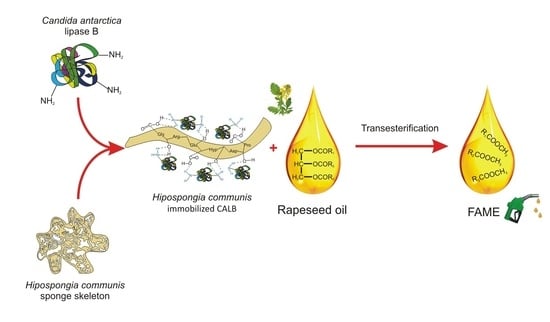
Spongin-Based Scaffolds from Hippospongia communis Demosponge as an Effective Support for Lipase Immobilization
Abstract
:1. Introduction
2. Results and Discussion
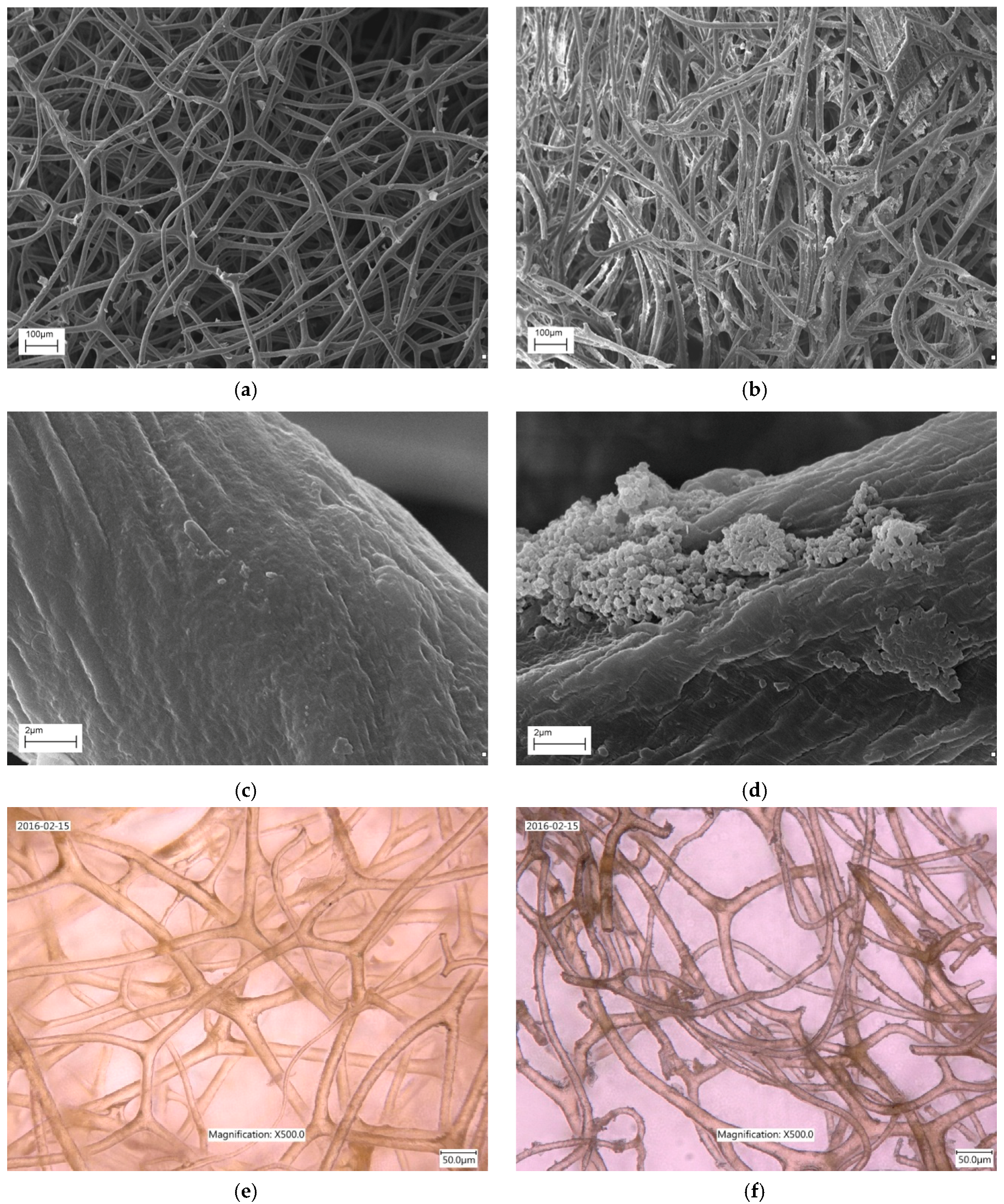
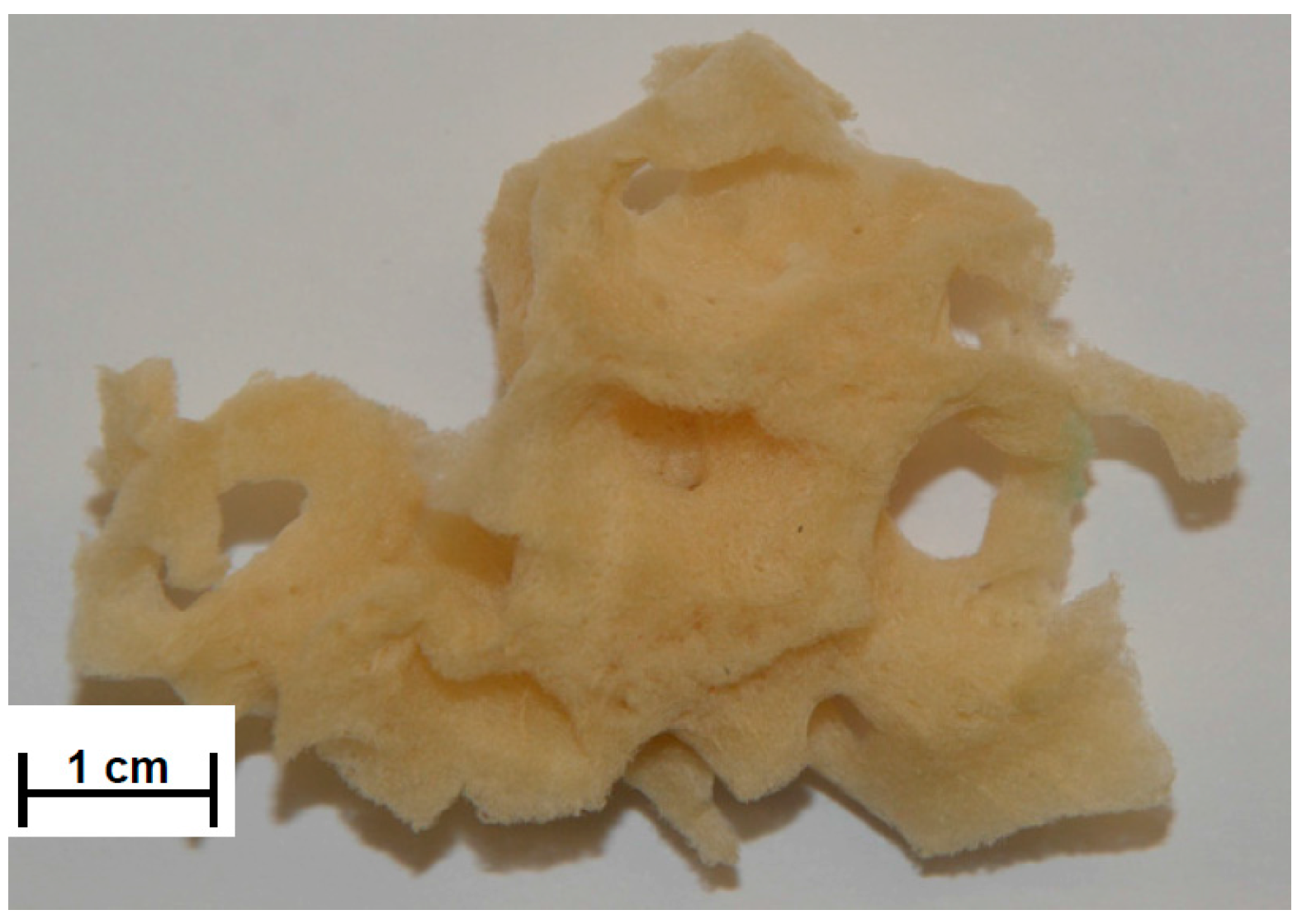
2.1. SEM and Digital Microscopy
2.2. FTIR Spectroscopy
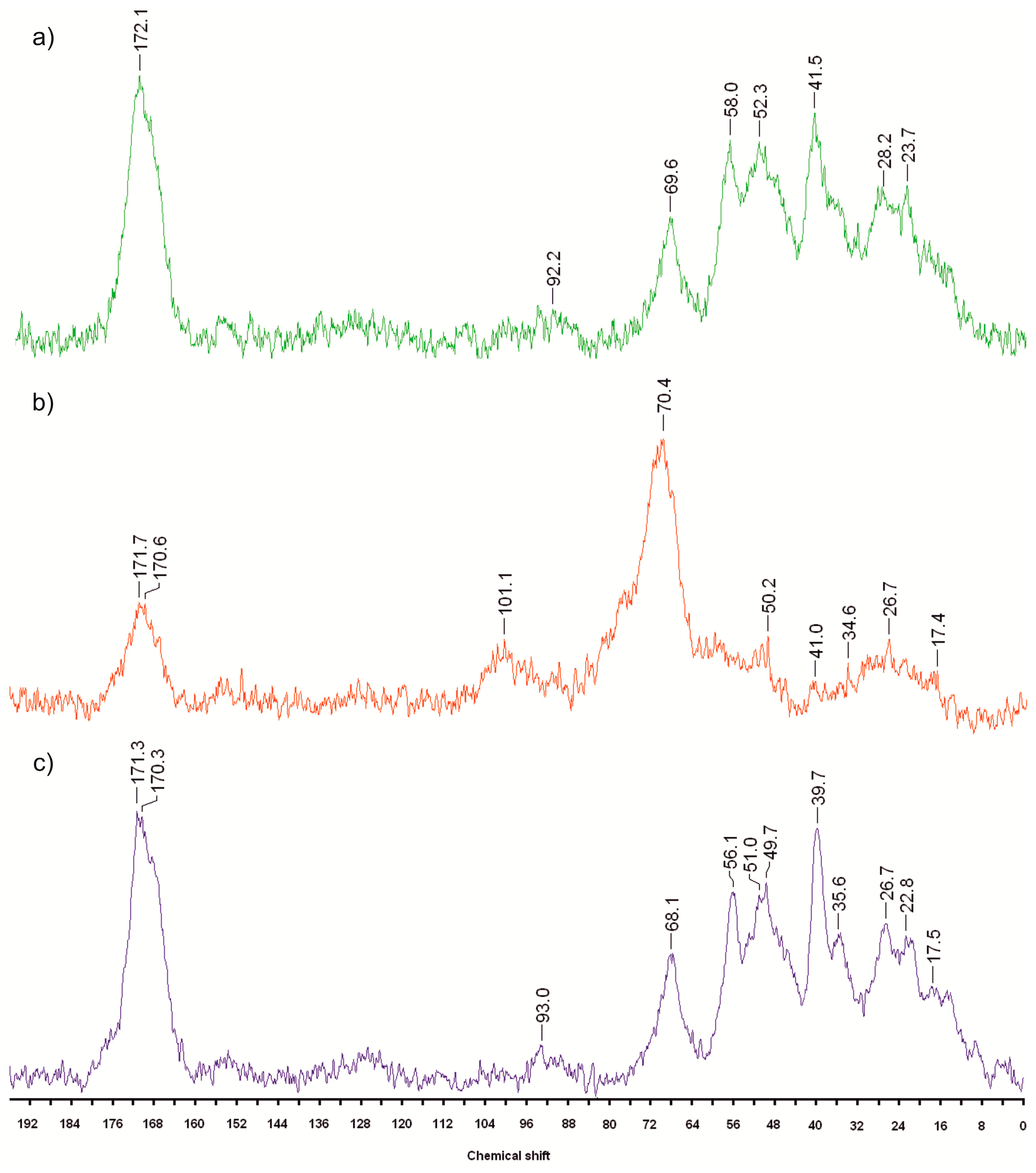
2.3. 13C CP MAS NMR Spectra
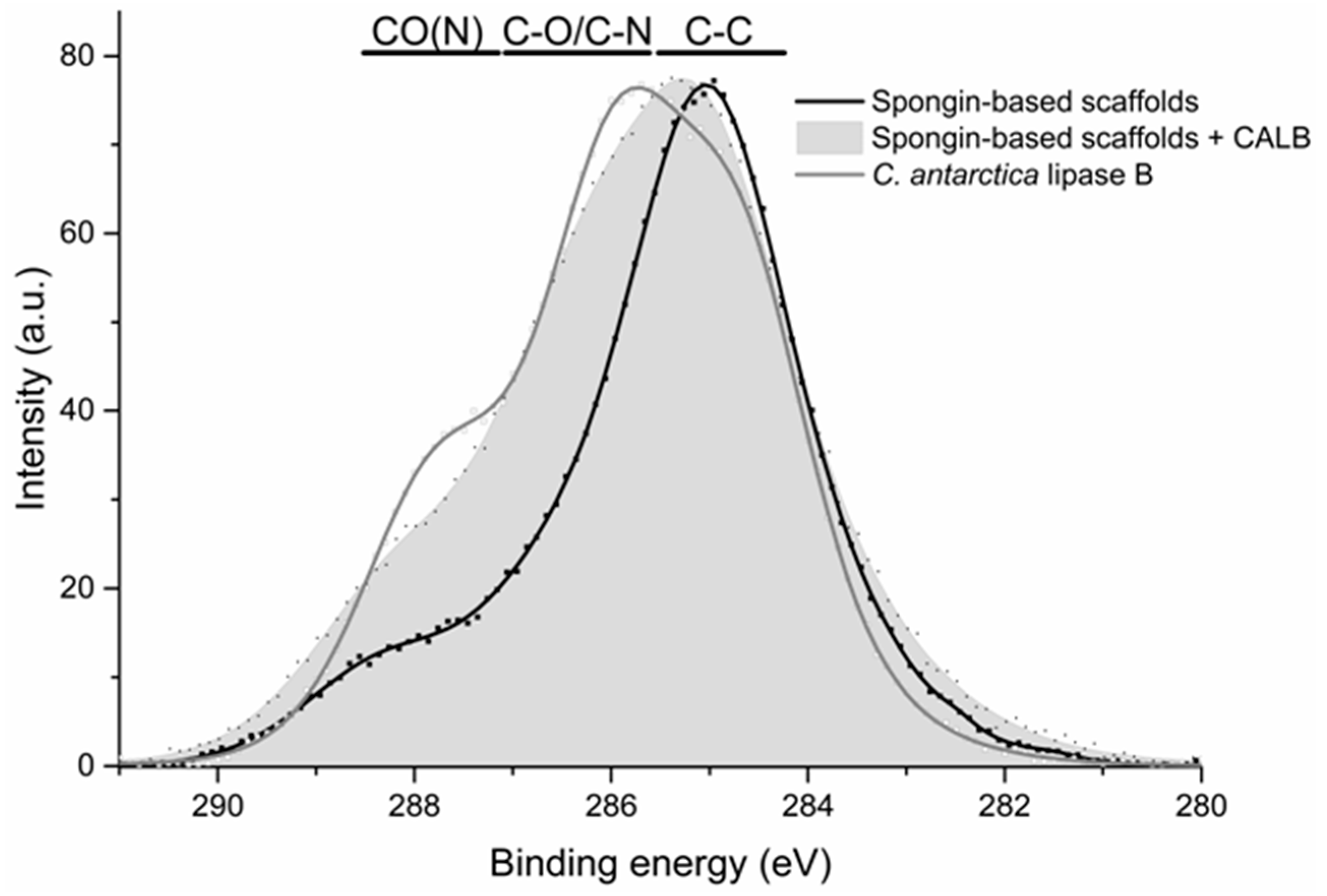
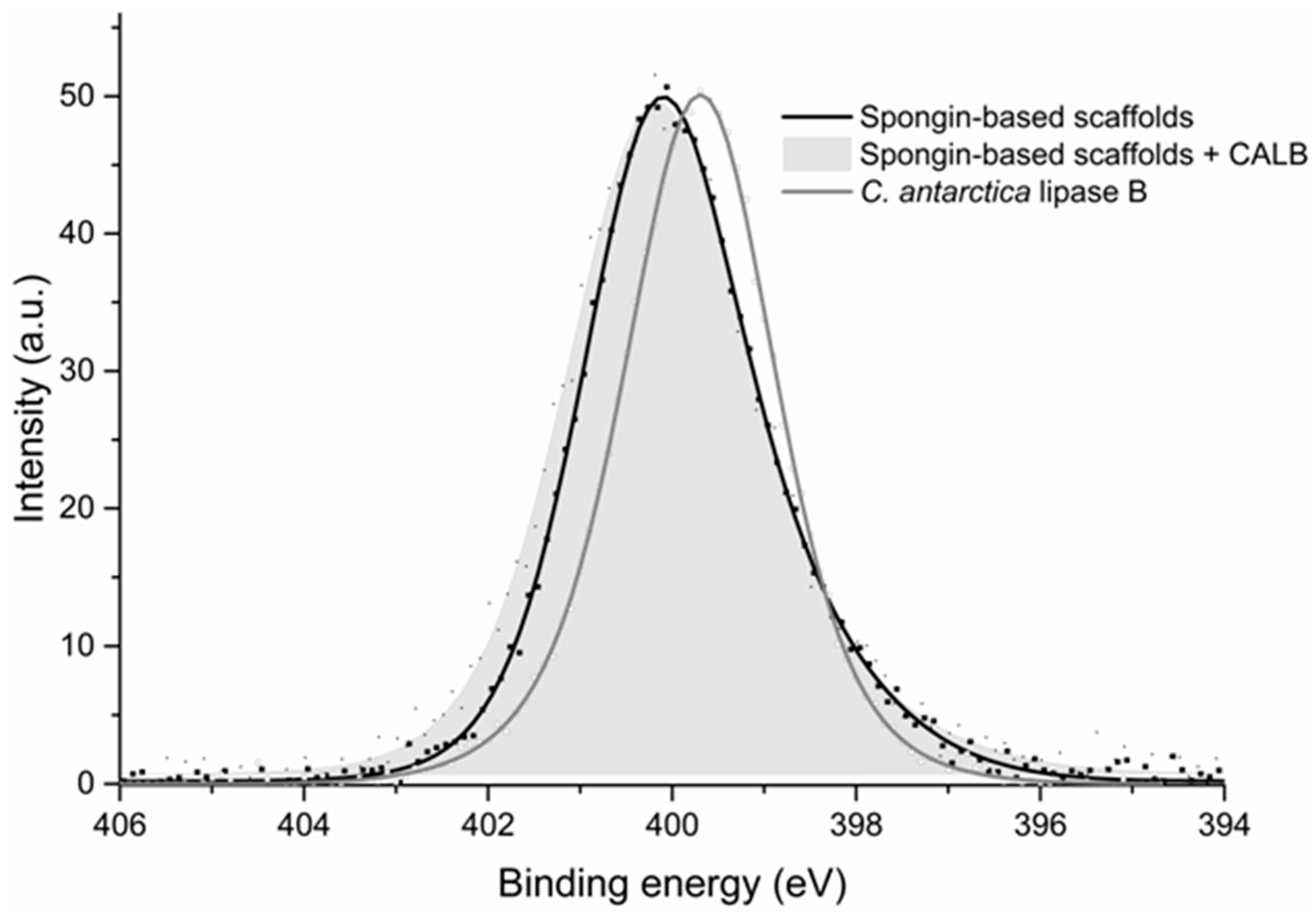
2.4. XPS Analysis
2.5. A Proposed Mechanism for CALB Attachment
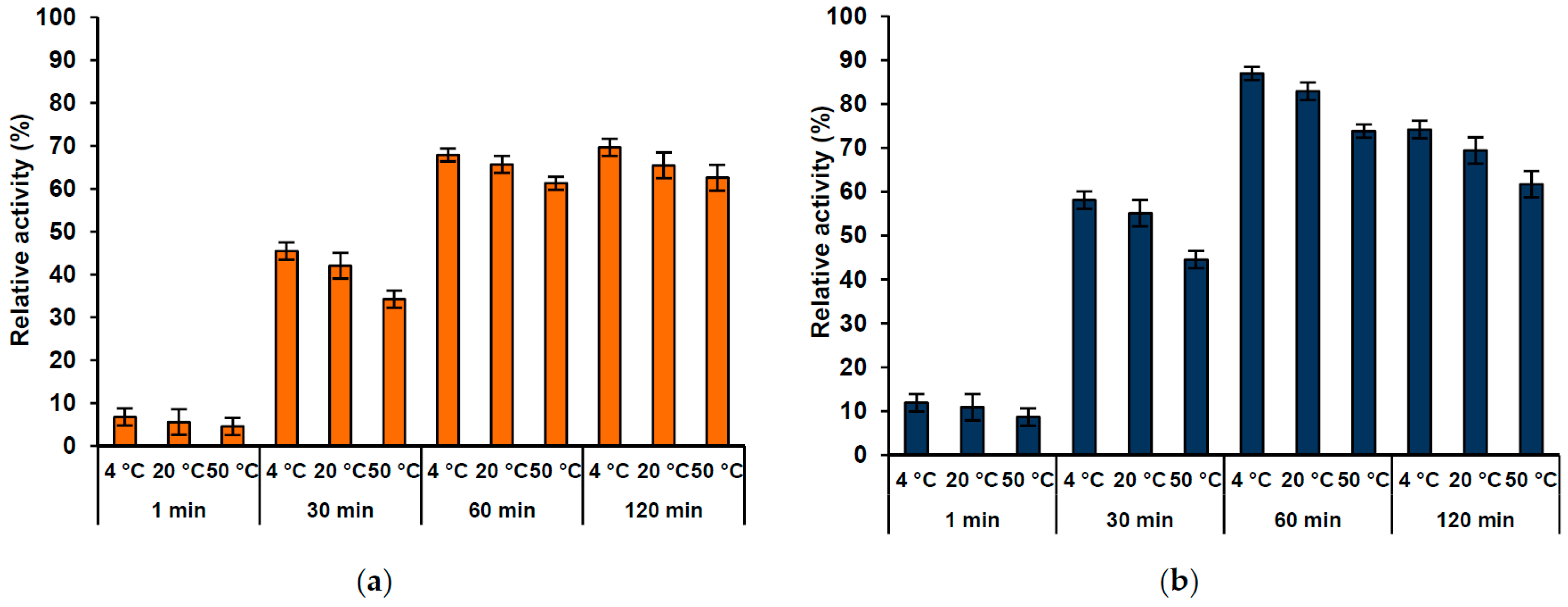
2.6. Lipase Activity Recovery
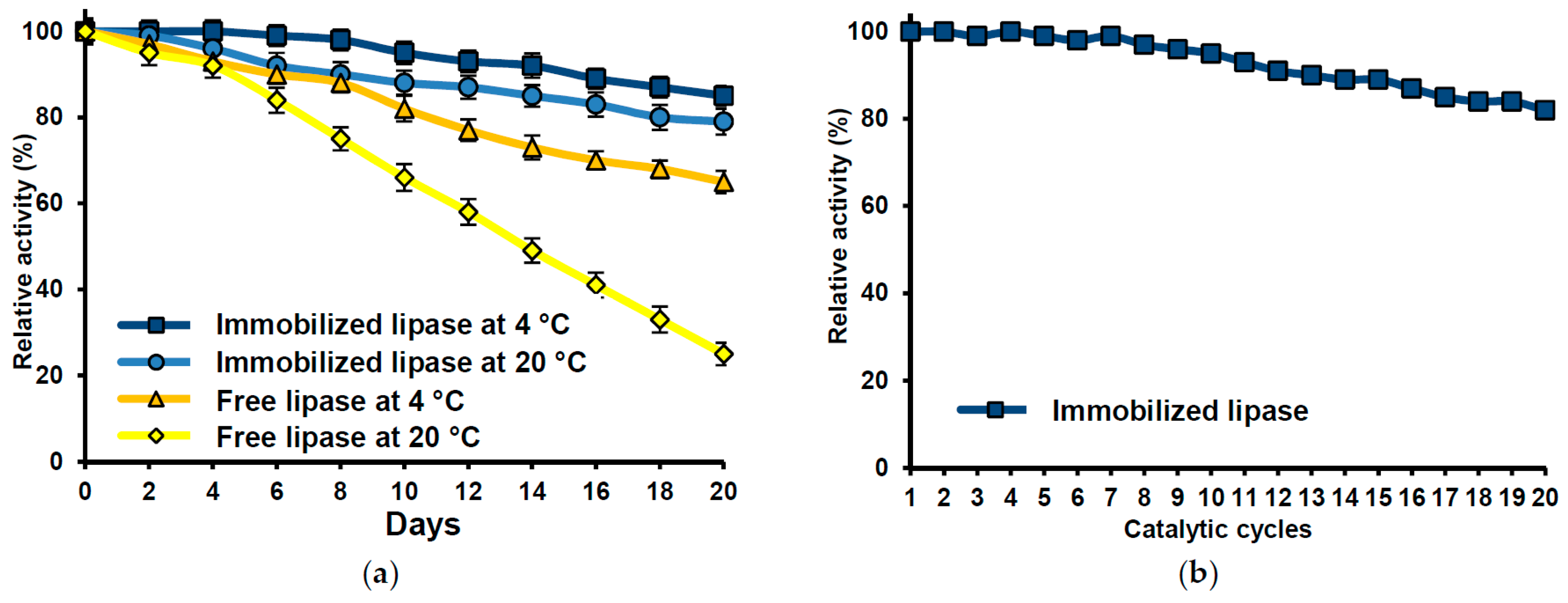
2.7. Immobilized Lipase Stability
2.8. Kinetic Parameters
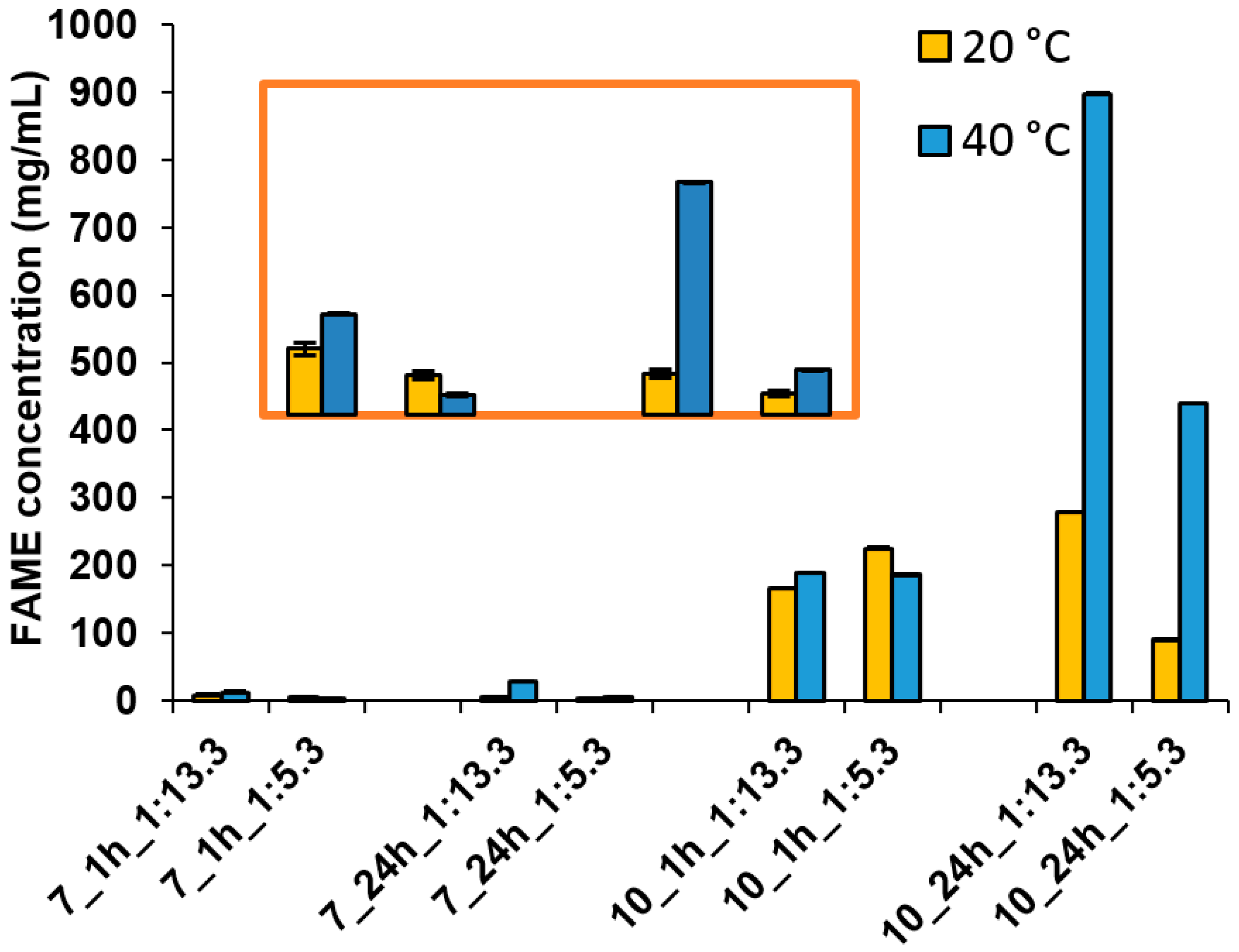
2.9. Rapeseed Oil Transesterification
3. Materials and Methods
3.1. Materials
3.2. Preparation of Spongin-Based Scaffolds from Hippospongia communis Demosponge
3.3. Lipase Immobilization
3.4. Analysis of Products Following Immobilization
3.5. Hydrolytic Activity
3.6. Stability of Immobilized Lipase
3.7. Kinetic Parameters
3.8. Rapeseed Oil Transesterification
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Rosevear, A. Immobilised biocatalysts—A critical review. J. Chem. Technol. Biotechnol. 1984, 34, 127–150. [Google Scholar] [CrossRef]
- Jesionowski, T.; Zdarta, J.; Krajewska, B. Enzyme immobilization by adsorption: A review. Adsorption 2014, 20, 801–821. [Google Scholar] [CrossRef]
- Guzik, U.; Hupert-Kocurek, K.; Wojcieszyńska, D. Immobilization as a strategy for improving enzyme properties-application to oxidoreductases. Molecules 2014, 19, 8995–9018. [Google Scholar] [CrossRef] [PubMed]
- Matosevic, S.; Szita, N.; Baganz, F. Fundamentals and applications of immobilized microfluidic enzymatic reactors. J. Chem. Technol. Biotechnol. 2011, 86, 325–334. [Google Scholar] [CrossRef]
- Nicolini, J.V.; de Resende, N.S.; Ferraz, H.C. Adsorption of horseradish peroxidase onto titanate nanowires. J. Chem. Technol. Biotechnol. 2015, 90, 739–746. [Google Scholar] [CrossRef]
- Wang, F.; Guo, C.; Liu, H.-Z.; Liu, C.-Z. Immobilization of Pycnoporus sanguineus laccase by metal affinity adsorption on magnetic chelator particles. J. Chem. Technol. Biotechnol. 2008, 83, 97–104. [Google Scholar] [CrossRef]
- Yewle, J.N.; Wei, Y.; Puleo, D.A.; Daunert, S.; Bachas, L.G. Oriented immobilization of proteins on hydroxyapatite surface using bifunctional bisphosphonates as linkers. Biomacromolecules 2012, 13, 1742–1749. [Google Scholar] [CrossRef] [PubMed]
