The Synergetic Effects of Combining Structural Biology and EPR Spectroscopy on Membrane Proteins
Abstract
:1. Introduction
2. Continuous Wave (cw) and Pulsed EPR Methods—An Overview
2.1. Site-Directed Spin Labeling
2.2. Spin Label Side Chain Mobility
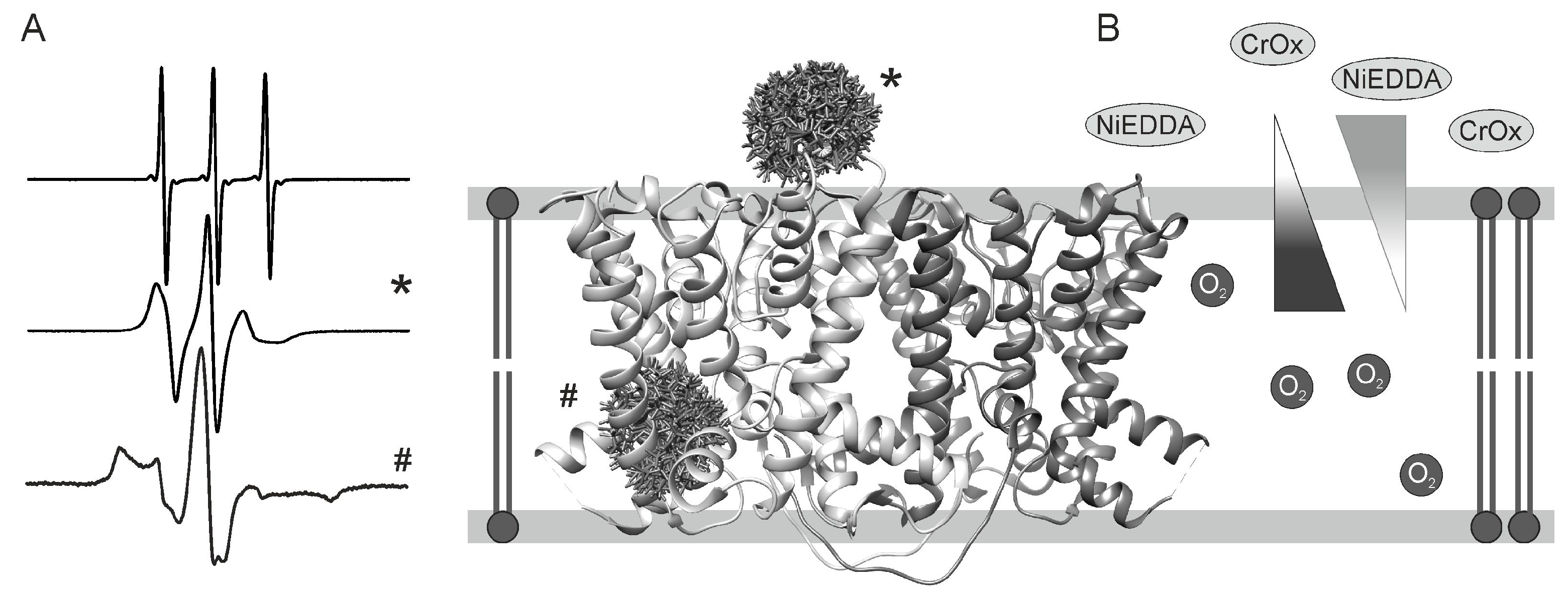
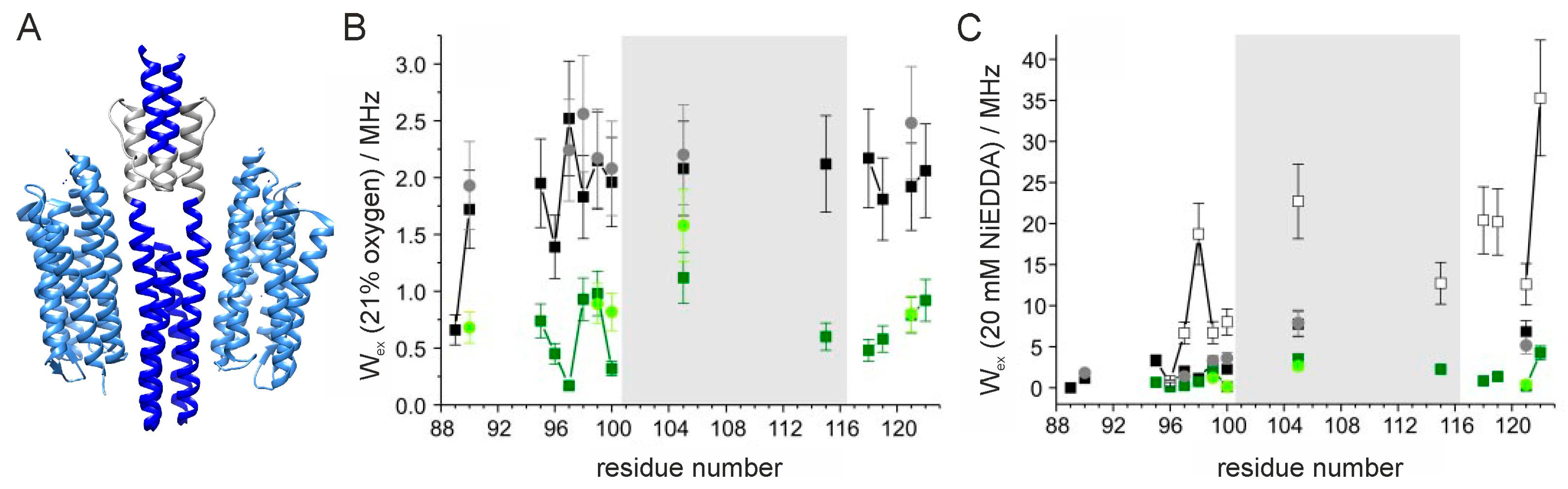
2.3. Accessibility of the Spin Label Side Chain
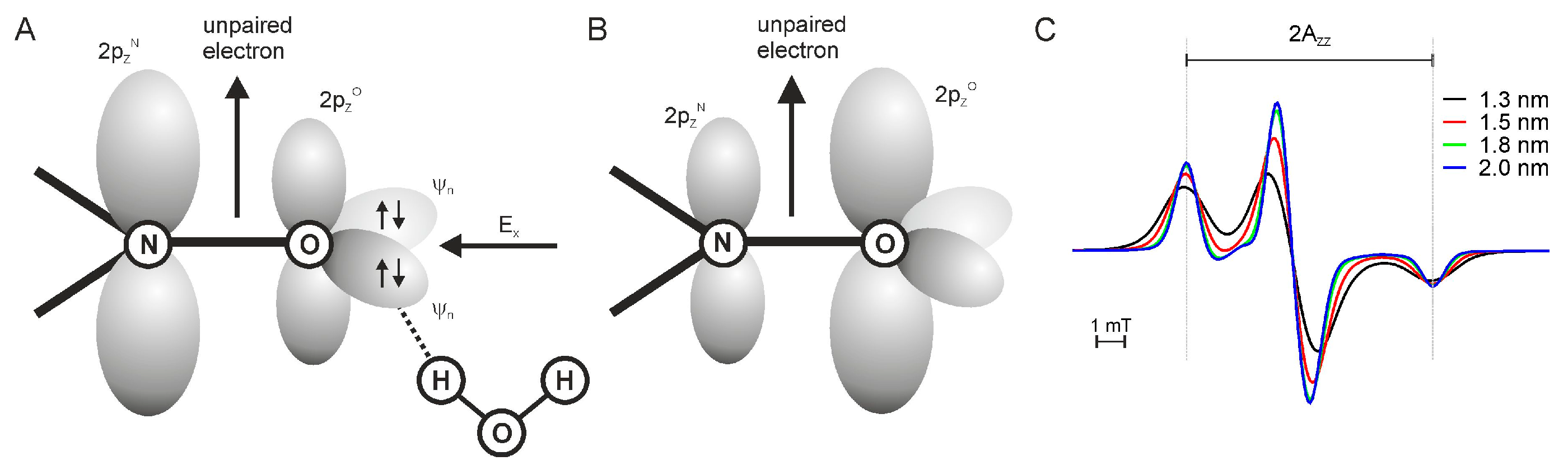
2.4. Polarity of the Spin Label Side Chain Environment
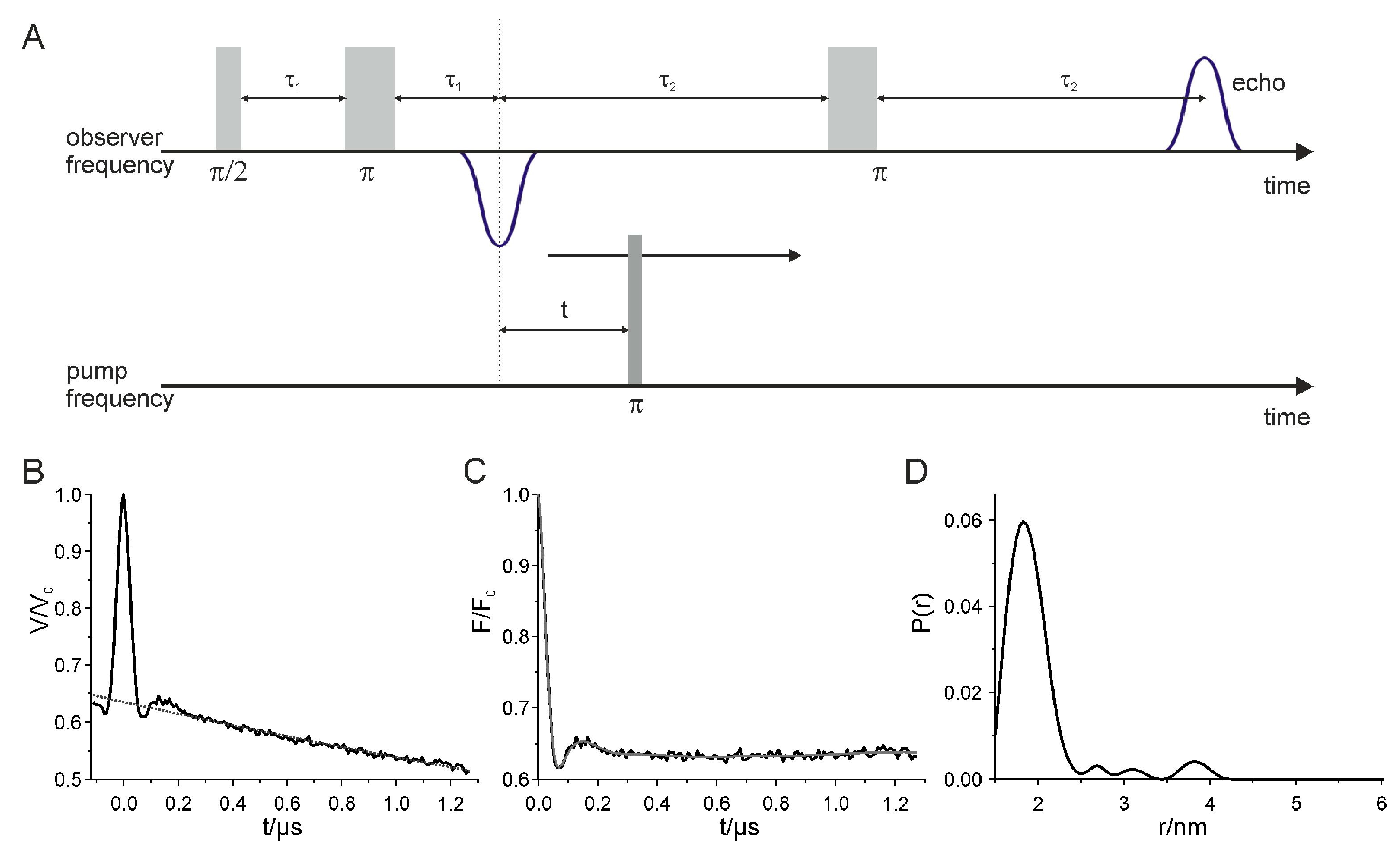
2.5. Interspin Distance Measurements
3. EPR Spectroscopy Applied to Different Classes of Membrane Proteins
3.1. Receptors
3.2. Ion Channels
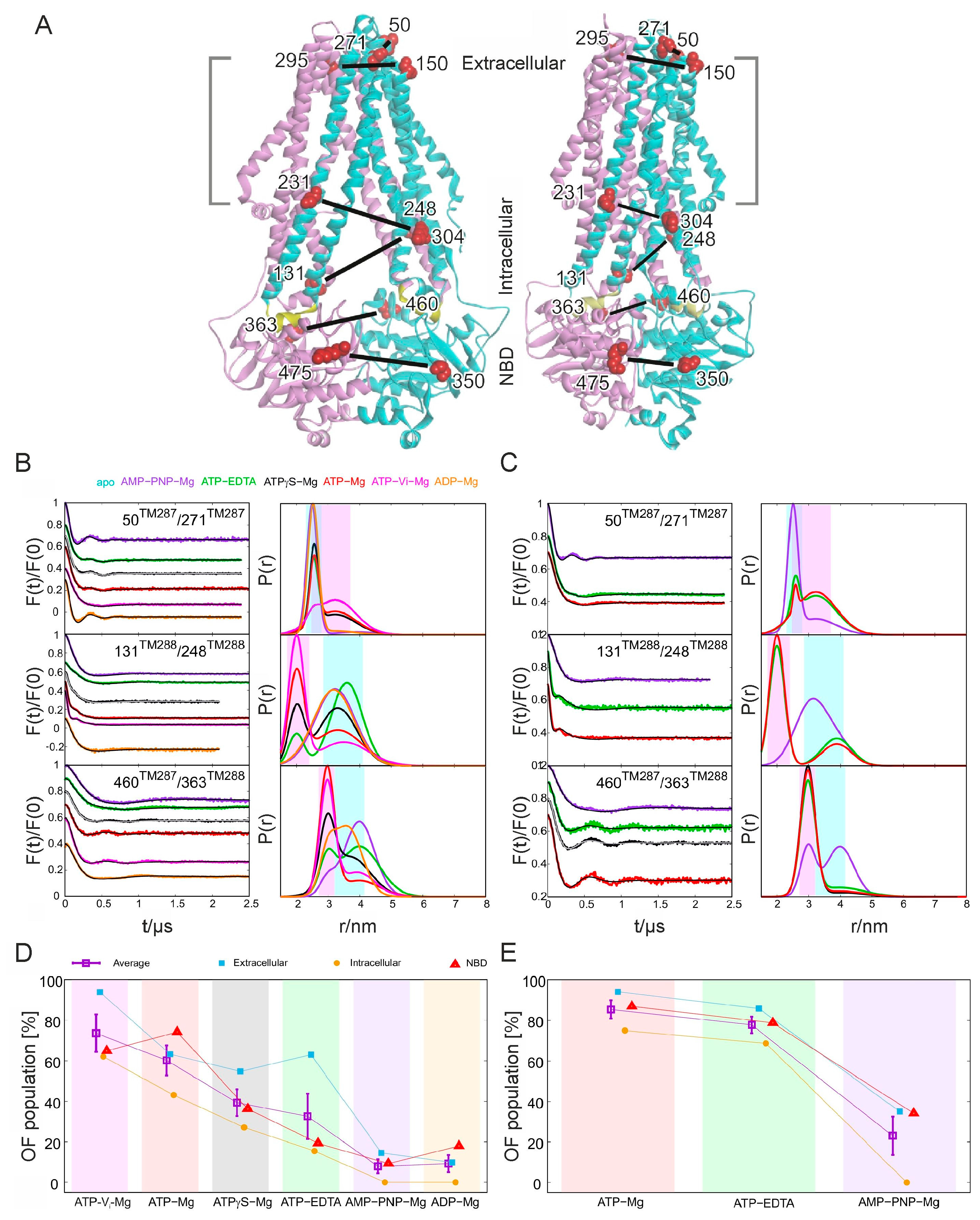
3.3. ABC Transporters
3.4. Secondary Active Proteins
3.5. New Developments
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Loll, B.; Kern, J.; Saenger, W.; Zouni, A.; Biesiadka, J. Towards complete cofactor arrangement in the 3.0 Å resolution structure of photosystem II. Nature 2005, 438, 1040–1044. [Google Scholar] [CrossRef] [PubMed]
- Guzenko, D.; Chernyatina, A.A.; Strelkov, S.V. Crystallographic studies of intermediate filament proteins. In Fibrous proteins: Structures and Mechanisms; Parry, D.A.D., Squire, J.M., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 151–170. [Google Scholar]
- Nissen, P.; Hansen, J.; Ban, N.; Moore, P.B.; Steitz, T.A. The structural basis of ribosome activity in peptide bond synthesis. Science 2000, 289, 920–930. [Google Scholar] [CrossRef] [PubMed]
- Bordignon, E.; Steinhoff, H.-J. Membrane protein structure and dynamics studied by site-directed spin-labeling ESR. In Esr Spectroscopy in Membrane Biophysics; Springer: Berlin, Germany, 2007; pp. 129–164. [Google Scholar]
- Prisner, T.; Rohrer, M.; MacMillan, F. Pulsed EPR spectroscopy: Biological applications. Annu. Rev. Phys. Chem. 2001, 52, 279–313. [Google Scholar] [CrossRef] [PubMed]
- Klug, C.S.; Feix, J.B. Methods and applications of site-directed spin labeling EPR spectroscopy. Methods Cell Biol. 2008, 84, 617–658. [Google Scholar] [PubMed]
- Klare, J.P. Site-directed spin labeling EPR spectroscopy in protein research. Biol. Chem. 2013, 394, 1281–1300. [Google Scholar] [CrossRef] [PubMed]
- Roser, P.; Schmidt, M.J.; Drescher, M.; Summerer, D. Site-directed spin labeling of proteins for distance measurements in vitro and in cells. Org. Biomol. Chem. 2016, 14, 5468–5476. [Google Scholar] [CrossRef] [PubMed]
- Berliner, L.J.; Grunwald, J.; Hankovszky, H.O.; Hideg, K. A novel reversible thiol-specific spin label: Papain active site labeling and inhibition. Anal. Biochem. 1982, 119, 450–455. [Google Scholar] [CrossRef]
- Kolb, H.C.; Finn, M.; Sharpless, K.B. Click chemistry: Diverse chemical function from a few good reactions. Angew. Chem. Int. Ed. 2001, 40, 2004–2021. [Google Scholar] [CrossRef]
- Jakobsen, U.; Shelke, S.A.; Vogel, S.; Sigurdsson, S.T. Site-directed spin-labeling of nucleic acids by click chemistry: Detection of abasic sites in duplex DNA by EPR spectroscopy. J. Am. Chem. Soc. 2010, 132, 10424–10428. [Google Scholar] [CrossRef] [PubMed]
- Ding, P.; Wunnicke, D.; Steinhoff, H.J.; Seela, F. Site-directed spin-labeling of DNA by the azide–alkyne ‘click’ reaction: Nanometer distance measurements on 7-deaza-2′-deoxyadenosine and 2′-deoxyuridine nitroxide conjugates spatially separated or linked to a ‘da-dt’ base pair. Chem. A Eur. J. 2010, 16, 14385–14396. [Google Scholar] [CrossRef] [PubMed]
- Griffith, O.; Jost, P.; Berliner, L. Spin Labeling: Theory and Applications; Academic Press: New York, NY, USA, 1976; Volume 1, p. 453. [Google Scholar]
- McHaourab, H.S.; Lietzow, M.A.; Hideg, K.; Hubbell, W.L. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry 1996, 35, 7692–7704. [Google Scholar] [CrossRef] [PubMed]
- Hubbell, W.L.; McHaourab, H.S.; Altenbach, C.; Lietzow, M.A. Watching proteins move using site-directed spin labeling. Structure 1996, 4, 779–783. [Google Scholar] [CrossRef]
- Steinhoff, H.-J.; Hubbell, W.L. Calculation of electron paramagnetic resonance spectra from brownian dynamics trajectories: Application to nitroxide side chains in proteins. Biophys. J. 1996, 71, 2201–2212. [Google Scholar] [CrossRef]
- Steinhoff, H.-J.; Müller, M.; Beier, C.; Pfeiffer, M. Molecular dynamics simulation and EPR spectroscopy of nitroxide side chains in bacteriorhodopsin. J. Mol. Liq. 2000, 84, 17–27. [Google Scholar] [CrossRef]
- Barnes, J.P.; Liang, Z.; McHaourab, H.S.; Freed, J.H.; Hubbell, W.L. A multifrequency electron spin resonance study of T4 lysozyme dynamics. Biophys. J. 1999, 76, 3298–3306. [Google Scholar] [CrossRef]
- Borbat, P.P.; Costa-Filho, A.J.; Earle, K.A.; Moscicki, J.K.; Freed, J.H. Electron spin resonance in studies of membranes and proteins. Science 2001, 291, 266–269. [Google Scholar] [CrossRef] [PubMed]
- Freed, J.H. Theory of slow tumbling ESR spectra for nitroxides. Spin Label. Theory Appl. 1976, 1, 53–132. [Google Scholar]
