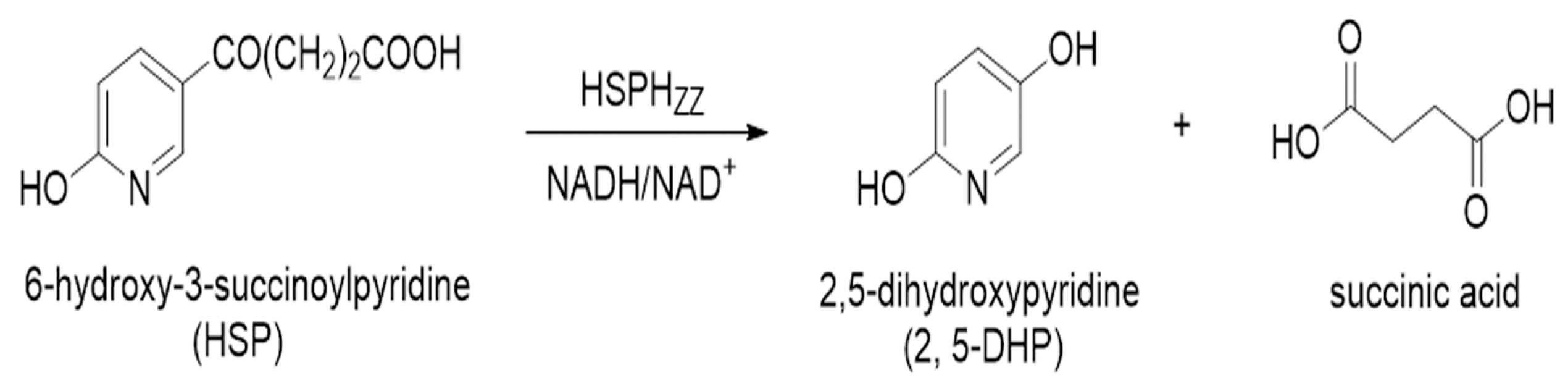
Characterization of a Novel Nicotine Hydroxylase from Pseudomonas sp. ZZ-5 That Catalyzes the Conversion of 6-Hydroxy-3-Succinoylpyridine into 2,5-Dihydroxypyridine
Abstract
:1. Introduction
2. Results and Discussion
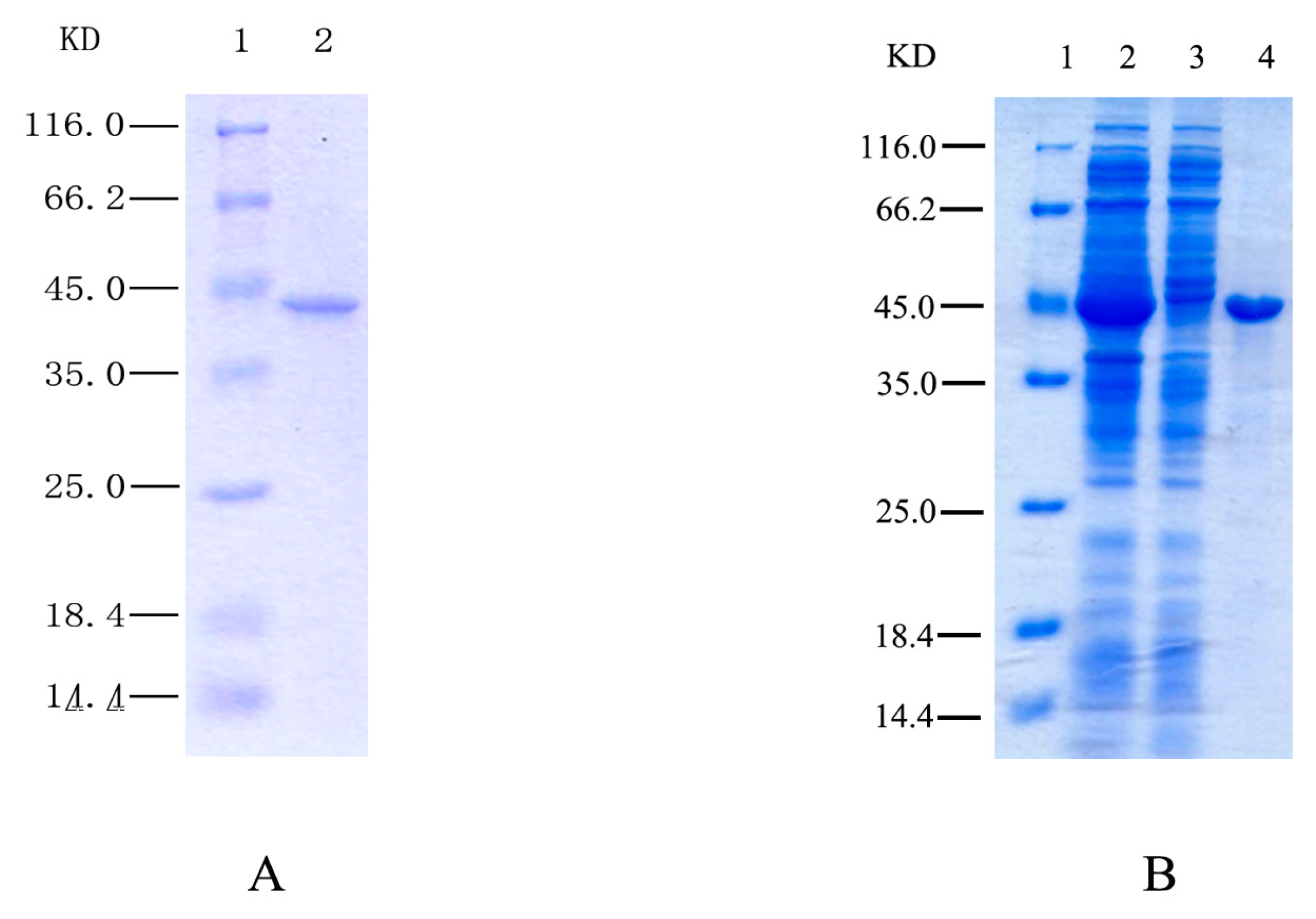
2.1. Purification and Identification of HSPHZZ
2.2. Gene Cloning and Sequence Analysis of HSPHZZ
2.3. Expression of Recombinant HSPHZZ in Escherichia coli BL21-Codon Plus (DE3)-RIL
2.4. Effect of Temperature and pH on Activity of the Recombinant HSPHZZ
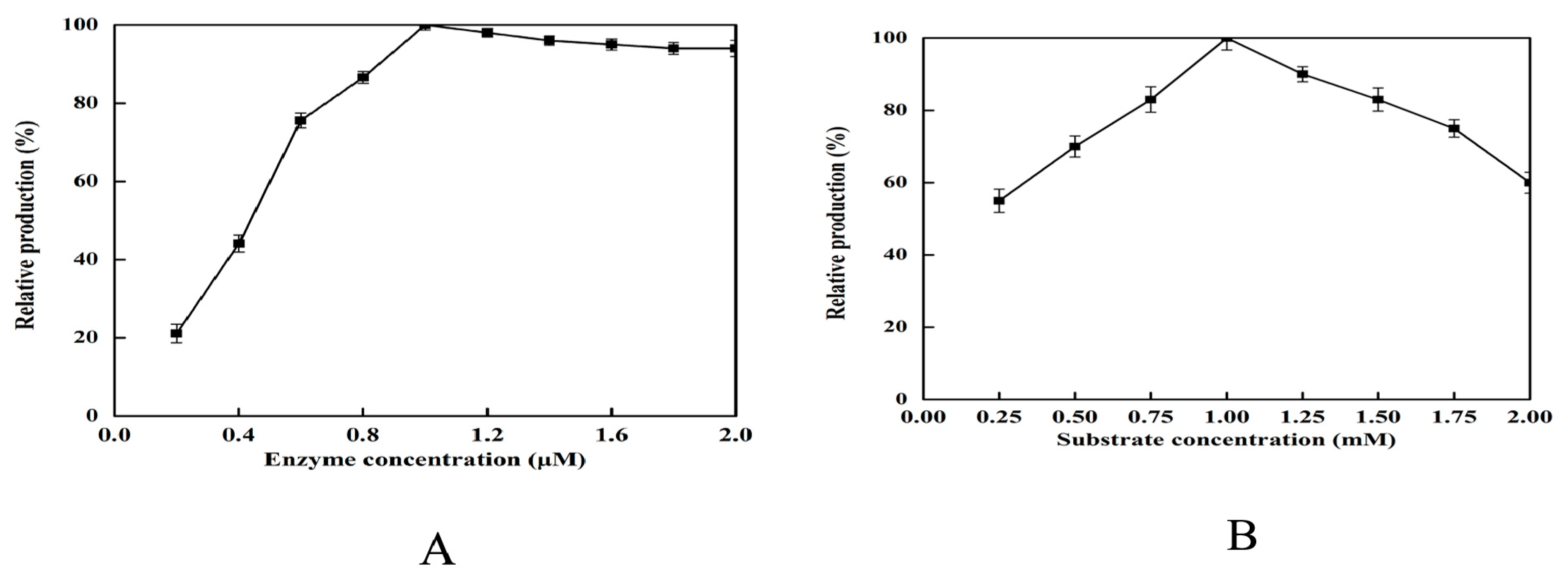
2.5. Effect of Enzyme and Substrate Concentration on 2,5-DHP Production
2.6. Effect of Metal Ions, Organic Solvents, and Detergents on Enzymatic Activity
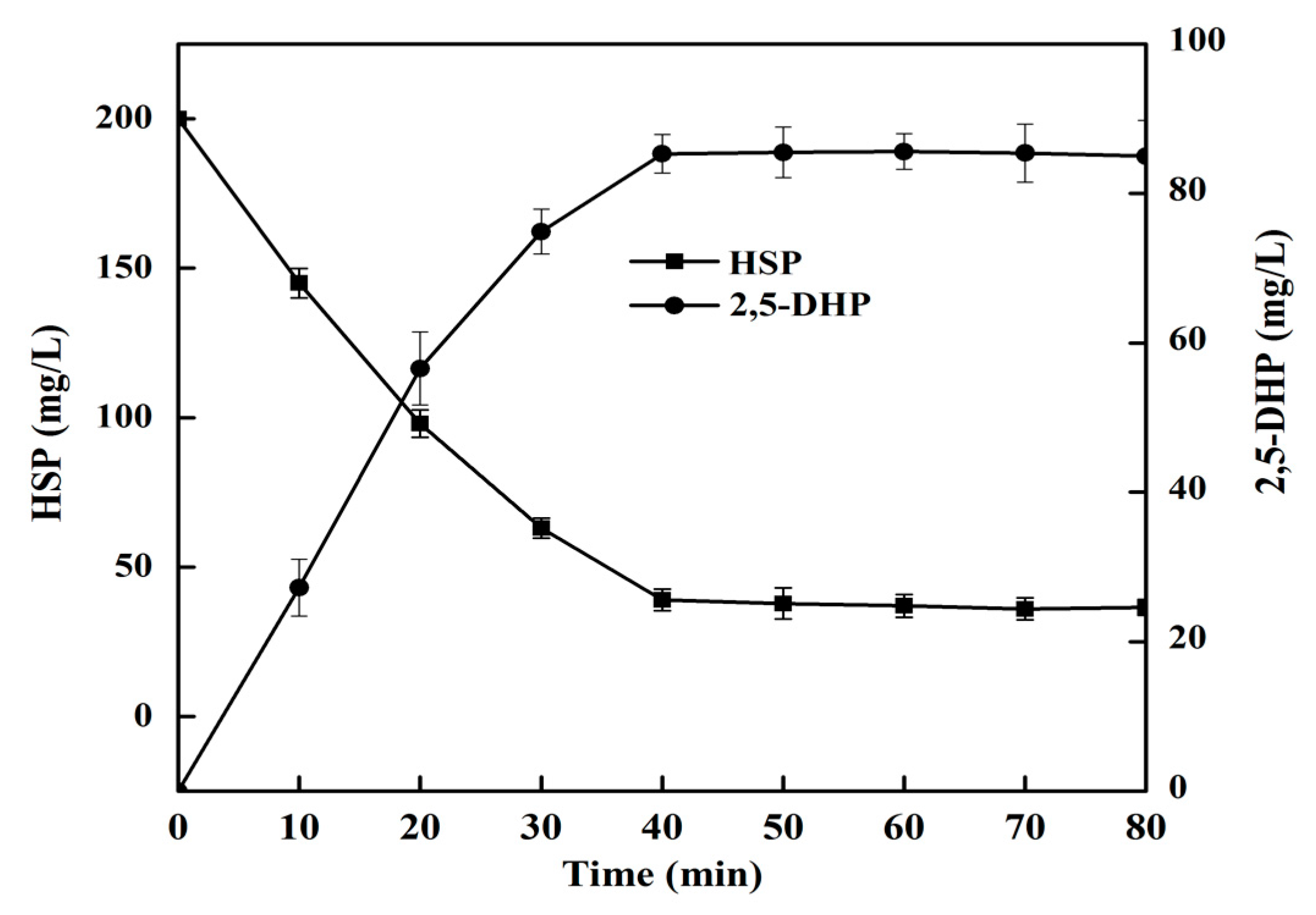
2.7. 2,5-DHP Production from HSP by HSPHZZ under Optimum Conditions
3. Experimental Section
3.1. Chemicals, Strains, and Plasmids
3.2. Purification of HSPHZZ
3.3. Enzymatic Activity Assay
3.4. N-Terminal Amino Acid Sequence of HSPHZZ
3.5. Gene Cloning and Construction of Expression Plasmid
3.6. Expression and Purification of Recombinant HSPHZZ
3.7. Catalytic Properties of Recombinant HSPHZZ
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Benowitz, N.L. Nicotine addiction. N. Engl. J. Med. 2010, 362, 2295–2303. [Google Scholar] [CrossRef] [PubMed]
- Hecht, S.S. Tobacco smoke carcinogens and lung cancer. J. Natl. Cancer Inst. 1999, 91, 1194–1210. [Google Scholar] [CrossRef] [PubMed]
- Civilini, M.; Domenis, C.; Sebastianutto, N.; de Bertoldi, M. Nicotine decontamination of tobacco agro-industrial waste and its degradation by micro-organisms. Waste Manag. Res. 1997, 15, 349–358. [Google Scholar] [CrossRef]