- Banjanac, K.; Mihailović, M.; Prlainović, N.; Stojanović, M.; Carević, M.; Marinković, A.; Bezbradica, D. Cyanuric chloride functionalized silica nanoparticles for covalent immobilization of lipase. J. Chem. Technol. Biotechnol. 2016, 91, 439–448. [Google Scholar] [CrossRef]
- Tavares, A.P.M.; Silva, C.G.; Drazic, G.; Silva, A.M.T.; Loureiro, J.M.; Faria, J.L. Laccase immobilization over multi-walled carbon nanotubes: Kinetic, thermodynamic and stability studies. J. Colloid Interface Sci. 2015, 454, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Mohamad, N.R.; Marzuki, N.H.C.; Buang, N.A.; Huyop, F.; Wahab, R.A. An overview of technologies for immobilization of enzymes and surface analysis techniques for immobilized enzymes. Biotechnol. Biotechnol. Equip. 2015, 29, 205–220. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.; Christena, L.R.; Rajaram, Y.R.S. Enzyme immobilization: An overview on techniques and support materials. 3 Biotech 2013, 3, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Krajewska, B. Application of chitin- and chitosan-based materials for enzyme immobilizations: A review. Enzym. Microb. Technol. 2004, 35, 126–139. [Google Scholar] [CrossRef]
- Saeed, A.; Iqbal, M. Loofa (Luffa cylindrica) sponge: Review of development of the biomatrix as a tool for biotechnological applications. Biotechnol. Prog. 2013, 29, 573–600. [Google Scholar] [CrossRef] [PubMed]
- Pallela, R.; Ehrlich, H. Marine Sponges: Chemicobiological and Biomedical Applications; Springer International Publishing AG: New York, NY, USA, 2016. [Google Scholar]
- Dahlmans, J.J.; Meijerink, T.A.J. Insoluble Carrier-Bound Enzymes. U.S. Patent 3,919,048, 11 November 1975. [Google Scholar]
- Chen, J.P.; Lin, T.C. Loofa sponge as a scaffold for culture of Rat hepatocytes. Biotechnol. Prog. 2008, 21, 315–319. [Google Scholar] [CrossRef] [PubMed]
- Annunciado, T.R.; Sydenstricker, T.H.D.; Amico, S.C. Experimental investigation of various vegetable fibers as sorbent materials for oil spills. Mar. Pollut. Bull. 2005, 50, 1340–1346. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, H.; Maldonado, M.; Hanke, T.; Meissner, H.; Born, R. Spongins: Nanostructural investigations and development of biominimetic material model. VDI Berichte 2003, 1803, 287–292. [Google Scholar]