- Beier, C.; Steinhoff, H.-J. A structure-based simulation approach for electron paramagnetic resonance spectra using molecular and stochastic dynamics simulations. Biophys. J. 2006, 91, 2647–2664. [Google Scholar] [CrossRef] [PubMed]
- DeSensi, S.C.; Rangel, D.P.; Beth, A.H.; Lybrand, T.P.; Hustedt, E.J. Simulation of nitroxide electron paramagnetic resonance spectra from brownian trajectories and molecular dynamics simulations. Biophys. J. 2008, 94, 3798–3809. [Google Scholar] [CrossRef] [PubMed]
- Hubbell, W.L.; Cafiso, D.S.; Altenbach, C. Identifying conformational changes with site-directed spin labeling. Nat. Struct. Mol. Biol. 2000, 7, 735–739. [Google Scholar] [CrossRef] [PubMed]
- Altenbach, C.; Froncisz, W.; Hemker, R.; McHaourab, H.; Hubbell, W.L. Accessibility of nitroxide side chains: Absolute heisenberg exchange rates from power saturation EPR. Biophys. J. 2005, 89, 2103–2112. [Google Scholar] [CrossRef] [PubMed]
- Altenbach, C.; Greenhalgh, D.A.; Khorana, H.G.; Hubbell, W.L. A collision gradient method to determine the immersion depth of nitroxides in lipid bilayers: Application to spin-labeled mutants of bacteriorhodopsin. Proc. Natl. Acad. Sci. USA 1994, 91, 1667–1671. [Google Scholar] [CrossRef] [PubMed]
- Marsh, D.; Dzikovski, B.G.; Livshits, V.A. Oxygen profiles in membranes. Biophys. J. 2006, 90, L49–L51. [Google Scholar] [CrossRef] [PubMed]
- Mchaourab, H.S.; Hyde, J.S. Dependence of the multiple-quantum EPR signal on the spin-lattice relaxation time. Effect of oxygen in spin-labeled membranes. J. Magn. Reson. Ser. B 1993, 101, 178–184. [Google Scholar] [CrossRef]
- Percival, P.; Hyde, J.S. Pulsed EPR spectrometer, II. Rev. Sci. Instrum. 1975, 46, 1522–1529. [Google Scholar] [CrossRef]
- Nielsen, R.D.; Canaan, S.; Gladden, J.A.; Gelb, M.H.; Mailer, C.; Robinson, B.H. Comparing continuous wave progressive saturation EPR and time domain saturation recovery EPR over the entire motional range of nitroxide spin labels. J. Magn. Reson. 2004, 169, 129–163. [Google Scholar] [CrossRef] [PubMed]
- Plato, M.; Steinhoff, H.-J.; Wegener, C.; Törring, J.T.; Savitsky, A.; Möbius, K. Molecular orbital study of polarity and hydrogen bonding effects on the g and hyperfine tensors of site directed no spin labelled bacteriorhodopsin. Mol. Phys. 2002, 100, 3711–3721. [Google Scholar] [CrossRef]
- Steinhoff, H.J.; Radzwill, N.; Thevis, W.; Lenz, V.; Brandenburg, D.; Antson, A.; Dodson, G.; Wollmer, A. Determination of interspin distances between spin labels attached to insulin: Comparison of electron paramagnetic resonance data with the X-ray structure. Biophys. J. 1997, 73, 3287–3298. [Google Scholar] [CrossRef]
- Cooke, J.A.; Brown, L.J. Distance measurements by continuous wave EPR spectroscopy to monitor protein folding. In Protein Folding, Misfolding, and Disease: Methods and Protocols; Hill, A.F., Barnham, K.J., Bottomley, S.P., Cappai, R., Eds.; Humana Press: Totowa, NJ, USA, 2011; pp. 73–96. [Google Scholar]
- Schiemann, O.; Prisner, T.F. Long-range distance determinations in biomacromolecules by EPR spectroscopy. Q. Rev. Biophys. 2007, 40, 1–53. [Google Scholar] [CrossRef] [PubMed]
- Berliner, L.J.; Eaton, S.S.; Eaton, G.R. Distance Measurements in Biological Systems by EPR; Springer Science & Business Media: New York, NY, USA, 2006; Volume 19. [Google Scholar]
- Rabenstein, M.D.; Shin, Y.K. Determination of the distance between two spin labels attached to a macromolecule. Proc. Natl. Acad. Sci. USA 1995, 92, 8239–8243. [Google Scholar] [CrossRef] [PubMed]
- Likhtenshtein, G.; Wiley, J. Spin labelling methods in molecular biology. FEBS Lett. 1977, 83, 1. [Google Scholar]
- Radzwill, N.; Gerwert, K.; Steinhoff, H.J. Time-resolved detection of transient movement of helices F and G in doubly spin-labeled bacteriorhodopsin. Biophys. J. 2001, 80, 2856–2866. [Google Scholar] [CrossRef]
- Fiori, W.R.; Millhauser, G.L. Exploring the peptide 310-helix⇆ α–helix equilibrium with double label electron spin resonance. Biopolymers 1995, 37, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Pannier, M.; Veit, S.; Godt, A.; Jeschke, G.; Spiess, H.W. Dead-time free measurement of dipole-dipole interactions between electron spins. J. Magn. Reson. 2000, 142, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Jeschke, G.; Polyhach, Y. Distance measurements on spin-labelled biomacromolecules by pulsed electron paramagnetic resonance. Phys. Chem. Chem. Phys. 2007, 9, 1895–1910. [Google Scholar] [CrossRef] [PubMed]
- Jeschke, G.; Panek, G.; Godt, A.; Bender, A.; Paulsen, H. Data analysis procedures for pulse ELDOR measurements of broad distance distributions. Appl. Magn. Reson. 2004, 26, 223. [Google Scholar] [CrossRef]
- Chiang, Y.-W.; Borbat, P.P.; Freed, J.H. The determination of pair distance distributions by pulsed ESR using tikhonov regularization. J. Magn. Reson. 2005, 172, 279–295. [Google Scholar] [CrossRef] [PubMed]
- Jeschke, G. Deeranalysis2006—A comprehensive software package for analyzing pulsed ELDOR data. Appl. Magn. Reson. 2006, 30, 473–498. [Google Scholar] [CrossRef]
