Expression, Purification, Crystallization, and X-ray Structural Analysis of CRISPR-Associated Protein Cas6 from Methanocaldococcus jannaschii
Abstract
:1. Introduction
2. Results and Discussion
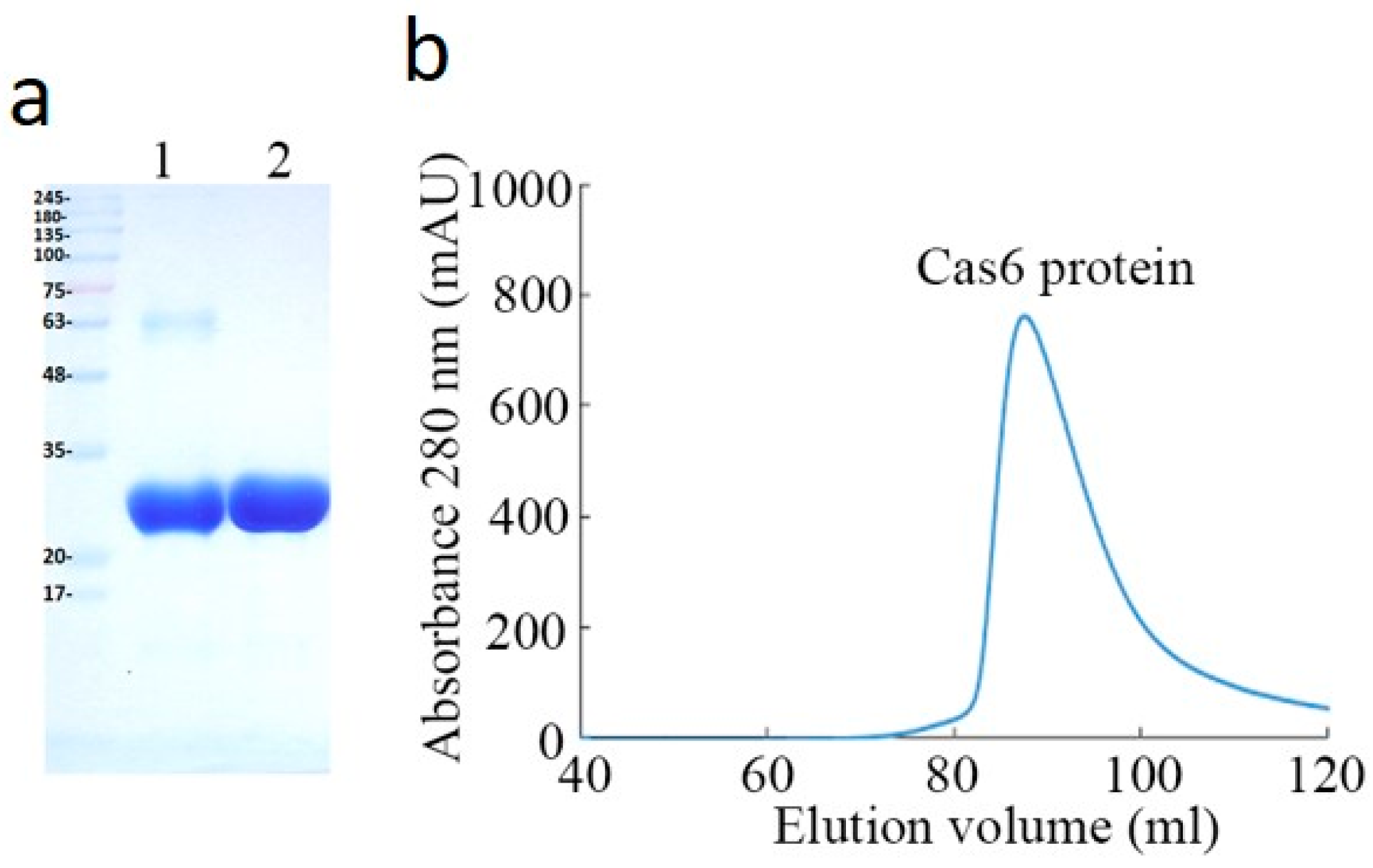
2.1. Purification and Characterizaction of MjCas6
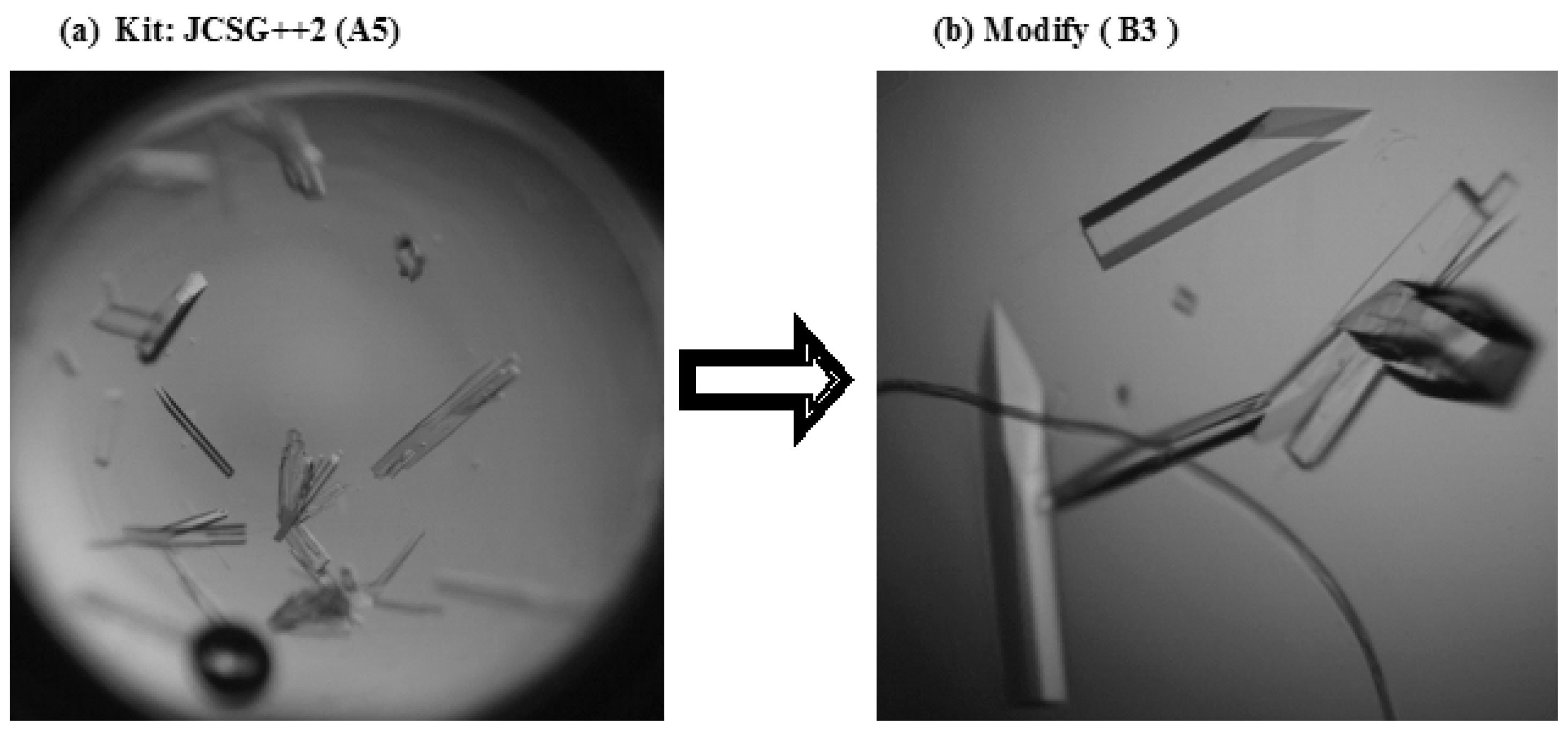
2.2. Crystallization of the MjCas6 Protein
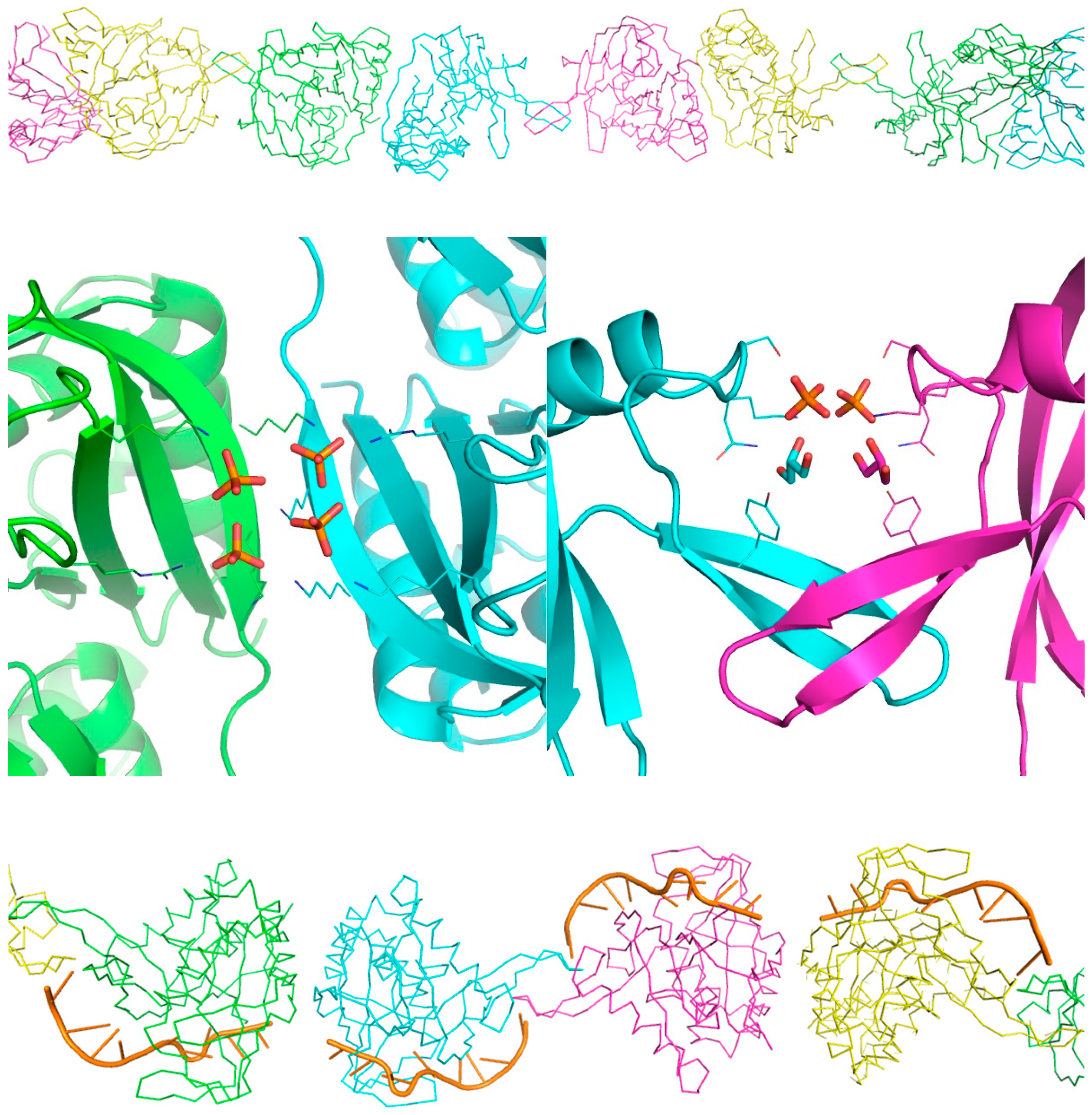
2.3. X-ray Analysis of the MjCas6 Crystal
2.4. Functional Implications of the MjCas6 Structure
3. Materials and Methods
3.1. Protein Production
3.2. Crystallization
3.3. Data Collection
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Terns, M.P.; Terns, R.M. CRISPR-based adaptive immune systems. Curr. Opin. Microbiol. 2011, 14, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Sorek, R.; Lawrence, C.M.; Wiedenheft, B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu. Rev. Biochem. 2013, 82, 237–266. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Doudna, J.A. The structural biology of CRISPR-Cas systems. Curr. Opin. Struct. Biol. 2015, 30, 100–111. [Google Scholar] [CrossRef] [PubMed]
- Jackson, R.N.; Wiedenheft, B. A conserved structural chassis for mounting versatile CRISPR RNA-guided immune responses. Mol. Cell 2015, 58, 722–728. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.J.; Charpentier, E.; Haft, D.H.; et al. An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 2015, 13, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Bhaya, D.; Davison, M.; Barrangou, R. CRISPR-cas systems in bacteria and archaea: Versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 2011, 45, 273–297. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.; van Embden, J.D.A.; Gaastra, W.; Schouls, L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002, 43, 1565–1575. [Google Scholar] [CrossRef] [PubMed]
- Brouns, S.J.J.; Jore, M.M.; Lundgren, M.; Westra, E.R.; Slijkhuis, R.J.H.; Snijders, A.P.L.; Dickman, M.J.; Makarova, K.S.; Koonin, E.V.; van der Oost, J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 2008, 321, 960–964. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Makarova, K.S.; Zhang, F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017, 37, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Haft, D.H.; Selengut, J.; Mongodin, E.F.; Nelson, K.E. A guild of 45 CRISPR-Associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 2005, 1, e60. [Google Scholar] [CrossRef] [PubMed]
- Vestergaard, G.; Garrett, R.A.; Shah, S.A. CRISPR adaptive immune systems of Archaea. RNA Biol. 2014, 11, 156–167. [Google Scholar] [CrossRef] [PubMed]
- Hale, C.; Kleppe, K.; Terns, R.M.; Terns, M.P. Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus. RNA 2008, 14, 2572–2579. [Google Scholar] [CrossRef] [PubMed]
- Niewoehner, O.; Jinek, M.; Doudna, J.A. Evolution of CRISPR RNA recognition and processing by Cas6 endonucleases. Nucleic Acids Res. 2014, 42, 1341–1353. [Google Scholar] [CrossRef] [PubMed]