- Kim, M.M.; Mendis, E.; Rajapakse, N.; Lee, S.H.; Kim, S.K. Effect of spongin derived from Hymeniacidon sinapium on bone mineralization. J. Biomed. Mater. Res. Part B 2009, 90B, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Green, D.W.; Howard, D.; Yang, X.; Kelly, M.; Oreffo, R.O. Natural marine sponge fiber skeleton: A biomimetic scaffold for human osteoprogenitor cell attachment, growth, and differentiation. Tissue Eng. 2003, 9, 1159–1166. [Google Scholar] [CrossRef] [PubMed]
- Green, D.W. Tissue bionics: Examples in biomimetic tissue engineering. Biomed. Mater. 2008, 3, 034010. [Google Scholar] [CrossRef] [PubMed]
- Green, D.W.; Padula, M.P.; Santos, J.; Chou, J.; Milthorpe, B.; Ben-Nissan, B. A therapeutic potential for marine skeletal proteins in bone regeneration. Mar. Drugs 2013, 11, 1203–1220. [Google Scholar] [CrossRef] [PubMed]
- Norman, M.; Bartczak, P.; Zdarta, J.; Tylus, W.; Szatkowski, T.; Stelling, A.L.; Ehrlich, H.; Jesionowski, T. Adsorption of C.I. Natural Red 4 onto spongin skeleton of marine demosponge. Materials 2015, 8, 96–116. [Google Scholar] [CrossRef]
- Derewenda, Z.S.; Derewenda, U.; Dodson, G.G. The crystal and molecular structure of the Rhizomucor miehei triacylglyceride lipase at 1.9 Å resolution. J. Mol. Biol. 1992, 227, 818–839. [Google Scholar] [CrossRef]
- Palomo, J.M.; Munoz, G.; Fernandez-Lorente, G.; Mateo, C.; Fernandez-Lafuente, R.; Guisan, J.M. Interfacial adsorption of lipases on very hydrophobic support (octadecyl-Sepabeads): Immobilization, hyperactivation and stabilization of the open form of lipases. J. Mol. Catal. B 2008, 19, 279–286. [Google Scholar] [CrossRef]
- Silva, N.C.A.; Mirandaa, J.S.; Bolina, I.C.A.; Silva, W.C.; Hiratac, D.B.; de Castro, H.F.; Mendes, A.A. Immobilization of porcine pancreatic lipase on poly-hydroxybutyrate particles for the production of ethyl esters from macaw palm oils and pineapple flavor. Biochem. Eng. J. 2014, 82, 139–149. [Google Scholar] [CrossRef]
- Hernández, K.; Garcia-Galan, C.; Fernández-Lafuente, R. Simple and efficient immobilization of lipase B from Candida antarctica on porous styrene–divinylbenzene beads. Enzym. Microb. Technol. 2011, 49, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Mendes, A.A.; Rodrigues, D.S.; Filice, M.; Fernández-Lafuente, R.; Guisán, J.M.; Palomo, J.M. Regioselective monohydrolysis of per-O-acetylated-1-substituted-β-glucopyranosides catalyzed by immobilized lipases. Tetrahedron 2008, 64, 10721–10727. [Google Scholar] [CrossRef]
