Study of Extraction and Enzymatic Properties of Cell-Envelope Proteinases from a Novel Wild Lactobacillus plantarum LP69
Abstract
:1. Introduction
2. Results
2.1. Optimization of Extraction Conditions of CEP by RSM
2.1.1. Box–Behnken Design and Response Surface Method
2.1.2. The Influence of Time, Temperature and Buffer pH on the Response Value
2.1.3. Model Validation
2.2. Enzymatic Properties of CEP
2.2.1. Effects of pH and Temperature on Enzyme Activity
2.2.2. Effect of Metal Ions and Inhibitors on Enzyme Activity
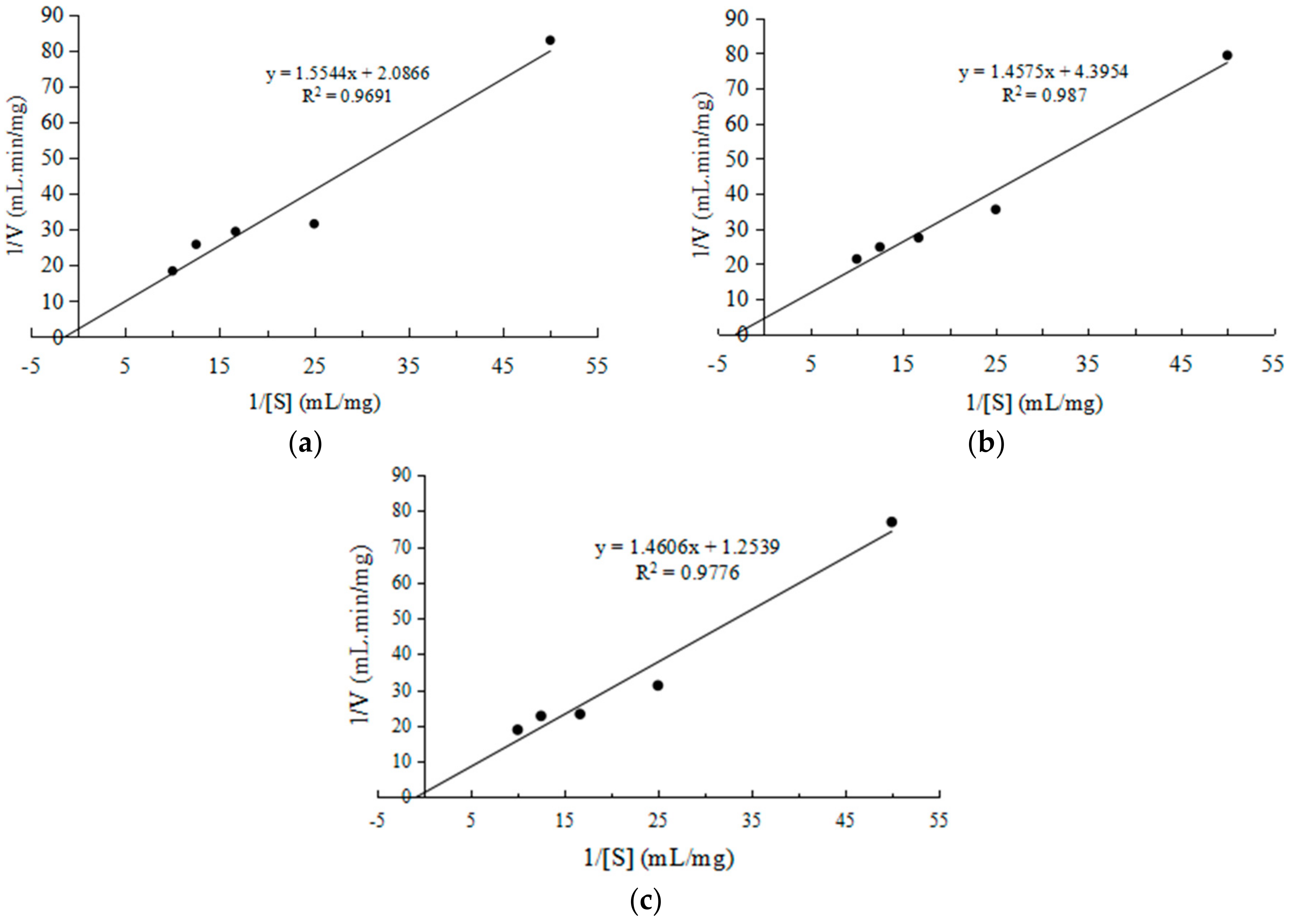
2.2.3. Kinetic Studies of CEP
3. Discussion
4. Materials and Methods
4.1. Strains and Chemicals
4.2. Preparation of Cell-Free Extracts of L. Plantarum LP69
4.3. Determination of Enzyme Activity
4.4. Determination of Protein Concentration
4.5. Specific Activity Assay
4.6. Experimental Design
4.7. Enzymatic Properties of CEP from L. plantarum LP69
4.7.1. Effect of pH on the Enzyme Activity
4.7.2. Temperature Optimum
4.7.3. Influence of Metal Ions and Inhibitors on Enzyme Activity
4.7.4. Kinetic Studies of CEP
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Guo, Y.X.; Pan, D.D.; Zeng, X.Y.; Tanokura, M. Purification and characterization of CEP from Lactococcus lactis ssp. Lactis. Food Chem. 2009, 112, 533–538. [Google Scholar] [CrossRef]
- Fung, W.Y.; Liong, M.T. Evaluation of proteolytic and ACE-inhibitory activity of Lactobacillus acidophilus in soy whey growth medium via response surface methodology. LWT Food Sci. Technol. 2010, 43, 563–567. [Google Scholar] [CrossRef]
- Espeche, T.M.B.; Savoy de Giori, G.; Hebert, E.M. Release of the cell-envelope-associated proteinase of Lactobacillus delbrueckii subspecies lactis CRL 581 is dependent upon pH and temperature. J. Agric. Food Chem. 2009, 57, 8607–8611. [Google Scholar] [CrossRef] [PubMed]
- Tsakalidou, E.; Anastasiou, R.; Vandenberghe, I.; van Beeumen, J.; Kalantzopoulos, G. Cell-wall-bound proteinase of Lactobacillus delbrueckii subsp. lactis ACA-DC 178: Characterization and specificity for β-casein. Appl. Environ. Microbiol. 1999, 65, 2035–2040. [Google Scholar] [PubMed]
- Sánchez, B.; Bressollier, P.; Chaignepain, S.; Schmitter, J.M.; Urdaci, M.C. Identification of surface-associated proteins in the probiotic bacterium Lactobacillus rhamnosus GG. Int. Dairy J. 2009, 19, 85–88. [Google Scholar] [CrossRef]
- Jarocki, P.; Podlesny, M.; Wasko, A.; Siuda, A.; Targonski, Z. Differentiation of three Lactobacillus rhamnosus strains (E/N, Oxy and Pen) by SDS-PAGE and two-dimensional electrophoresis of surface-associated proteins. J. Microbiol. Biotechnol. 2010, 20, 558–562. [Google Scholar] [PubMed]