- Polyhach, Y.; Bordignon, E.; Jeschke, G. Rotamer libraries of spin labelled cysteines for protein studies. Phys. Chem. Chem. Phys. 2011, 13, 2356–2366. [Google Scholar] [CrossRef] [PubMed]
- Puljung, M.C.; DeBerg, H.A.; Zagotta, W.N.; Stoll, S. Double electron-electron resonance reveals cAMP-induced conformational change in HCN channels. Proc. Natl. Acad. Sci. USA 2014, 111, 9816–9821. [Google Scholar] [CrossRef] [PubMed]
- Van Eps, N.; Caro, L.N.; Morizumi, T.; Ernst, O.P. Characterizing rhodopsin signaling by EPR spectroscopy: From structure to dynamics. Photochem. Photobiol. Sci. 2015, 14, 1586–1597. [Google Scholar] [CrossRef] [PubMed]
- Manglik, A.; Kim, T.H.; Masureel, M.; Altenbach, C.; Yang, Z.; Hilger, D.; Lerch, M.T.; Kobilka, T.S.; Thian, F.S.; Hubbell, W.L.; et al. Structural insights into the dynamic process of β2-adrenergic receptor signaling. Cell 2015, 161, 1101–1111. [Google Scholar] [CrossRef] [PubMed]
- Dror, R.O.; Mildorf, T.J.; Hilger, D.; Manglik, A.; Borhani, D.W.; Arlow, D.H.; Philippsen, A.; Villanueva, N.; Yang, Z.; Lerch, M.T.; et al. Structural basis for nucleotide exchange in heterotrimeric g proteins. Science 2015, 348, 1361–1365. [Google Scholar] [CrossRef] [PubMed]
- Sattig, T.; Rickert, C.; Bamberg, E.; Steinhoff, H.-J.; Bamann, C. Light-induced movement of the transmembrane helix B in channelrhodopsin-2. Angew. Chem. Int. Ed. 2013, 52, 9705–9708. [Google Scholar] [CrossRef] [PubMed]
- Krause, N.; Engelhard, C.; Heberle, J.; Schlesinger, R.; Bittl, R. Structural differences between the closed and open states of channelrhodopsin-2 as observed by EPR spectroscopy. FEBS Lett. 2013, 587, 3309–3313. [Google Scholar] [CrossRef] [PubMed]
- Wolff, E.K.; Bogomolni, R.A.; Scherrer, P.; Hess, B.; Stoeckenius, W. Color discrimination in halobacteria: Spectroscopic characterization of a second sensory receptor covering the blue-green region of the spectrum. Proc. Natl. Acad. Sci. USA 1986, 83, 7272–7276. [Google Scholar] [CrossRef] [PubMed]
- Klare, J.P.; Chizhov, I.; Engelhard, M. Microbial rhodopsins: Scaffolds for ion pumps, channels, and sensors. In Bioenergetics; Springer: Berlin, Germany, 2007; pp. 73–122. [Google Scholar]
- Spudich, J.L.; Yang, C.-S.; Jung, K.-H.; Spudich, E.N. Retinylidene proteins: Structures and functions from archaea to humans. Annu. Rev. Cell Dev. Biol. 2000, 16, 365–392. [Google Scholar] [CrossRef] [PubMed]
- Royant, A.; Nollert, P.; Edman, K.; Neutze, R.; Landau, E.M.; Pebay-Peyroula, E.; Navarro, J. X-ray structure of sensory rhodopsin II at 2.1-Å resolution. Proc. Natl. Acad. Sci. USA 2001, 98, 10131–10136. [Google Scholar] [CrossRef] [PubMed]
- Luecke, H.; Schobert, B.; Lanyi, J.K.; Spudich, E.N.; Spudich, J.L. Crystal structure of sensory rhodopsin II at 2.4 angstroms: Insights into color tuning and transducer interaction. Science 2001, 293, 1499–1503. [Google Scholar] [CrossRef] [PubMed]
- Aravind, L.; Ponting, C.P. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol. Lett. 1999, 176, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Appleman, J.A.; Stewart, V. Mutational analysis of a conserved signal-transducing element: The hamp linker of the escherichia coli nitrate sensor narx. J. Bacteriol. 2003, 185, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Spudich, J.L. Demonstration of 2:2 stoichiometry in the functional SRI–HtrI signaling complex in halobacterium membranes by gene fusion analysis. Biochemistry 2002, 41, 3891–3896. [Google Scholar] [CrossRef] [PubMed]
- Wegener, A.A.; Klare, J.P.; Engelhard, M.; Steinhoff, H.J. Structural insights into the early steps of receptor—Transducer signal transfer in archaeal phototaxis. EMBO J. 2001, 20, 5312–5319. [Google Scholar] [CrossRef] [PubMed]
- Hulko, M.; Berndt, F.; Gruber, M.; Linder, J.U.; Truffault, V.; Schultz, A.; Martin, J.; Schultz, J.E.; Lupas, A.N.; Coles, M. The hamp domain structure implies helix rotation in transmembrane signaling. Cell 2006, 126, 929–940. [Google Scholar] [CrossRef] [PubMed]
- Airola, M.V.; Watts, K.J.; Bilwes, A.M.; Crane, B.R. Structure of concatenated hamp domains provides a mechanism for signal transduction. Structure 2010, 18, 436–448. [Google Scholar] [CrossRef] [PubMed]
- Doebber, M.; Bordignon, E.; Klare, J.P.; Holterhues, J.; Martell, S.; Mennes, N.; Li, L.; Engelhard, M.; Steinhoff, H.-J. Salt-driven equilibrium between two conformations in the hamp domain from natronomonas pharaonis: The language of signal transfer? J. Biol. Chem. 2008, 283, 28691–28701. [Google Scholar] [CrossRef] [PubMed]
- Klare, J.P.; Bordignon, E.; Engelhard, M.; Steinhoff, H.-J. Transmembrane signal transduction in archaeal phototaxis: The sensory rhodopsin II-transducer complex studied by electron paramagnetic resonance spectroscopy. Eur. J. Cell Biol. 2011, 90, 731–739. [Google Scholar] [CrossRef] [PubMed]