- Sefcikova, J.; Roth, M.; Yu, G.; Li, H. Cas6 processes tight and relaxed repeat RNA via multiple mechanisms: A hypothesis. BioEssays 2017, 39, 1700019. [Google Scholar] [CrossRef] [PubMed]
- Matthews, B.W. Solvent content of protein crystals. J. Mol. Biol. 1968, 33, 491–497. [Google Scholar] [CrossRef]
- Vagin, A.; Teplyakov, A. Molecular replacement with MOLREP. Acta Crystallogr. D. Biol. Crystallogr. 2010, 66, 22–25. [Google Scholar] [CrossRef] [PubMed]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Storoni, L.C.; Read, R.J. Likelihood-enhanced fast translation functions. Acta Crystallogr. Sect. D Biol. Crystallogr. 2005, 61, 458–464. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.-W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Kielkopf, C.L.; Lücke, S.; Green, M.R. U2AF homology motifs: Protein recognition in the RRM world. Genes Dev. 2004, 18, 1513–1526. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Preamplume, G.; Terns, M.P.; Terns, R.M.; Li, H. Interaction of the Cas6 riboendonuclease with CRISPR RNAs: Recognition and cleavage. Structure 2011, 19, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Park, H.-M.; Shin, M.; Sun, J.; Kim, G.S.; Lee, Y.C.; Park, J.-H.; Kim, B.Y.; Kim, J.-S. Crystal structure of a Cas6 paralogous protein from pyrococcus furiosus. Proteins Struct. Funct. Bioinform. 2012, 80, 1895–1900. [Google Scholar] [CrossRef] [PubMed]
- Carte, J.; Wang, R.; Li, H.; Terns, R.M.; Terns, M.P. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008, 22, 3489–3496. [Google Scholar] [CrossRef] [PubMed]