- Verdugo, C.; Luna, D.; Posadillo, A.; Sancho, E.D.; Rodriguez, S.; Bautista, F.; Luque, R.; Marinasa, J.M.; Romero, A.A. Production of a new second generation biodiesel with a low cost lipase derived from Thermomyces lanuginosus: Optimization by response surface methodology. Catal. Today 2011, 167, 107–112. [Google Scholar] [CrossRef]
- Luna, D.; Posadillo, A.; Caballero, V.; Verdugo, C.; Bautista, F.M.; Romero, A.A.; Sancho, E.D.; Luna, C.; Calero, J. New biofuel integrating glycerol into its composition through the use of covalent immobilized pig pancreatic lipase. Int. J. Mol. Sci. 2012, 13, 10091–10112. [Google Scholar] [CrossRef] [PubMed]
- Caballero, V.; Bautista, F.M.; Campelo, J.M.; Luna, D.; Marinas, J.M.; Romero, A.A.; Hidalgo, J.M.; Luque, R.; Macario, A.; Giordano, G. Sustainable preparation of a novel glycerol-free biofuel by using pig pancreatic lipase: Partial 1,3-regiospecific alcoholysis of sunflower oil. Process Biochem. 2009, 44, 334–342. [Google Scholar] [CrossRef]
- Szatkowski, T.; Wysokowski, M.; Lota, G.; Pęziak, D.; Bazhenov, V.V.; Nowaczyk, G.; Walter, J.; Molodtsov, S.L.; Stöcker, H.; Himcinschi, C.; et al. Novel nanostructured hematite—Spongin composite developed using an extreme biomimetic approach. RSC Adv. 2015, 5, 79031–79040. [Google Scholar] [CrossRef]
- Norman, M.; Bartczak, P.; Zdarta, J.; Ehrlich, H.; Jesionowski, T. Anthocyanin dye conjugated with Hippospongia communis marine demosponge skeleton and its antiradical activity. Dyes Pigment. 2016, 134, 541–552. [Google Scholar] [CrossRef]
- Zdarta, J.; Jesionowski, T. Luffa cylindrica sponges as a thermally and chemically stable support for Aspergillus niger lipase. Biotechnol. Prog. 2016, 32, 657–665. [Google Scholar] [CrossRef] [PubMed]
- Wong, P.T.T.; Nong, R.K.; Caputo, T.A.; Godwin, T.A.; Rigas, B. Infrared spectroscopy of exfoliated human cervical cells: Evidence of extensive structural changes during carcinogenesis. Proc. Natl. Acad. Sci. USA 1991, 88, 10988–10992. [Google Scholar] [CrossRef] [PubMed]
- Portaccio, M.; Della Ventura, B.; Mita, D.G.; Manolova, N.; Stoilova, O.; Rashkov, I.; Lepore, M. FT-IR microscopy characterization of sol–gel layers prior and after glucose oxidase immobilization for biosensing applications. J. Sol-Gel Sci. Technol. 2011, 57, 204–211. [Google Scholar] [CrossRef]
- Beinert, W.D.; Ruterjans, H.; Muller, F.; Bacher, A. Nuclear magnetic resonance studies of the old yellow enzyme: 2. 13C NMR of the enzyme recombined with 13C-labeled flavin mononucleotides. Eur. J. Biochem. 1985, 152, 581–587. [Google Scholar] [CrossRef] [PubMed]