- Doyle, D.A.; Cabral, J.M.; Pfuetzner, R.A.; Kuo, A.; Gulbis, J.M.; Cohen, S.L.; Chait, B.T.; MacKinnon, R. The structure of the potassium channel: Molecular basis of K+ conduction and selectivity. Science 1998, 280, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Cordero-Morales, J.F.; Cuello, L.G.; Zhao, Y.; Jogini, V.; Cortes, D.M.; Roux, B.; Perozo, E. Molecular determinants of gating at the potassium-channel selectivity filter. Nat. Struct. Mol. Biol. 2006, 13, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Focke, P.J.; Matulef, K.; Bian, X.; Moënne-Loccoz, P.; Valiyaveetil, F.I.; Lockless, S.W. Ion-binding properties of a K+ channel selectivity filter in different conformations. Proc. Natl. Acad. Sci. USA 2015, 112, 15096–15100. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-S.; Sompornpisut, P.; Perozo, E. Structure of the KcsA channel intracellular gate in the open state. Nat. Struct. Mol. Biol. 2001, 8, 883–887. [Google Scholar] [CrossRef] [PubMed]
- Cortes, D.M.; Cuello, L.G.; Perozo, E. Molecular architecture of full-length KcsA. Role Cytoplasmic Domains Ion Permeat. Act. Gating 2001, 117, 165–180. [Google Scholar]
- Raghuraman, H.; Islam, S.M.; Mukherjee, S.; Roux, B.; Perozo, E. Dynamics transitions at the outer vestibule of the KcsA potassium channel during gating. Proc. Natl. Acad. Sci. USA 2014, 111, 1831–1836. [Google Scholar] [CrossRef] [PubMed]
- Matthies, D.; Dalmas, O.; Borgnia, M.J.; Dominik, P.K.; Merk, A.; Rao, P.; Reddy, B.G.; Islam, S.; Bartesaghi, A.; Perozo, E.; et al. Cryo-em structures of the magnesium channel cora reveal symmetry break upon gating. Cell 2016, 164, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Dalmas, O.; Sompornpisut, P.; Bezanilla, F.; Perozo, E. Molecular mechanism of Mg2+-dependent gating in cora. Nat. Commun. 2014, 5, 3590. [Google Scholar] [CrossRef] [PubMed]
- Dalmas, O.; Cuello, L.G.; Jogini, V.; Cortes, D.M.; Roux, B.; Perozo, E. Structural dynamics of the magnesium-bound conformation of cora in a lipid bilayer. Structure 2010, 18, 868–878. [Google Scholar] [CrossRef] [PubMed]
- Hänelt, I. Membrane region M2C2 in subunit KtrB of the K+ uptake system KtrAB from vibrio alginolyticus forms a flexible gate controlling K+ flux: An electron paramagnetic resonance study. J. Biol. Chem. 2010, 285, 28210–28219. [Google Scholar] [CrossRef] [PubMed]
- Perozo, E.; Cortes, D.M.; Sompornpisut, P.; Kloda, A.; Martinac, B. Open channel structure of Mscl and the gating mechanism of mechanosensitive channels. Nature 2002, 418, 942–948. [Google Scholar] [CrossRef] [PubMed]
- Perozo, E.; Kloda, A.; Cortes, D.M.; Martinac, B. Site-directed spin-labeling analysis of reconstituted Mscl in the closed state. J. Gen. Physiol. 2001, 118, 193–206. [Google Scholar] [CrossRef] [PubMed]
- DeBerg, H.A.; Brzovic, P.S.; Flynn, G.E.; Zagotta, W.N.; Stoll, S. Structure and energetics of allosteric regulation of HCN2 ion channels by cyclic nucleotides. J. Biol. Chem. 2016, 291, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Bagnéris, C.; DeCaen, P.G.; Hall, B.A.; Naylor, C.E.; Clapham, D.E.; Kay, C.W.; Wallace, B.A. Role of the C-terminal domain in the structure and function of tetrameric sodium channels. Nat. Commun. 2013, 4, 2465. [Google Scholar] [CrossRef] [PubMed]
- Dellisanti, C.D.; Ghosh, B.; Hanson, S.M.; Raspanti, J.M.; Grant, V.A.; Diarra, G.M.; Schuh, A.M.; Satyshur, K.; Klug, C.S.; Czajkowski, C. Site-directed spin labeling reveals pentameric ligand-gated ion channel gating motions. PLoS Biol. 2013, 11, e1001714. [Google Scholar] [CrossRef] [PubMed]
- Chakrapani, S. Chapter fourteen—EPR studies of gating mechanisms in ion channels. In Methods in Enzymology; Arun, K.S., Ed.; Academic Press: New York, NY, USA, 2015; Volume 557, pp. 279–306. [Google Scholar]
- Dalmas, O.; Hyde, H.C.; Hulse, R.E.; Perozo, E. Symmetry-constrained analysis of pulsed double electron–electron resonance (DEER) spectroscopy reveals the dynamic nature of the KcsA activation gate. J. Am. Chem. Soc. 2012, 134, 16360–16369. [Google Scholar] [CrossRef] [PubMed]
- Sine, S.M.; Engel, A.G. Recent advances in cys-loop receptor structure and function. Nature 2006, 440, 448–455. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.J.; Lester, H.A.; Lummis, S.C.R. The structural basis of function in cys-loop receptors. Q. Rev. Biophys. 2010, 43, 449–499. [Google Scholar] [CrossRef] [PubMed]
- Corringer, P.-J.; Poitevin, F.; Prevost, M.S.; Sauguet, L.; Delarue, M.; Changeux, J.-P. Structure and pharmacology of pentameric receptor channels: From bacteria to brain. Structure 2012, 20, 941–956. [Google Scholar] [CrossRef] [PubMed]
- Da Costa, C.J.B.; Baenziger, J.E. Gating of pentameric ligand-gated ion channels: Structural insights and ambiguities. Structure 2013, 21, 1271–1283. [Google Scholar] [CrossRef] [PubMed]