- Fira, D.; Kojic, M.; Banina, A.; Spasojevic, I.; Strahinic, I.; Topisirovic, L. Characterization of cell envelope-associated proteinases of Thermophilic lactobacilli. J. Appl. Microbiol. 2001, 90, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Laloi, P.; Atlan, D.; Blanc, B.; Gilbert, C.; Portalier, R. Cell-wall-associated proteinase of Lactobacillus delbrueckii subsp. bulgaricus CNRZ397: differential extraction, purification and properties of the enzyme. Appl. Microbiol. Biotechnol. 1991, 36, 196–204. [Google Scholar]
- MartÍn-Hernández, M.C.; Alting, A.C.; Exterkate, F.A. Purification and characterization of the mature, membrane-associated cell-envelope proteinase of Lactobacillus helveticus L89. Appl. Microbiol. Biotechnol. 1994, 40, 828–834. [Google Scholar] [CrossRef]
- Hébert, E.M.; Raya, R.; de Giori, G.S. Characterization of a cell membrane-associated proteinase from Lactobacillus helveticus CRL 581. Curr. Microbiol. 1997, 35, 161–164. [Google Scholar] [CrossRef]
- Bhowmik, T.; Johnson, M.C.; Ray, B. Isolation and partial characterization of the surface protein of Lactobacillus acidophilus strains. Int. J. Food Microbiol. 1985, 2, 311–321. [Google Scholar] [CrossRef]
- Atlan, D.; Laloi, P.; Portalier, R. X-prolyl-dipeptidylamino peptidase of Lactobacillus delbrueckii subsp. bulgaricus: Characterization of the enzyme and isolation of deficient mutants. Appl. Environ. Microbiol. 1990, 56, 2174–2179. [Google Scholar] [PubMed]
- Agyei, D.; Lim, W.; Zass, M.; Tan, D.; Danquah, M.K. Bioanalytical evaluation of Lactobacillus delbrueckii subsp. lactis 313 cell-envelope proteinase extraction. Chem. Eng. Sci. 2013, 95, 323–330. [Google Scholar]
- Sadat-Mekmene, L.; Genay, M.; Atlan, D.; Lortal, S.; Gagnaire, V. Original features of cell-envelope proteinases of Lactobacillus helveticus: A review. Int. J. Food Microbiol. 2011, 146, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Stefanovic, E.; Kilcawley, K.N.; Rea, M.C.; Fitzgerald, G.F.; McAuliffe, O. Genetic, enzymatic and metabolite profiling of the Lactobacillus casei group reveals strain biodiversity and potential applications for flavour diversification. J. Appl. Microbiol. 2017, 122, 1245–1261. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhang, L.; Zhang, L.; Feng, Z.; Shigwedha, N. Screening, purification, and characterization of proteinase from 3 Lactobacillus delbrueckii subsp. bulgaricus. RSC Adv. 2015, 5, 93733–93738. [Google Scholar] [CrossRef]
- Gobbetti, M.; Smacchi, E.; Corsetti, A. The proteolytic system of Lactobacillus sanfrancisco CB1: purification and characterization of a proteinase, a dipeptidase, and an aminopeptidase. Appl. Environ. Microbiol. 1996, 62, 3220–3226. [Google Scholar] [PubMed]
- Chen, H.; Ji, Z.; Shu, G.W.; Xing, H. Effect of probiotic Lactobacillus strains on Angiotensin-I Converting enzyme inhibitory activity from fermented goat milk. Adv. Mater. Res. 2012, 531, 442–445. [Google Scholar] [CrossRef]
- Shu, G.W.; Yang, H.; Chen, H.; Zhang, Q.H.; Tian, Y. Effect of incubation time, inoculum size, temperature, pasteurization time, goat milk powder and whey powder on ACE-inhibitory activity in fermented milk by L. plantarum LP69. Acta Sci. Pol. Technol. Aliment. 2015, 14, 107–116. [Google Scholar] [PubMed]
- Shu, G.W.; Huang, J.; Chen, L.; Lei, N.; Chen, H. Characterization of Goat Milk Hydrolyzed by Cell Envelope Proteinases from Lactobacillus plantarum LP69: Proteolytic System Optimization, Bioactivity, and Storage Stability Evaluation. Molecules 2018, 23, 1317. [Google Scholar] [CrossRef] [PubMed]
- Mehrnoush, A.; Mustafa, S.; Zaidul, I.S.M.D.; Mohd, Y.A.M. Optimization of the Conditions for Extraction of Serine Protease from Kesinai Plant (Streblus asper) Leaves Using Response Surface Methodology. Molecules 2011, 16, 9245–9260. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Pan, D.D. Purification and Enzymatic Properties of Cell Envelope Protease from Lactobacillus fermentum. Food Sci. 2011, 32, 262–268. [Google Scholar]
- Yamamoto, N.; Akino, A.; Takano, T. Purification and specificity of a cell wall-associated proteinase from Lactobacillus helveticus CP790. Int. J. Biochem. 1993, 114, 740–750. [Google Scholar] [CrossRef]
- Fernandez de Palenzia, P.; Pelaez, C.; Martin-Hernandez, M.C. Purification and characterization of the cell wall proteinase of Lactobacillus casei subsp. casei IFPL731 isolated from raw goat’s milk cheese. J. Agric. Food Chem. 1997, 45, 3401–3405. [Google Scholar]
- Genov, N.; Filippi, B.; Dolashka, P.; Wilson, K.S.; Betzel, C. Stability of subtilisins and related proteinases (subtilases). Int. J. Pept. Protein Res. 1995, 45, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez Patino, J.M.; Miňones Conde, J.; Linares, H.M.; Pedroche Jiménez, J.J.; Car-rera Sánchez, C.; Pizones, V.; Rodríguez, F.M. Interfacial and foamingproperties of enzyme-induced hydrolysis of sunflower protein isolate. Food Hydrocoll. 2007, 21, 782–793. [Google Scholar] [CrossRef]
- Corrochano, A.R.; Buckin, V.K.; Phil, M.; Giblin, L. Invited review: Whey proteins as antioxidants and promoters of cellular antioxidant pathways. J. Dairy Sci. 2018, 101, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Wang, C.N.; Guo, M.G. Changes in structure and antioxidant activity of β-lactoglobulin by ultrasound and enzymatic treatment. Ultrason. Sonochem. 2018, 43, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Rezvan, K.; Asghar, T.K.; Ahmad, M.; Reihane, K. Allergenicity reduction of bovine milk β-lactoglobulin by proteolytic activity of lactococcus lactis BMC12C and BMC19H isolated from Iranian dairy products. Int. J. Biol. Macromol. 2018, 112, 876–881. [Google Scholar]
- Kumar, D.; Chatli, M.K.; Singh, R.; Mehta, N.; Kumar, P. Antioxidant and antimicrobial activity of camel milk caseinhydrolysates and its fractions. Small Ruminant Res. 2016, 139, 20–25. [Google Scholar] [CrossRef]
- Carter, D.; He, X.; Munson, S.; Twigg, P.; Gernert, K.; Broom, M.; Miller, T. Three-dimensional structure of human serum albumin. Science 1989, 244, 1195–1198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agyei, D.; He, L.Z. Evaluation of cross-linked enzyme aggregates of lactobacillus cell-envelope proteinases, for protein degradation. Food Bioprod. Process. 2015, 94, 59–69. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Ngo, L.T.A.; Pham, T.L.; Le, V.V.M. Purification of Endopolygalacturonase from submerged culture of Aspergillus awamori L1 using a two-step procedure: Enzyme precipitation and gel filtration. J. Food Res. Int. 2008, 15, 135–140. [Google Scholar]
- Liu, Z.L.; Pan, J.H. A practical method for extending the biuret assay to protein determination of corn-based products. Food Chem. 2017, 224, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Lineweaver, H.; Burk, D. The determination of enzyme dissociation constants. J. Am. Chem. Soc. 1934, 56, 658–666. [Google Scholar] [CrossRef]






| Runs | Variables | Enzyme Activity (Y1, U/mL) | Specific Activity (Y2, U/mg) | ||
|---|---|---|---|---|---|
| Time (min, X1) | Temperature (°C, X2) | pH (X3) | |||
| 1 | 1 (90) | 0 (39) | 1 (7.0) | 17.6 ± 0.52 | 1.31 ± 0.02 |
| 2 | 0 (75) | 0 | 0 (6.5) | 20.64 ± 0.73 | 1.35 ± 0.04 |
| 3 | 1 | −1 (37) | 0 | 15.84 ± 0.59 | 1.22 ± 0.06 |
| 4 | −1 (60) | 0 | 1 | 11.75 ± 0.66 | 1.31 ± 0.02 |
| 5 | −1 | 0 | −1 (6.0) | 14.34 ± 0.71 | 1.27 ± 0.03 |
| 6 | 0 | 0 | 0 | 22.21 ± 0.67 | 1.39 ± 0.07 |
| 7 | 1 | 1 (41) | 0 | 21.42 ± 0.78 | 1.18 ± 0.06 |
| 8 | 0 | 1 | 1 | 13.43 ± 0.87 | 1.28 ± 0.03 |
| 9 | 1 | 0 | −1 | 22.16 ± 0.76 | 1.30 ± 0.10 |
| 10 | 0 | 1 | −1 | 16.29 ± 0.81 | 1.20 ± 0.03 |
| 11 | 0 | −1 | −1 | 13.72 ± 0.52 | 1.29 ± 0.07 |
| 12 | −1 | 1 | 0 | 11.21 ± 0.54 | 1.21 ± 0.11 |
| 13 | 0 | −1 | 1 | 16.43 ± 0.57 | 1.27 ± 0.08 |
| 14 | 0 | 0 | 0 | 20.77 ± 0.75 | 1.37 ± 0.04 |
| 15 | −1 | −1 | 0 | 15.94 ± 0.69 | 1.21 ± 0.05 |
| Source | DF | Enzyme Activity (Y1, U/mL) | Specific Activity (Y2, U/mg) | ||||
|---|---|---|---|---|---|---|---|
| MS | F | Pr > F | MS | F | Pr > F | ||
| Model | 9 | 20.73 | 10.99 | 0.0084 * | 0.0062 | 23.47 | 0.0014 * |
| X1 | 1 | 70.69 | 37.47 | 0.0017 * | <0.0001 | 0.047 | 0.8366 |
| X2 | 1 | 0.022 | 0.012 | 0.9181 | 0.0018 | 6.79 | 0.0479 * |
| X3 | 1 | 6.66 | 3.53 | 0.1190 | 0.0015 | 5.71 | 0.0625 |
| X1X2 | 1 | 26.57 | 14.09 | 0.0133 * | 0.0004 | 1.51 | 0.2739 |
| X1X3 | 1 | 0.97 | 0.51 | 0.5054 | 0.0002 | 0.85 | 0.3991 |
| X2X3 | 1 | 7.76 | 4.11 | 0.0984 | 0.0025 | 9.43 | 0.0277 * |
| X12 | 1 | 12.02 | 6.37 | 0.0529 | 0.015 | 56.63 | 0.0007 * |
| X22 | 1 | 40.20 | 21.31 | 0.0058 * | 0.038 | 142.84 | <0.0001 * |
| X32 | 1 | 31.91 | 16.91 | 0.0092 * | 0.0002 | 1.07 | 0.3490 |
| Residual | 5 | 1.89 | 0.0002 | ||||
| Lack of fit | 3 | 2.64 | 3.47 | 0.2315 | 0.0001 | 0.44 | 0.7506 |
| Pure error | 2 | 0.76 | 0.0004 | ||||
| R2 | 0.9519 | 0.9769 | |||||
| R2adj | 0.8652 | 0.9352 | |||||
| Inhibitors and Ions a | Relative Enzyme Activity (%) b | |
|---|---|---|
| Compound Concentration (mM) | ||
| 1.00 | 10.00 | |
| None | 100.00 | 100.00 |
| Ca2+ | 105.96 | 122.82 |
| Na+ | 91.42 | 93.75 |
| K+ | 94.23 | 106.01 |
| Zn2+ | 53.59 | 42.34 |
| PMSF | 40.94 | 25.73 |
| EDTA | 45.20 | 34.93 |
| Factors | Symbol | Coded Levels | ||
|---|---|---|---|---|
| −1 | 0 | 1 | ||
| Time (min) | X1 | 60 | 75 | 90 |
| Temperature (°C) | X2 | 37 | 39 | 41 |
| Buffer pH | X3 | 6.0 | 6.5 | 7.0 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, H.; Huang, J.; Cao, B.; Chen, L.; Song, N.; Lei, N. Study of Extraction and Enzymatic Properties of Cell-Envelope Proteinases from a Novel Wild Lactobacillus plantarum LP69. Catalysts 2018, 8, 325. https://doi.org/10.3390/catal8080325
Chen H, Huang J, Cao B, Chen L, Song N, Lei N. Study of Extraction and Enzymatic Properties of Cell-Envelope Proteinases from a Novel Wild Lactobacillus plantarum LP69. Catalysts. 2018; 8(8):325. https://doi.org/10.3390/catal8080325
Chicago/Turabian StyleChen, He, Jie Huang, Binyun Cao, Li Chen, Na Song, and Ni Lei. 2018. "Study of Extraction and Enzymatic Properties of Cell-Envelope Proteinases from a Novel Wild Lactobacillus plantarum LP69" Catalysts 8, no. 8: 325. https://doi.org/10.3390/catal8080325