- Zdarta, J.; Klapiszewski, Ł.; Wysokowski, M.; Norman, M.; Kołodziejczak-Radzimska, A.; Moszyński, D.; Ehrlich, H.; Maciejewski, H.; Stelling, A.L.; Jesionowski, T. Chitin-lignin material as a novel matrix for enzyme immobilization. Mar. Drugs 2015, 13, 2424–2446. [Google Scholar] [CrossRef] [PubMed]
- Tomizuka, N.; Ota, Y.; Yamada, K. Studies on lipase from Candida cylindracea. Agric. Biol. Chem. 1966, 30, 1090–1096. [Google Scholar]
- Uppenberg, J.; Hansen, M.T.; Patkar, S.; Jones, T.A. The sequence, crystal structure determination and refinement of two crystal forms of lipase B from Candida antarctica. Structure 1994, 2, 293–308. [Google Scholar] [CrossRef]
- Rouxhet, P.G.; Genet, M.J. XPS analysis of bio-organic systems. Surf. Interface Anal. 2011, 43, 1453–1470. [Google Scholar] [CrossRef]
- Porté-Durrieu, M.C.; Labrugere, F.; Villars, F.; Lefebvre, F.; Dutova, S.; Guette, A.; Bordenave, L.; Baquey, C. Development of RGD peptides grafted onto silica surfaces: XPS characterization and human endothelial cell interactions. J. Biomed. Mater. Res. Part A 1999, 46, 368–375. [Google Scholar] [CrossRef]
- Stevems, J.S.; Luca, M.; Pelendritis, M.; Terenghi, G.; Downes, S.; Schroeder, S.L.M. Quantitative analysis of complex amino acids and RGD peptides by X-ray photoelectron spectroscopy (XPS). Surf. Interface Anal. 2013, 45, 1238–1246. [Google Scholar] [CrossRef]
- Kerber, S.J.; Bruckner, J.J.; Wozniak, K.; Seal, S.; Hardcastle, S.; Barr, T.L. The nature of hydrogen in X-ray photoelectron spectroscopy: General patterns from hydroxides to hydrogen bonding. J. Vac. Sci. Technol. A 1996, 14, 1314–1320. [Google Scholar] [CrossRef]
- Garrone, R. The collagen of the Porifera. In Biology of Invertebrate and Lower Vertebrate Collagens; Bairati, A., Ed.; Plenum Press: New York, NY, USA, 1985; pp. 157–175. [Google Scholar]
- Svendsen, A. Lipase protein engineering. Biochim. Biophys. Acta 2000, 1543, 223–238. [Google Scholar] [CrossRef]
- Macario, A.; Moliner, M.; Diaz, U.; Jorda, J.L.; Corma, A.; Giordano, G. Biodiesel production by immobilized lipase on zeolites and related materials. Stud. Surface Sci. Catal. 2008, 174, 1011–1016. [Google Scholar]
- Zhang, R.; Xiao, X.; Tai, Q.; Huang, H.; Yang, J.; Hu, Y. Preparation of lignin–silica hybrids and its application in intumescent flame-retardant poly(lactic acid) system. High Perform. Polym. 2012, 24, 738–745. [Google Scholar] [CrossRef]