- Hibbs, R.E.; Gouaux, E. Principles of activation and permeation in an anion-selective cys-loop receptor. Nature 2011, 474, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Miller, P.S.; Aricescu, A.R. Crystal structure of a human GABAA receptor. Nature 2014, 512, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Hassaine, G.; Deluz, C.; Grasso, L.; Wyss, R.; Tol, M.B.; Hovius, R.; Graff, A.; Stahlberg, H.; Tomizaki, T.; Desmyter, A.; et al. X-ray structure of the mouse serotonin 5-HT3 receptor. Nature 2014, 512, 276–281. [Google Scholar] [CrossRef] [PubMed]
- Bocquet, N.; Nury, H.; Baaden, M.; Le Poupon, C.; Changeux, J.-P.; Delarue, M.; Corringer, P.-J. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature 2009, 457, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Hilf, R.J.C.; Dutzler, R. Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature 2009, 457, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Hilf, R.J.C.; Dutzler, R. X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature 2008, 452, 375–379. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, I.; Dutzler, R. Ligand activation of the prokaryotic pentameric ligand-gated ion channel elic. PLoS Biol. 2011, 9, e1001101. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Chen, Q.; Willenbring, D.; Yoshida, K.; Tillman, T.; Kashlan, O.B.; Cohen, A.; Kong, X.-P.; Xu, Y.; Tang, P. Structure of the pentameric ligand-gated ion channel ELIC cocrystallized with its competitive antagonist acetylcholine. Nat. Commun. 2012, 3, 714. [Google Scholar] [CrossRef] [PubMed]
- Spurny, R.; Ramerstorfer, J.; Price, K.; Brams, M.; Ernst, M.; Nury, H.; Verheij, M.; Legrand, P.; Bertrand, D.; Bertrand, S.; et al. Pentameric ligand-gated ion channel ELIC is activated by GABA and modulated by benzodiazepines. Proc. Natl. Acad. Sci. USA 2012, 109, E3028–E3034. [Google Scholar] [CrossRef] [PubMed]
- Ulens, C.; Spurny, R.; Thompson, A.J.; Alqazzaz, M.; Debaveye, S.; Han, L.; Price, K.; Villalgordo, J.M.; Tresadern, G.; Lynch, J.W.; et al. The prokaryote ligand-gated ion channel ELIC captured in a pore blocker-bound conformation by the alzheimer’s disease drug memantine. Structure 2014, 22, 1399–1407. [Google Scholar] [CrossRef] [PubMed]
- Schmandt, N.; Velisetty, P.; Chalamalasetti, S.V.; Stein, R.A.; Bonner, R.; Talley, L.; Parker, M.D.; Mchaourab, H.S.; Yee, V.C.; Lodowski, D.T.; et al. A chimeric prokaryotic pentameric ligand–gated channel reveals distinct pathways of activation. J. Gen. Physiol. 2015, 146, 323–340. [Google Scholar] [CrossRef] [PubMed]
- Velisetty, P.; Chalamalasetti, S.V.; Chakrapani, S. Structural basis for allosteric coupling at the membrane-protein interface in gloeobacter violaceus ligand-gated ion channel (GLIC). J. Biol. Chem. 2014, 289, 3013–3025. [Google Scholar] [CrossRef] [PubMed]
- Sauguet, L.; Shahsavar, A.; Poitevin, F.; Huon, C.; Menny, A.; Nemecz, À.; Haouz, A.; Changeux, J.-P.; Corringer, P.-J.; Delarue, M. Crystal structures of a pentameric ligand-gated ion channel provide a mechanism for activation. Proc. Natl. Acad. Sci. USA 2014, 111, 966–971. [Google Scholar] [CrossRef] [PubMed]
- Ter Beek, J.; Guskov, A.; Slotboom, D.J. Structural diversity of ABC transporters. J. Gen. Physiol. 2014, 143, 419–435. [Google Scholar] [CrossRef] [PubMed]
- Locher, K.P. Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat. Struct. Mol. Biol. 2016, 23, 487–493. [Google Scholar] [CrossRef] [PubMed]
- Böhm, S.; Licht, A.; Wuttge, S.; Schneider, E.; Bordignon, E. Conformational plasticity of the type I maltose ABC importer. Proc. Natl. Acad. Sci. USA 2013, 110, 5492–5497. [Google Scholar] [CrossRef] [PubMed]
- Grote, M.; Polyhach, Y.; Jeschke, G.; Steinhoff, H.-J.; Schneider, E.; Bordignon, E. Transmembrane signaling in the maltose ABC transporter MalFGK2-E: Periplasmic MalF-P2 loop communicates substrate availability to the ATP-bound MalK dimer. J. Biol. Chem. 2009, 284, 17521–17526. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Qasim, S.; Davidson, A.L. Uncoupling substrate transport from ATP hydrolysis in the Escherichia coli maltose transporter. J. Biol. Chem. 2010, 285, 39986–39993. [Google Scholar] [CrossRef] [PubMed]
- Orelle, C.; Ayvaz, T.; Everly, R.M.; Klug, C.S.; Davidson, A.L. Both maltose-binding protein and ATP are required for nucleotide-binding domain closure in the intact maltose ABC transporter. Proc. Natl. Acad. Sci. USA 2008, 105, 12837–12842. [Google Scholar] [CrossRef] [PubMed]
- Rice, A.J.; Harrison, A.; Alvarez, F.J.D.; Davidson, A.L.; Pinkett, H.W. Small substrate transport and mechanism of a molybdate ATP binding cassette transporter in a lipid environment. J. Biol. Chem. 2014, 289, 15005–15013. [Google Scholar] [CrossRef] [PubMed]
- Rice, A.J.; Alvarez, F.J.D.; Davidson, A.L.; Pinkett, H.W. Effects of lipid environment on the conformational changes of an ABC importer. Channels 2014, 8, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Sippach, M.; Weidlich, D.; Klose, D.; Abé, C.; Klare, J.; Schneider, E.; Steinhoff, H.-J. Conformational changes of the histidine ATP-binding cassette transporter studied by double electron–electron resonance spectroscopy. Biochim. Biophys. Acta Biomembr. 2014, 1838, 1760–1768. [Google Scholar] [CrossRef] [PubMed]