- Calvin, K.; Li, H. RNA-splicing endonuclease structure and function. Cell. Mol. Life Sci. 2008, 65, 1176–1185. [Google Scholar] [CrossRef] [PubMed]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar] [PubMed]


| Data Collection | |
| Space group | C2 |
| Cell dimensions | |
| a, b, c (Å) | 200.84, 85.26, 100.06 |
| α, β, γ (°) | 90.00, 118.47, 90.00 |
| Resolution (Å) | 30.0–1.85 (1.92–1.85) |
| Rmeas (%) | 8.4 (58.6) |
| I/σ (I) | 17.3 (2.0) |
| Completeness (%) | 99.7 (99.7) |
| Redundancy | 3.7 (3.7) |
| Refinement | |
| Resolution (Å) | 29.1–1.85 (1.92–1.85) |
| No. reflections | 125,321 (12,110) |
| Rwork/Rfree | 0.167/0.194 |
| B (Å2)/No. atoms | |
| Protein | 29.8/8038 |
| Ligand + ion | 67.9/154 |
| Water | 43.3/1098 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.011 |
| Bond angles (°) | 1.02 |
| Source Organism | Methanocaldococcus jannaschii DSM 2661 |
|---|---|
| DNA source | ATCC: The Global Bioresource center |
| Forward primer † | 5′-GGGAATTCCATATGATGAGGGAGAGTATGAGAA-3′ |
| Reverse primer ‡ | 5′-CCGCTCGAGAATTTTTGTTTTGAGTTTTTTATT-3′ |
| Expression vector | Modified pET21b |
| Expression host | E. coli BL21(DE3) |
| Complete amino-acid sequence of the construct produced | MRESMRIELELQTDNFTVIPYNHQYYLASAIYNKIHSANPAYAKRLHNYQKFKFFTFSLLQIRKRVIRKEGIETIDGKAYLYISSPNNEFIENFVAGLLEDGKLRVGNVEFFVRKAKILPIPKKFNILKTISPIYLKTMIETEDGLKTYDLLPNNSKFYENLKNNLKKKYEAFYNEKCDMNFEFEVLKFRPKRMRIKNDIYCRCSEMVFKVWGDYDLIKFGYECGFGEKNSMGFGMVVNVEDKNQKNKKLKTKILEHHHHHH |
| Method | Sitting-Drop Vapor Diffusion |
|---|---|
| Plate type | Intelli-plate 48-3 |