- Zhang, D.H.; Zhang, Y.F.; Zhi, G.Y.; Xie, Y.L. Effect of hydrophobic/hydrophilic characteristics of magnetic microspheres on the immobilization of BSA. Colloids Surfaces B 2011, 82, 302–306. [Google Scholar] [CrossRef] [PubMed]
- Narwal, S.K.; Saun, N.K.; Gupta, R. Characterization and catalytic properties of free and silica-bound lipase: A comparative study. J. Oleo Sci. 2014, 63, 599–605. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.T.; Ren, X.Y.; Liu, Y.M.; Wei, Y.; Qing, L.S.; Liao, X. Covalent immobilization of porcine pancreatic lipase on carboxyl-activated magnetic nanoparticles: Characterization and application for enzymatic inhibition assays. Mater. Sci. Eng. 2014, 38, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Cabrera-Padilla, R.Y.; Lisboa, M.C.; Fricks, A.T.; Franceschi, E.; Lima, A.S.; Silva, D.P.; Soares, C.M.F. Immobilization of Candida rugose lipase on poly (3-hydroxybutyrate-co-hydroxyvalerate): A new eco-friendly support. J. Ind. Microbiol. Biotechnol. 2012, 39, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Elnashar, M.M.M.; Mostafa, H.; Morsy, N.A.; Awad, G.E.A. Biocatalysts: Isolation, identification, and immobilization of thermally stable lipase onto three novel biopolymeric supports. Ind. Eng. Chem. Res. 2013, 52, 14760–14767. [Google Scholar] [CrossRef]
- Dong, L.; Ge, C.; Qin, P.; Chen, Y.; Xu, Q. Immobilization and catalytic properties of Candida lipolytic lipase on surface of organic intercalated and modified MgAl-LDHs. Solid State Sci. 2014, 31, 8–15. [Google Scholar] [CrossRef]
- Bencze, L.C.; Bartha-Vari, B.H.; Katona, G.; Tos, M.I.; Paizs, C.; Irimie, F.D. Nanobioconjugates of Candida antarctica lipase B and single-walled carbon nanotubes in biodiesel production. Bioresour. Technol. 2016, 200, 853–860. [Google Scholar] [CrossRef] [PubMed]
- Gremos, S.; Kekos, D.; Kolisis, F. Supercritical carbon dioxide biocatalysis as a novel and green methodology for the enzymatic acylation of fibrous cellulose in one step. Bioresour. Technol. 2012, 115, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Tutar, H.; Yilmaz, E.; Pehlivan, E.; Yilmaz, M. Immobilization of Candida rugosa lipase on sporopollenin from Lycopodium clavatum. Int. J. Biolog. Macromol. 2009, 45, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Talbert, J.N.; Wang, L.S.; Duncan, B.; Jeong, Y.; Andler, S.M.; Rotello, V.M.; Goddard, J.M. Immobilization and stabilization of lipase (CaLB) through hierarchical interfacial assembly. Biomacromolecules 2014, 15, 3915–3922. [Google Scholar] [CrossRef] [PubMed]