- Hvorup, R.N.; Goetz, B.A.; Niederer, M.; Hollenstein, K.; Perozo, E.; Locher, K.P. Asymmetry in the structure of the ABC transporter-binding protein complex BtuCD-BtuF. Science 2007, 317, 1387–1390. [Google Scholar] [CrossRef] [PubMed]
- Goetz, B.A.; Perozo, E.; Locher, K.P. Distinct gate conformations of the ABC transporter BtuCD revealed by electron spin resonance spectroscopy and chemical cross-linking. FEBS Lett. 2009, 583, 266–270. [Google Scholar] [CrossRef] [PubMed]
- Joseph, B.; Jeschke, G.; Goetz, B.A.; Locher, K.P.; Bordignon, E. Transmembrane gate movements in the type II ATP-binding cassette (ABC) importer BtuCD-F during nucleotide cycle. J. Biol. Chem. 2011, 286, 41008–41017. [Google Scholar] [CrossRef] [PubMed]
- Joseph, B.; Korkhov, V.M.; Yulikov, M.; Jeschke, G.; Bordignon, E. Conformational cycle of the vitamin B12 ABC importer in liposomes detected by double electron-electron resonance (DEER). J. Biol. Chem. 2014, 289, 3176–3185. [Google Scholar] [CrossRef] [PubMed]
- Majsnerowska, M.; Hänelt, I.; Wunnicke, D.; Schäfer, L.V.; Steinhoff, H.-J.; Slotboom, D.J. Substrate-induced conformational changes in the S-component ThiT from an energy coupling factor transporter. Structure 2013, 21, 861–867. [Google Scholar] [CrossRef] [PubMed]
- Finkenwirth, F.; Sippach, M.; Landmesser, H.; Kirsch, F.; Ogienko, A.; Grunzel, M.; Kiesler, C.; Steinhoff, H.-J.; Schneider, E.; Eitinger, T. ATP-dependent conformational changes trigger substrate capture and release by an ECF-type biotin transporter. J. Biol. Chem. 2015, 290, 16929–16942. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Verhalen, B.; Stein, R.A.; Wen, P.-C.; Tajkhorshid, E.; McHaourab, H.S. Conformational dynamics of the nucleotide binding domains and the power stroke of a heterodimeric ABC transporter. eLife 2014, 3, e02740. [Google Scholar] [CrossRef] [PubMed]
- Nöll, A.; Thomas, C.; Herbring, V.; Zollmann, T.; Barth, K.; Mehdipour, A.R.; Tomasiak, T.M.; Brüchert, S.; Joseph, B.; Abele, R.; et al. Crystal structure and mechanistic basis of a functional homolog of the antigen transporter TAP. Proc. Natl. Acad. Sci. USA 2017, 114, E438–E447. [Google Scholar] [CrossRef] [PubMed]
- Mittal, A.; Böhm, S.; Grütter, M.G.; Bordignon, E.; Seeger, M.A. Asymmetry in the homodimeric ABC transporter MsbA recognized by a darpin. J. Biol. Chem. 2012, 287, 20395–20406. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Yang, G.; Mchaourab, H.S. Structural basis of energy transduction in the transport cycle of MsbA. Science 2005, 308, 1023–1028. [Google Scholar] [CrossRef] [PubMed]
- Zou, P.; Bortolus, M.; McHaourab, H.S. Conformational cycle of the ABC transporter MsbA in liposomes: Detailed analysis using double electron–electron resonance spectroscopy. J. Mol. Biol. 2009, 393, 586–597. [Google Scholar] [CrossRef] [PubMed]
- Hohl, M.; Hürlimann, L.M.; Böhm, S.; Schöppe, J.; Grütter, M.G.; Bordignon, E.; Seeger, M.A. Structural basis for allosteric cross-talk between the asymmetric nucleotide binding sites of a heterodimeric ABC exporter. Proc. Natl. Acad. Sci. USA 2014, 111, 11025–11030. [Google Scholar] [CrossRef] [PubMed]
- Timachi, M.H.; Hutter, C.A.J.; Hohl, M.; Assafa, T.; Böhm, S.; Mittal, A.; Seeger, M.A.; Bordignon, E. Exploring conformational equilibria of a heterodimeric ABC transporter. eLife 2017, 6, e20236. [Google Scholar] [CrossRef] [PubMed]
- Wen, P.-C.; Verhalen, B.; Wilkens, S.; Mchaourab, H.S.; Tajkhorshid, E. On the origin of large flexibility of P-glycoprotein in the inward-facing state. J. Biol. Chem. 2013, 288, 19211–19220. [Google Scholar] [CrossRef] [PubMed]
- Van Wonderen, J.H.; McMahon, R.M.; O’Mara, M.L.; McDevitt, C.A.; Thomson, A.J.; Kerr, I.D.; MacMillan, F.; Callaghan, R. The central cavity of ABCB1 undergoes alternating access during ATP hydrolysis. FEBS J. 2014, 281, 2190–2201. [Google Scholar] [CrossRef] [PubMed]
- Herget, M.; Baldauf, C.; Schölz, C.; Parcej, D.; Wiesmüller, K.-H.; Tampé, R.; Abele, R.; Bordignon, E. Conformation of peptides bound to the transporter associated with antigen processing (TAP). Proc. Natl. Acad. Sci. USA 2011, 108, 1349–1354. [Google Scholar] [CrossRef] [PubMed]
- Hellmich, U.A.; Lyubenova, S.; Kaltenborn, E.; Doshi, R.; van Veen, H.W.; Prisner, T.F.; Glaubitz, C. Probing the ATP hydrolysis cycle of the ABC multidrug transporter LmrA by pulsed EPR spectroscopy. J. Am. Chem. Soc. 2012, 134, 5857–5862. [Google Scholar] [CrossRef] [PubMed]
- Borbat, P.P.; Surendhran, K.; Bortolus, M.; Zou, P.; Freed, J.H.; McHaourab, H.S. Conformational motion of the ABCc transporter MsbA induced by ATP hydrolysis. PLoS Biol. 2007, 5, e271. [Google Scholar] [CrossRef] [PubMed]
- George, A.M.; Jones, P.M. Perspectives on the structure–function of ABCc transporters: The switch and constant contact models. Prog. Biophys. Mol. Biol. 2012, 109, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Higgins, C.F.; Linton, K.J. The ATP switch model for ABC transporters. Nat. Struct. Mol. Biol. 2004, 11, 918–926. [Google Scholar] [CrossRef] [PubMed]