- Novotny, T.E.; Zhao, F. Consumption and production waste: Another externality of tobacco use. Tob. Control 1999, 8, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Seckar, J.A.; Stavanja, M.S.; Harp, P.R.; Yi, Y.; Garner, C.D.; Doi, J. Environmental fate and effects of nicotine released during cigarette production. Environ. Toxicol. Chem. 2008, 27, 1505–1514. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.H.; He, H.Z.; Wang, M.Z.; Wang, S.; Zhang, J.; Wei, W.; Xu, H.X.; Lu, Z.M.; Shen, D.S. Bioaugmentation of activated sludge with Acinetobacter sp. TW enhances nicotine degradation in a synthetic tobacco wastewater treatment system. Bioresour. Technol. 2013, 142, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Zhong, W.; Zhu, C.; Shu, M.; Sun, K.; Zhao, L.; Wang, C.; Ye, Z.; Chen, J. Degradation of nicotine in tobacco waste extract by newly isolated Pseudomonas sp. ZUTSKD. Bioresour. Technol. 2010, 101, 6935–6941. [Google Scholar] [CrossRef] [PubMed]
- Ruan, A.D.; Min, H.; Zhu, W. Studies on biodegradation of nicotine by Arthrobacter sp. strain HF-2. J. Environ. Sci. Health B 2006, 41, 1159–1170. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.N.; Xu, P.; Tang, H.Z.; Meng, J.; Liu, X.L.; Huang, J.; Chen, H.; Du, Y.; Blankespoor, H.D. Biodegradation and detoxification of nicotine in tobacco solid waste by a Pseudomonas sp. Biotechnol. Lett. 2004, 26, 1493–1496. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.Z.; Yang, G.Q.; Min, H.; Lv, Z.M. A novel nicotine catabolic plasmid pMH1 in Pseudomonas sp. strain HF-1. Can. J. Microbiol. 2009, 55, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.G.; Ma, Y.; Wen, Y.Z.; Chen, L.S.; Wu, L.F.; Liu, W.P. Functional identification of two novel genes from Pseudomonas sp. strain HZN6 involved in the catabolism of nicotine. Appl. Environ. Microbiol. 