- Landarani-Isfahani, A.; Taheri-Kafrani, A.; Amini, M.; Mirkhani, V.; Moghadam, M.; Soozanipour, A.; Razmjou, A. Xylanase immobilized on novel multifunctional hyperbranched polyglycerol-grafted magnetic nanoparticles: An efficient and robust biocatalyst. Langmuir 2015, 31, 9219–9227. [Google Scholar] [CrossRef] [PubMed]
- Zivkovic, L.T.; Zivkovic, L.S.; Babic, B.M.; Kokunesoski, M.J.; Jokic, B.M.; Karadzi, I.M. Immobilization of Candida rugosa lipase by adsorption onto biosafe meso/macroporous silica and zirconia. Biochem. Eng. J. 2015, 93, 73–83. [Google Scholar] [CrossRef]
- Calero, J.; Verdugo, C.; Luna, D.; Sancho, E.D.; Luna, C.; Posadillo, A.; Bautista, F.M.; Romero, A.A. Selective ethanolysis of sunflower oil with Lipozyme RM IM, an immobilized Rhizomucor miehei lipase, to obtain a biodiesel-like biofuel, which avoids glycerol production through the monoglyceride formation. New Biotechnol. 2014, 31, 596–601. [Google Scholar] [CrossRef] [PubMed]
- Dizge, N.; Keskinler, B. Enzymatic production of biodiesel from canola oil using immobilized lipase. Biomass Bioenergy 2008, 32, 1274–1278. [Google Scholar] [CrossRef]
- Xu, Y.; Nordblad, M.; Nielsen, P.M.; Brask, J.; Woodley, J.M. In situ visualization and effect of glycerol in lipase-catalyzed ethanolysis of rapeseed oil. J. Mol. Catal. B 2011, 72, 213–219. [Google Scholar] [CrossRef]
- Zdarta, J.; Sałek, K.; Kołodziejczak-Radzimska, A.; Siwińska-Stefańska, K.; Szwarc-Rzepka, K.; Norman, M.; Klapiszewski, Ł.; Bartczak, P.; Kaczorek, E.; Jesionowski, T. Immobilization of Amano Lipase A onto Stöber silica surface: Process characterization and kinetic studies. Open Chem. 2015, 13, 138–148. [Google Scholar] [CrossRef]
- Bradford, M.M. Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
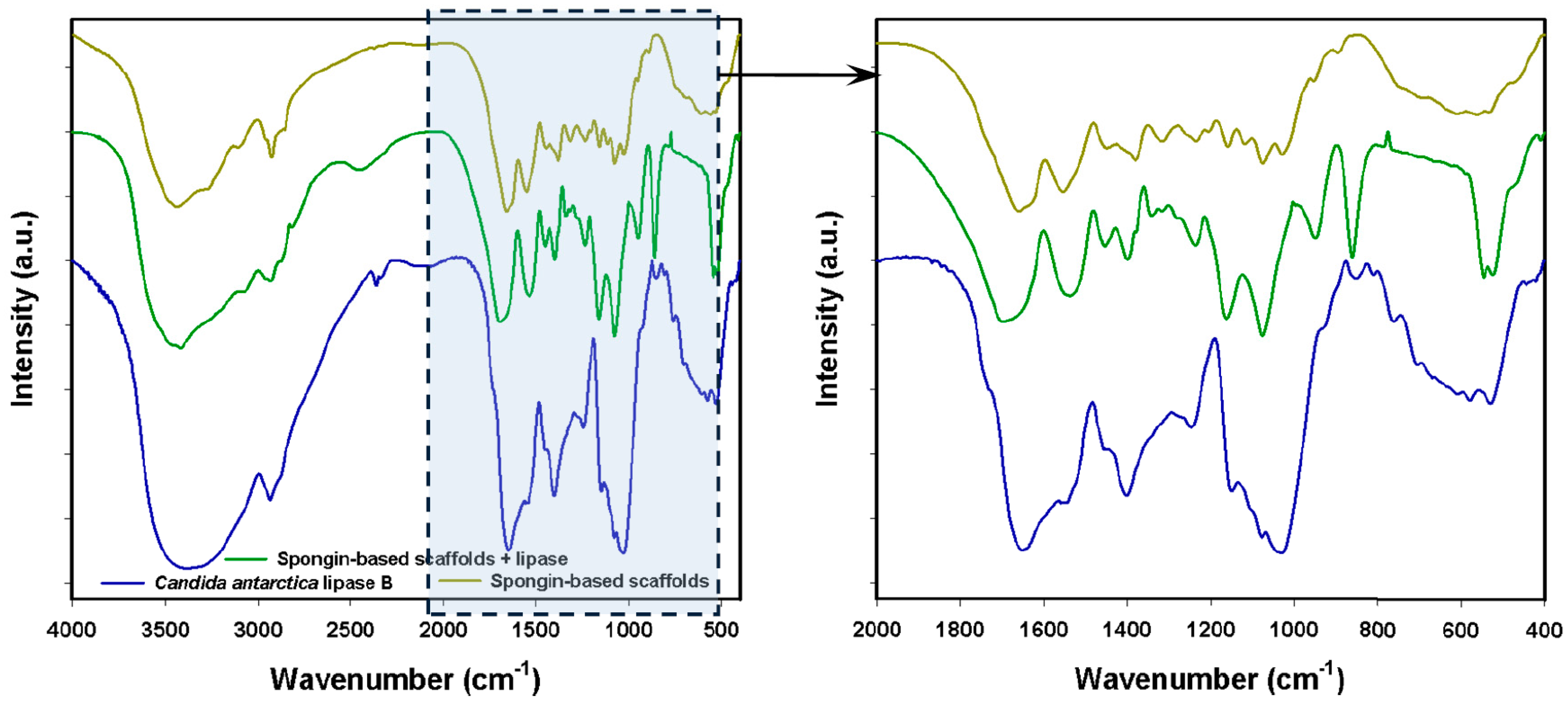


| Candida antarctica Lipase B | Spongin | Product Following Immobilization | Vibrational Assignment |
|---|---|---|---|
| 3480 | 3435 | 3445 | –OH stretching |
| 3325 | 3292 | 3295 | –NH stretching |
| 2937 | 2927 | 2929 | C–H stretching |
| 1643 | 1647 | 1655 | –NH deformational (amide I) |
| 1542 | 1546 | 1538 | –NH stretching (amide II) |
| 1394 | 1375 | 1390 | C–OH bending |
| 1256 | 1243 | 1240 | C–N stretching (amide III) |
| 1026 | 1154, 1073 | 1150, 1065 | C–O–C stretching |
| 815 | - | 820 | –NH2 deformational |
| 645–535 | 640–530 | 580–530 | C–C bending |
| Sample Name | Atomic % | Ratio of | |||||||
|---|---|---|---|---|---|---|---|---|---|
| C | O | N | Ca | Si | Na | P | N/C | O/C | |