- Shintre, C.A.; Pike, A.C.; Li, Q.; Kim, J.-I.; Barr, A.J.; Goubin, S.; Shrestha, L.; Yang, J.; Berridge, G.; Ross, J. Structures of ABCB10, a human ATP-binding cassette transporter in apo-and nucleotide-bound states. Proc. Natl. Acad. Sci. USA 2013, 110, 9710–9715. [Google Scholar] [CrossRef] [PubMed]
- Ward, A.; Reyes, C.L.; Yu, J.; Roth, C.B.; Chang, G. Flexibility in the ABC transporter MsbA: Alternating access with a twist. Proc. Natl. Acad. Sci. USA 2007, 104, 19005–19010. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Yang, J.G.; Zhitnitsky, D.; Lewinson, O.; Rees, D.C. Structural basis for heavy metal detoxification by an ATM1-type ABC exporter. Science 2014, 343, 1133–1136. [Google Scholar] [CrossRef] [PubMed]
- Dawson, R.J.P.; Locher, K.P. Structure of a bacterial multidrug ABC transporter. Nature 2006, 443, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, H.G.; Tong, Z.; Mathavan, I.; Li, Y.; Iwata, S.; Zirah, S.; Rebuffat, S.; van Veen, H.W.; Beis, K. Structure of an antibacterial peptide ATP-binding cassette transporter in a novel outward occluded state. Proc. Natl. Acad. Sci. USA 2014, 111, 9145–9150. [Google Scholar] [CrossRef] [PubMed]
- Sauna, Z.E.; Ambudkar, S.V. Characterization of the catalytic cycle of atp hydrolysis by human P-glycoprotein: The two ATP hydrolysis events in a single catalytic cycle are kinetically similar but affect different functional outcomes. J. Biol. Chem. 2001, 276, 11653–11661. [Google Scholar] [CrossRef] [PubMed]
- Sauna, Z.E.; Ambudkar, S.V. About a switch: How P-glycoprotein (ABCB1) harnesses the energy of ATP binding and hydrolysis to do mechanical work. Mol. Cancer Ther. 2007, 6, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Basso, C.; Vergani, P.; Nairn, A.C.; Gadsby, D.C. Prolonged nonhydrolytic interaction of nucleotide with CFTR’s NH2-terminal nucleotide binding domain and its role in channel gating. J. Gen. Physiol. 2003, 122, 333–348. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.-F.; Li, M.; Hwang, T.-C. Stable ATP binding mediated by a partial NBD dimer of the CFTR chloride channel. J. Gen. Physiol. 2010, 135, 399–414. [Google Scholar] [CrossRef] [PubMed]
- Hohl, M.; Briand, C.; Grütter, M.G.; Seeger, M.A. Crystal structure of a heterodimeric ABC transporter in its inward-facing conformation. Nat. Struct. Mol. Biol. 2012, 19, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Forrest, L.R.; Krämer, R.; Ziegler, C. The structural basis of secondary active transport mechanisms. Biochim. Biophys. Acta Bioenerg. 2011, 1807, 167–188. [Google Scholar] [CrossRef] [PubMed]
- Masureel, M.; Martens, C.; Stein, R.A.; Mishra, S.; Ruysschaert, J.-M.; McHaourab, H.S.; Govaerts, C. Protonation drives the conformational switch in the multidrug transporter LmrP. Nat. Chem. Biol. 2014, 10, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Martens, C.; Stein, R.A.; Masureel, M.; Roth, A.; Mishra, S.; Dawaliby, R.; Konijnenberg, A.; Sobott, F.; Govaerts, C.; Mchaourab, H.S. Lipids modulate the conformational dynamics of a secondary multidrug transporter. Nat. Struct. Mol. Biol. 2016, 23, 744–751. [Google Scholar] [CrossRef] [PubMed]
- Claxton, D.P.; Quick, M.; Shi, L.; de Carvalho, F.D.; Weinstein, H.; Javitch, J.A.; McHaourab, H.S. Ion/substrate-dependent conformational dynamics of a bacterial homolog of neurotransmitter:Sodium symporters. Nat. Struct. Mol. Biol. 2010, 17, 822–829. [Google Scholar] [CrossRef] [PubMed]
- Kazmier, K.; Sharma, S.; Quick, M.; Islam, S.M.; Roux, B.; Weinstein, H.; Javitch, J.A.; McHaourab, H.S. Conformational dynamics of ligand-dependent alternating access in LeuT. Nat. Struct. Mol. Biol. 2014, 21, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Kazmier, K.; Sharma, S.; Islam, S.M.; Roux, B.; Mchaourab, H.S. Conformational cycle and ion-coupling mechanism of the Na+/Hydantoin transporter Mhp1. Proc. Natl. Acad. Sci. USA 2014, 111, 14752–14757. [Google Scholar] [CrossRef] [PubMed]
- Bracher, S.; Schmidt, C.C.; Dittmer, S.I.; Jung, H. Core transmembrane domain 6 plays a pivotal role in the transport cycle of the sodium/proline symporter PutP. J. Biol. Chem. 2016, 291, 26208–26215. [Google Scholar] [CrossRef] [PubMed]
- Raba, M.; Dunkel, S.; Hilger, D.; Lipiszko, K.; Polyhach, Y.; Jeschke, G.; Bracher, S.; Klare, J.P.; Quick, M.; Jung, H.; et al. Extracellular loop 4 of the proline transporter PutP controls the periplasmic entrance to ligand binding sites. Structure 2014, 22, 769–780. [Google Scholar] [CrossRef] [PubMed]
- Hilger, D.; Polyhach, Y.; Jung, H.; Jeschke, G. Backbone structure of transmembrane domain IX of the Na+/proline transporter PutP of Escherichia coli. Biophys. J. 2009, 96, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Hilger, D.; Polyhach, Y.; Padan, E.; Jung, H.; Jeschke, G. High-resolution structure of a Na+/H+ antiporter dimer obtained by pulsed electron paramagnetic resonance distance measurements. Biophys. J. 2007, 93, 3675–3683. [Google Scholar] [CrossRef] [PubMed]