2012, 78, 2154–2160. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.J.; Lu, L.L.; Gu, G.F.; Xiao, M. A novel pathway for nicotine degradation by Aspergillus oryzae 112822 isolated from tobacco leaves. Res. Microbiol. 2010, 161, 626–633. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.G.; Li, N.; Lu, Z.M.; Yang, Y.J.; Ma, Y.; Niu, L.L.; He, J.; Liu, W.P. Conversion of nornicotine to 6-hydroxy-nornicotine and 6-hydroxy-myosmine by Shinella sp. strain HZN7. Appl. Microbiol. Biotechnol. 2016, 100, 10019–10029. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.Z.; Yang, G.Q.; Wang, X.; Yao, Y.L.; Min, H.; Lu, Z.M. Nicotine degradation by two novel bacterial isolates of Acinetobacter sp. TW and Sphingomonas sp. TY and their responses in the presence of neonicotinoid insecticides. World J. Microbiol. Biotechnol. 2011, 27, 1633–1640. [Google Scholar] [CrossRef]
- Enamorado, M.F.; Ondachi, P.W.; Comins, D.L. A five-step synthesis of (S)-macrostomine from (S)-nicotine. Org. Lett. 2010, 12, 4513–4515. [Google Scholar] [CrossRef] [PubMed]
- Ondachi, P.W.; Comins, D.L. Synthesis of fused-ring nicotine derivatives from (S)-nicotine. J. Org. Chem. 2010, 75, 1706–1716. [Google Scholar] [CrossRef] [PubMed]
- Kagarlitskii, A.D.; Iskakova, M.K.; Turmukhambetov, A.Z. Catalytic conversion of nicotine into nicotinonitrile–a pharmaceutical intermediate product. Pharm. Chem. J. 2002, 36, 26–27. [Google Scholar] [CrossRef]
- Fananas, F.J.; Arto, T.; Mendoza, A.; Rodriguez, F. Synthesis of 2,5-dihydropyridine derivatives by gold-catalyzed reactions of β-ketoesters and propargylamines. Org. Lett. 2011, 13, 4184–4187. [Google Scholar] [CrossRef] [PubMed]