| C. antarctica lipase B | 58.2 | 30.7 | 11.1 | - | - | - | - | 0.19 | 0.53 |
| Spongin-based scaffolds | 73.4 | 17.4 | 7.3 | 0.6 | 1.3 | - | - | 0.10 | 0.24 |
| Spongin-based scaffolds + CALB | 53.9 | 31.7 | 9.7 | 0.8 | - | 1.3 | 2.6 | 0.18 | 0.59 |
| Sample Number | Temperature (°C) | pH | no/nm | Time (h) | Conversion (%) | TG (% w/w) | FAME (% w/w) | Glycerol (% w/w) |
|---|---|---|---|---|---|---|---|---|
| 1 | 20 | 7 | 1:13.3 | 1 | 0.9 ± 0.1 | 99.1 ± 0.1 | 0.8 ± 0.1 | 0.1 ± 0.1 |
| 2 | 24 | 0.6 ± 0.1 | 99.4 ± 0.1 | 0.6 ± 0.1 | 0.0 ± 0.1 | |||
| 3 | 1:5.3 | 1 | 0.7 ± 0.1 | 99.3 ± 0.1 | 0.7 ± 0.1 | 0.0 ± 0.1 | ||
| 4 | 24 | 0.4 ± 0.1 | 99.6 ± 0.1 | 0.4 ± 0.1 | 0.0 ± 0.1 | |||
| 5 | 10 | 1:13.3 | 1 | 18.5 ± 0.5 | 81.5 ± 0.5 | 16.9 ± 0.5 | 1.6 ± 0.1 | |
| 6 | 24 | 31.1 ± 0.7 | 68.9 ± 0.7 | 28.4 ± 0.7 | 2.7 ± 0.2 | |||
| 7 | 1:5.3 | 1 | 13.4 ± 0.4 | 66.7 ± 0.7 | 31.1 ± 0.7 | 2.2 ± 0.1 | ||
| 8 | 24 | 33.3 ± 0.7 | 86.6 ± 0.4 | 12.5 ± 0.7 | 0.9 ± 0.1 | |||
| 9 | 40 | 7 | 1:13.3 | 1 | 1.4 ± 0.1 | 98.6 ± 0.1 | 1.3 ± 0.1 | 0.1 ± 0.1 |
| 10 | 24 | 3.1 ± 0.2 | 96.9 ± 0.2 | 2.8 ± 0.21 | 0.3 ± 0.1 | |||
| 11 | 1:5.3 | 1 | 0.4 ± 0.1 | 99.6 ± 0.1 | 0.4 ± 0.1 | 0.0 ± 0.1 | ||
| 12 | 24 | 0.8 ± 0.1 | 99.2 ± 0.1 | 0.8 ± 0.1 | 0.0 ± 0.1 | |||
| 13 | 10 | 1:13.3 | 1 | 21.1 ± 0.4 | 78.9 ± 0.4 | 19.3 ± 0.4 | 1.8 ± 0.2 | |
| 14 | 24 | 99.1 ± 0.9 | 0.0 ± 0.9 | 91.4 ± 0.9 | 8.6 ± 0.5 | |||
| 15 | 1:5.3 | 1 | 27.5 ± 0.6 | 72.5 ± 0.6 | 25.7 ± 0.6 | 1.8 ± 0.2 | ||
| 16 | 24 | 64.9 ± 0.8 | 35.1 ± 0.8 | 60.7 ± 0.8 | 4.2 ± 0.3 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zdarta, J.; Norman, M.; Smułek, W.; Moszyński, D.; Kaczorek, E.; Stelling, A.L.; Ehrlich, H.; Jesionowski, T. Spongin-Based Scaffolds from Hippospongia communis Demosponge as an Effective Support for Lipase Immobilization. Catalysts 2017, 7, 147. https://doi.org/10.3390/catal7050147
Zdarta J, Norman M, Smułek W, Moszyński D, Kaczorek E, Stelling AL, Ehrlich H, Jesionowski T. Spongin-Based Scaffolds from Hippospongia communis Demosponge as an Effective Support for Lipase Immobilization. Catalysts. 2017; 7(5):147. https://doi.org/10.3390/catal7050147
Chicago/Turabian StyleZdarta, Jakub, Małgorzata Norman, Wojciech Smułek, Dariusz Moszyński, Ewa Kaczorek, Allison L. Stelling, Hermann Ehrlich, and Teofil Jesionowski. 2017. "Spongin-Based Scaffolds from Hippospongia communis Demosponge as an Effective Support for Lipase Immobilization" Catalysts 7, no. 5: 147. https://doi.org/10.3390/catal7050147