- Dastvan, R.; Fischer, A.W.; Mishra, S.; Meiler, J.; Mchaourab, H.S. Protonation-dependent conformational dynamics of the multidrug transporter EmrE. Proc. Natl. Acad. Sci. USA 2016, 113, 1220–1225. [Google Scholar] [CrossRef] [PubMed]
- Khantwal, C.M.; Abraham, S.J.; Han, W.; Jiang, T.; Chavan, T.S.; Cheng, R.C.; Elvington, S.M.; Liu, C.W.; Mathews, I.I.; Stein, R.A.; et al. Revealing an outward-facing open conformational state in a CLC Cl−/H+ exchange transporter. eLife 2016, 5, e11189. [Google Scholar] [CrossRef] [PubMed]
- Hänelt, I.; Wunnicke, D.; Bordignon, E.; Steinhoff, H.-J.; Slotboom, D.J. Conformational heterogeneity of the aspartate transporter GltPh. Nat. Struct. Mol. Biol. 2013, 20, 210–214. [Google Scholar] [CrossRef] [PubMed]
- Georgieva, E.R.; Borbat, P.P.; Ginter, C.; Freed, J.H.; Boudker, O. Conformational ensemble of the sodium-coupled aspartate transporter. Nat. Struct. Mol. Biol. 2013, 20, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.-H.; Hazell, A.S. Excitotoxic mechanisms and the role of astrocytic glutamate transporters in traumatic brain injury. Neurochem. Int. 2006, 48, 394–403. [Google Scholar] [CrossRef] [PubMed]
- Fontana, A.C.K. Current approaches to enhance glutamate transporter function and expression. J. Neurochem. 2015, 134, 982–1007. [Google Scholar] [CrossRef] [PubMed]
- Danbolt, N.C. Glutamate uptake. Prog. Neurobiol. 2001, 65, 1–105. [Google Scholar] [CrossRef]
- Teichman, S.; Qu, S.; Kanner, B.I. The equivalent of a thallium binding residue from an archeal homolog controls cation interactions in brain glutamate transporters. Proc. Natl. Acad. Sci. USA 2009, 106, 14297–14302. [Google Scholar] [CrossRef] [PubMed]
- Groeneveld, M.; Slotboom, D.J. Na+: Aspartate coupling stoichiometry in the glutamate transporter homologue Glt(Ph). Biochemistry 2010, 49, 3511–3513. [Google Scholar] [CrossRef] [PubMed]
- Zerangue, N.; Kavanaugh, M.P. Flux coupling in a neuronal glutamate transporter. Nature 1996, 383, 634–637. [Google Scholar] [CrossRef] [PubMed]
- Raunser, S. Structure and function of prokaryotic glutamate transporters from Escherichia coli and Pyrococcus horikoshii. Biochemistry 2006, 45, 12796–12805. [Google Scholar] [CrossRef] [PubMed]
- Reyes, N.; Ginter, C.; Boudker, O. Transport mechanism of a bacterial homologue of glutamate transporters. Nature 2009, 462, 880–885. [Google Scholar] [CrossRef] [PubMed]
- Groeneveld, M.; Slotboom, D.J. Rigidity of the subunit interfaces of the trimeric glutamate transporter GltT during translocation. J. Mol. Biol. 2007, 372, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Verdon, G.; Boudker, O. Crystal structure of an asymmetric trimer of a bacterial glutamate transporter homolog. Nat. Struct. Mol. Biol. 2012, 19, 355–357. [Google Scholar] [CrossRef] [PubMed]
- Boudker, O.; Ryan, R.M.; Yernool, D.; Shimamoto, K.; Gouaux, E. Coupling substrate and ion binding to extracellular gate of a sodium-dependent aspartate transporter. Nature 2007, 445, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Yernool, D.; Boudker, O.; Jin, Y.; Gouaux, E. Structure of a glutamate transporter homologue from Pyrococcus horikoshii. Nature 2004, 431, 811–818. [Google Scholar] [CrossRef] [PubMed]
- Focke, P.J.; Moenne-Loccoz, P.; Larsson, H.P. Opposite movement of the external gate of a glutamate transporter homolog upon binding cotransported sodium compared with substrate. J. Neurosci. 2011, 31, 6255–6262. [Google Scholar] [CrossRef] [PubMed]
- Singer, S.; Nicolson, G.L. The fluid mosaic model of the structure of cell membranes. Science 1972, 175, 720–731. [Google Scholar] [CrossRef] [PubMed]
- Von Heijne, G. The distribution of positively charged residues in bacterial inner membrane proteins correlates with the trans-membrane topology. EMBO J. 1986, 5, 3021. [Google Scholar]
- Phillips, R.; Ursell, T.; Wiggins, P.; Sens, P. Emerging roles for lipids in shaping membrane-protein function. Nature 2009, 459, 379–385. [Google Scholar] [CrossRef] [PubMed]
- Spector, A.A.; Yorek, M.A. Membrane lipid composition and cellular function. J. Lipid Res. 1985, 26, 1015–1035. [Google Scholar] [PubMed]
- Lee, A.G. How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta Biomembr. 2004, 1666, 62–87. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, F.J.D.; Orelle, C.; Davidson, A.L. Functional reconstitution of an ABC transporter in nanodiscs for use in electron paramagnetic resonance spectroscopy. J. Am. Chem. Soc. 2010, 132, 9513–9515. [Google Scholar] [CrossRef] [PubMed]
- Bayburt, T.H.; Sligar, S.G. Membrane protein assembly into nanodiscs. FEBS Lett. 2010, 584, 1721–1727. [Google Scholar] [CrossRef] [PubMed]
- Davidson, W.S.; Hilliard, G.M. The spatial organization of apolipoprotein AI on the edge of discoidal high density lipoprotein particles a mass spectrometry study. J. Biol. Chem. 2003, 278, 27199–27207. [Google Scholar] [CrossRef] [PubMed]
- Gorshkova, I.N.; Liu, T.; Kan, H.-Y.; Chroni, A.; