simulation. J. Phys. Chem. 2012, 116, 2575–2585. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Russell, A.J. The role of hydration in enzyme activity and stability: 1. Water adsorption by alcohol dehydrogenase in a continuous gas phase reactor. Biotechnol. Bioeng. 1996, 49, 700–708. [Google Scholar] [CrossRef]
- Yang, L.; Dordick, J.S.; Garde, S. Hydration of enzyme in nonaqueous media is consistent with solvent dependence of its activity. Biophys. J. 2004, 87, 812–821. [Google Scholar] [CrossRef] [PubMed]
- Kuper, J.; Tee, K.L.; Wilmanns, M.; Roccatano, D.; Schwaneberg, U.; Wong, T.S. The role of active-site Phe87 in modulating the organic co-solvent tolerance of cytochrome P450 BM3 monooxygenase. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2012, 68, 1013–1017. [Google Scholar] [CrossRef] [PubMed]
- Kuper, J.; Wong, T.S.; Roccatano, D.; Wilmanns, M.; Schwaneberg, U. Understanding a mechanism of organic cosolvent inactivation in heme monooxygenase P450 BM-3. J. Am. Chem. Soc. 2007, 129, 5786–5787. [Google Scholar] [CrossRef] [PubMed]
- Roccatano, D.; Wong, T.S.; Schwaneberg, U.; Zacharias, M. Toward understanding the inactivation mechanism of monooxygenase P450 BM-3 by organic cosolvents: A molecular dynamics simulation study. Biopolymers 2006, 83, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Stepankova, V.; Bidmanova, S.; Koudelakova, T.; Prokop, Z.; Chaloupkova, R.; Damborsky, J. Strategies for stabilization of enzymes in organic solvents. ACS Catal. 2013, 3, 2823–2836. [Google Scholar] [CrossRef]
- Chakravorty, D.; Parameswaran, S.; Dubey, V.K.; Patra, S. Unraveling the rationale behind organic solvent stability of lipases. Appl. Biochem. Biotechnol. 2012, 167, 439–461. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Kardashliev, T.; Ruff, A.J.; Bocola, M.; Schwaneberg, U. Lessons from diversity of directed evolution experiments by an analysis of 3000 mutations. Biotechnol. Bioeng. 2014, 111, 2380–2389. [Google Scholar] [CrossRef] [PubMed]
- Frauenkron-Machedjou, V.J.; Fulton, A.; Zhu, L.; Anker, C.; Bocola, M.; Jaeger, K.E.; Schwaneberg, U. Towards understanding directed evolution: More than half of all amino acid positions contribute to ionic liquid resistance of Bacillus subtilis lipase A. ChemBioChem 2015, 16, 937–945. [Google Scholar] [CrossRef] [PubMed]
- Fulton, A.; Frauenkron-Machedjou, V.J.; Skoczinski, P.; Wilhelm, S.; Zhu, L.; Schwaneberg, U.; Jaeger, K.E. Exploring the protein stability landscape: Bacillus subtilis lipase A as a model for detergent tolerance. ChemBioChem 2015, 16, 930–936. [Google Scholar] [CrossRef] [PubMed]
- Secundo, F.; Fialà, S.; Fraaije, M.W.; de Gonzalo, G.; Meli, M.; Zambianchi, F.; Ottolina, G. Effects of water miscible organic solvents on the activity and conformation of the baeyer–villiger monooxygenases from Thermobifida fusca and Acinetobacter calcoaceticus: A comparative study. Biotechnol. Bioeng. 2011, 108, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Uematsu, M.; Frank, E. Static dielectric constant of water and steam. J. Phys. Chem. Ref. Data 1980, 9, 1291–1306. [Google Scholar] [CrossRef]
- MacGregor, W.S. The chemical and physical properties of DMSO. Ann. N. Y. Acad. Sci. 1967, 141, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Korman, T.P.; Sahachartsiri, B.; Charbonneau, D.M.; Huang, G.L.; Beauregard, M.; Bowie, J.U. Dieselzymes: Development of a stable and methanol tolerant lipase for biodiesel production by directed evolution. Biotechnol. Biofuels 2013, 6, 70. [Google Scholar] [CrossRef] [PubMed]
- Mundhada, H.; Marienhagen, J.; Scacioc, A.; Schenk, A.; Roccatano, D.; Schwaneberg, U. SeSaM-Tv-II generates a protein sequence space that is unobtainable by epPCR. ChemBioChem 2011, 12, 1595–1601. [Google Scholar] [CrossRef] [PubMed]
- Dror, A.; Shemesh, E.; Dayan, N.; Fishman, A. Protein engineering by random mutagenesis and structure-guided consensus of Geobacillus stearothermophilus lipase T6 for enhanced stability in methanol. Appl. Environ. Microbiol. 2014, 80, 1515–1527. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Bagchi, B. Comparative study of protein unfolding in aqueous urea and dimethyl sulfoxide solutions: Surface polarity, solvent specificity, and sequence of secondary structure melting. J. Phys. Chem. 2014, 118, 5691–5697. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Joo, J.C.; Park, K.; Kim, Y.H.; Yoo, Y.J. Prediction of the solvent affecting site and the computational design of stable Candida antarctica lipase B in a hydrophilic organic solvent. J. Biotechnol. 2013, 163, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Sears, P.; Schuster, M.; Wang, P.; Witte, K.; Wong, C.H. Engineering subtilisin for peptide coupling: Studies on the effects of counterions and site-specific modifications on the stability and specificity of the enzyme. J. Am. Chem. Soc. 1994, 116, 6521–6530. [Google Scholar] [CrossRef]
- Martinez, P.; Arnold, F.H. Surface charge substitutions increase the stability of. alpha.-lytic protease in organic solvents. J. Am. Chem. Soc. 1991, 113, 6336–6337. [Google Scholar] [CrossRef]
- Reetz, M.T.; Soni, P.; Fernández, L.; Gumulya, Y.; Carballeira, J.D. Increasing the stability of an enzyme toward hostile organic solvents by directed evolution based on iterative saturation mutagenesis using the B-FIT method. Chem. Commun. 2010, 46, 8657–8658. [Google Scholar] [CrossRef] [PubMed]
- Studier, F.W. Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 2005, 41, 207–234. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Dassault Systèmes BIOVIA. Discovery Studio Modeling Environment, 4th ed.; Dessault Systèmes BIOVIA: San Diego, CA, USA, 2015. [Google Scholar]
- van Pouderoyen, G.; Eggert, T.; Jaeger, K.E.; Dijkstra, B.W. The crystal structure of Bacillus subtilis lipase: A minimal α/β hydrolase fold enzyme. J. Mol. Biol. 2001, 309, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Krieger, E.; Koraimann, G.; Vriend, G. Increasing the precision of comparative models with YASARA NOVA – a self-parameterizing force field. Proteins Struct. Funct. Bioinform. 2002, 47, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Van Durme, J.; Delgado, J.; Stricher, F.; Serrano, L.; Schymkowitz, J.; Rousseau, F. A graphical interface for the FoldX forcefield. Bioinformatics 2011, 27, 1711–1712. [Google Scholar] [CrossRef] [PubMed]
- Guerois, R.; Nielsen, J.E.; Serrano, L. Predicting changes in the stability of proteins and protein complexes: A study of more than 1000 mutations. J. Mol. Biol. 2002, 320, 369–387. [Google Scholar] [CrossRef]
- Donald, J.E.; Kulp, D.W.; DeGrado, W.F. Salt bridges: Geometrically specific, designable interactions. Proteins Struct. Funct. Bioinform. 2011, 79, 898–915. [Google Scholar] [CrossRef] [PubMed]
- The PyMOL Molecular Graphics System, version 1.3r1; Llc Schrödinger: New York, NY, USA, 2010.

| Organic Cosolvent | c (M) (% v/v) | RWT (%) | σ (%) |
|---|---|---|---|
| DOX | 2.6 (≈ 22) | 31 | 9.6 |
| TFE | 2.9 (≈ 12) | 30 | 12.0 |
| DMSO | 8.4 (≈ 60) | 29 | 10.5 |
| Diversity Generation Method | DOX | (%) | TFE | (%) | DMSO | (%) |
|---|---|---|---|---|---|---|
| BSLA SSM | 159 | (100) | 181 | (100) | 371 | (100) |
| epPCR-low | 12 | (8) | 15 | (8) | 27 | (7) |
| epPCR-high | 14 | (9) | 21 | (12) | 35 | (9) |
| SeSaM | 16 | (10) | 15 | (8) | 40 | (11) |
| epPCR-low + SeSaM | 22 | (14) | 24 | (13) | 54 | (15) |
| epPCR-high + SeSaM | 26 | (16) | 28 | (15) | 60 | (16) |
| epPCR-low + epPCR-high | 17 | (11) | 24 | (13) | 43 | (12) |
| epPCR-low + epPCR-high + SeSaM | 27 | (17) | 30 | (17) | 65 | (18) |
| Organic Cosolvent | Diversity Generation Method | Location of Beneficial Substitutions in BSLA | |||
|---|---|---|---|---|---|
| Exposed | (%) | Buried | (%) | ||
| DOX | SSM | 138 | (87) | 21 | (13) |
| epPCR-low | 10 | (83) | 2 | (17) | |
| epPCR-high | 12 | (86) | 2 | (14) | |
| SeSaM | 14 | (88) | 2 | (12) | |
| TFE | SSM | 152 | (84) | 29 | (16) |
| epPCR-low | 13 | (87) | 2 | (13) | |
| epPCR-high | 19 | (90) | 2 | (10) | |
| SeSaM | 14 | (93) | 1 | (7) | |
| DMSO | SSM | 275 | (74) | 96 | (26) |
| epPCR-low | 18 | (67) | 9 | (33) | |
| epPCR-high | 27 | (77) | 8 | (23) | |
| SeSaM | 31 | (78) | 9 | (22) | |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Markel, U.; Zhu, L.; Frauenkron-Machedjou, V.J.; Zhao, J.; Bocola, M.; Davari, M.D.; Jaeger, K.-E.; Schwaneberg, U. Are Directed Evolution Approaches Efficient in Exploring Nature’s Potential to Stabilize a Lipase in Organic Cosolvents? Catalysts 2017, 7, 142. https://doi.org/10.3390/catal7050142
Markel U, Zhu L, Frauenkron-Machedjou VJ, Zhao J, Bocola M, Davari MD, Jaeger K-E, Schwaneberg U. Are Directed Evolution Approaches Efficient in Exploring Nature’s Potential to Stabilize a Lipase in Organic Cosolvents? Catalysts. 2017; 7(5):142. https://doi.org/10.3390/catal7050142
Chicago/Turabian StyleMarkel, Ulrich, Leilei Zhu, Victorine Josiane Frauenkron-Machedjou, Jing Zhao, Marco Bocola, Mehdi D. Davari, Karl-Erich Jaeger, and Ulrich Schwaneberg. 2017. "Are Directed Evolution Approaches Efficient in Exploring Nature’s Potential to Stabilize a Lipase in Organic Cosolvents?" Catalysts 7, no. 5: 142. https://doi.org/10.3390/catal7050142