- Abbas, H.A.S.; El Sayed, W.A.; Fathy, N.M. Synthesis and antitumor activity of new dihydropyridine thioglycosides and their corresponding dehydrogenated forms. Eur. J. Med. Chem. 2010, 45, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Briede, J.; Stivrina, M.; Vigante, B.; Stoldere, D.; Duburs, G. Acute effect of antidiabetic 1,4-dihydropyridine compound cerebrocrast on cardiac function and glucose metabolism in the isolated, perfused normal rat heart. Cell Biochem. Chem. Funct. 2008, 26, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Kappe, C.O. Biologically active dihydropyrimidones of the Biginelli-type—A literature survey. Eur. J. Med. Chem. 2000, 35, 1043–1052. [Google Scholar] [CrossRef]
- Hilgeroth, A. Dimeric 4-aryl-1,4-dihydropyridines: Development of a third class of nonpeptidic HIV-1 protease inhibitors. Mini-Rev. Med. Chem. 2002, 2, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.Z.; Yao, Y.X.; Zhang, D.K.; Meng, X.Z.; Wang, L.J.; Yu, H.; Ma, L.Y.; Xu, P. A novel NADH-dependent and FAD-containing hydroxylase is crucial for nicotine degradation by Pseudomonas putida. J. Biol. Chem. 2011, 286, 39179–39187. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Huang, H.; Xie, K.; Xu, P. Identification of nicotine biotransformation intermediates by Agrobacterium tumefaciens strain S33 suggests a novel nicotine degradation pathway. Appl. Microbiol. Biotechnol. 2012, 95, 1567–1578. [Google Scholar] [CrossRef] [PubMed]
- Eppink, M.H.; Schreuder, H.A.; Van Berkel, W.J. Identification of a novel conserved sequence motif in flavoprotein hydroxylases with a putative dual function in FAD/NAD(P)H binding. Protein Sci. 1997, 6, 2454–2458. [Google Scholar] [CrossRef] [PubMed]
- Li, H.L.; Xie, K.B.; Huang, H.Y.; Wang, S.N. 6-Hydroxy-3-Succinoylpyridine hydroxylase catalyzes a central step of nicotine degradation in Agrobacterium tumefaciens S33. PLoS ONE 2014, 9, e103324. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Huang, S.; Zang, J.; Jia, C.X.; Mao, D.B. Cloning, expression and characterization of a novel fructosyltransferase from Aspergillus oryzae ZZ-01 for the synthesis of sucrose 6-acetate. Catalysts 2016, 6, 67. [Google Scholar] [CrossRef]
- Yang, Z.; Pan, W.B. Ionic liquids: Green solvents for nonaqueous biocatalysis. Enzym. Microb. Technol. 2005, 37, 19–28. [Google Scholar] [CrossRef]
- Wang, S.N.; Liu, Z.; Tang, H.Z.; Meng, J.; Xu, P. Characterization of environmentally friendly nicotine degradation by Pseudomonas putida biotype A strain S16. Microbiology 2007, 153, 1556–1565. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Hausinger, R.P.; Tang, H.Z.; Xu, P. Mechanism of the 6-Hydroxy-3-succinoyl-pyridine 3-monooxygenase flavoprotein from Pseudomonas putida S16. J. Biol. Chem. 2014, 289, 29158–29170. [Google Scholar] [CrossRef] [PubMed]

| Step | Total Protein (mg) | Total Activity (U) | Specific Activity (U/mg) | Fold | Yield (%) |
|---|---|---|---|---|---|
| Crude cell extract | 672.6 | 195.4 | 0.29 | 1 | 100 |
| (NH4)2SO4 precipitation | 542.3 | 173.5 | 0.31 | 1.1 | 88.8 |
| DEAE sepharose Phenyl sepharose | 75.2 22.3 | 100.3 54.5 | 1.3 2.4 | 4.6 8.4 | 51.3 27.8 |
| Superdex-200 | 5.1 | 26.1 | 5.1 | 17.6 | 13.4 |
| Substrate | kcat(s−1) | Km(mM) | kcat/Km(s−1 mM−1) |
|---|---|---|---|
| HSP | 2.1 ± 0.3 | 0.18 ± 0.04 | 11.7 ± 1.5 |
| NADH | 1.3 ± 0.2 | 0.23 ± 0.03 | 5.7 ± 1.1 |
| Metals or Inhibitors | Concentration | Relative Activity (%) |
|---|---|---|
| None | - | 100 |
| Mg2+ | 5 mM | 86 ± 3 |
| Zn2+ | 5 mM | 27 ± 2 |
| Cu2+ | 5 mM | 23 ± 1 |
| Ca2+ | 5 mM | 92 ± 2 |
| Mn2+ | 5 mM | 87 ± 4 |
| Fe2+ | 5 mM | 13 ± 2 |
| Ni2+ | 5 mM | 81 ± 3 |
| Co2+ | 5 mM | 31 ± 3 |
| K+ | 5 mM | 101 ± 5 |
| Na+ | 5 mM | 98 ± 2 |
| EDTA | 0.5 mM 5 mM | 79 ± 2 85 ± 2 |
| Metals or Inhibitors | Concentration (%) | Relative Activity (%) |
|---|---|---|
| None | — | 100 |
| Methanol | 20 (v/v) | 61 ± 2 |
| Acetone | 20 (v/v) | 59 ± 4 |
| Chloroform | 20 (v/v) | 33 ± 2 |
| Formaldehyde | 20 (v/v) | 23 ± 3 |
| Toluene | 20 (v/v) | 28 ± 5 |
| DMF | 20 (v/v) | 65 ± 2 |
| DMSO | 20 (v/v) | 93 ± 5 |
| Tween 20 | 1 (w/v) | 86 ± 5 |
| Tween 40 | 1 (w/v) | 110 ± 4 |
| Tween 80 | 1 (w/v) | 91 ± 1 |
| Triton X-100 | 1 (w/v) | 48 ± 3 |
| Span 20 | 1 (w/v) | 12 ± 4 |
| Span 40 | 1 (w/v) | 17 ± 5 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, T.; Zang, J.; Zheng, Y.; Tang, H.; Huang, S.; Mao, D. Characterization of a Novel Nicotine Hydroxylase from Pseudomonas sp. ZZ-5 That Catalyzes the Conversion of 6-Hydroxy-3-Succinoylpyridine into 2,5-Dihydroxypyridine. Catalysts 2017, 7, 257. https://doi.org/10.3390/catal7090257
Wei T, Zang J, Zheng Y, Tang H, Huang S, Mao D. Characterization of a Novel Nicotine Hydroxylase from Pseudomonas sp. ZZ-5 That Catalyzes the Conversion of 6-Hydroxy-3-Succinoylpyridine into 2,5-Dihydroxypyridine. Catalysts. 2017; 7(9):257. https://doi.org/10.3390/catal7090257
Chicago/Turabian StyleWei, Tao, Jie Zang, Yadong Zheng, Hongzhi Tang, Sheng Huang, and Duobin Mao. 2017. "Characterization of a Novel Nicotine Hydroxylase from Pseudomonas sp. ZZ-5 That Catalyzes the Conversion of 6-Hydroxy-3-Succinoylpyridine into 2,5-Dihydroxypyridine" Catalysts 7, no. 9: 257. https://doi.org/10.3390/catal7090257