| Temperature (K) | 277.15 |
| Protein concentration (mg mL−1) | 14.3 |
| Buffer composition of protein solution | 50 mM Tris pH 8.0, 100 Mm NaCl, 5% Glycerol, 2 mM TCEP |
| Composition of reservoir solution | 100 mM HEPES Sodium Salt pH 7.5, 600 mM Sodium dihydrogen Phosphate, 600 mM Potassium dihydrogen Phosphate |
| Volume (μL) and ratio of drop | 2 (1:1 ratio of protein and reservoir) |
| Volume of reservoir (μL) | 180 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, M.-C.; Tseng, S.-T.; Yang, J.-C.; Hsieh, T.-J.; Wu, S.-C.; Kuan, S.-M.; Chen, M.-J.; Chang, M.-C.; Wang, C.-C.; Chen, H.-L.; et al. Expression, Purification, Crystallization, and X-ray Structural Analysis of CRISPR-Associated Protein Cas6 from Methanocaldococcus jannaschii. Crystals 2017, 7, 344. https://doi.org/10.3390/cryst7110344
Lee M-C, Tseng S-T, Yang J-C, Hsieh T-J, Wu S-C, Kuan S-M, Chen M-J, Chang M-C, Wang C-C, Chen H-L, et al. Expression, Purification, Crystallization, and X-ray Structural Analysis of CRISPR-Associated Protein Cas6 from Methanocaldococcus jannaschii. Crystals. 2017; 7(11):344. https://doi.org/10.3390/cryst7110344
Chicago/Turabian StyleLee, Ming-Chang, Shih-Ting Tseng, Juan-Cheng Yang, Tung-Ju Hsieh, Shang-Chuen Wu, Shu-Min Kuan, Ming-Jen Chen, Ming-Chiu Chang, Chun-Chiu Wang, Hsiu-Lin Chen, and et al. 2017. "Expression, Purification, Crystallization, and X-ray Structural Analysis of CRISPR-Associated Protein Cas6 from Methanocaldococcus jannaschii" Crystals 7, no. 11: 344. https://doi.org/10.3390/cryst7110344






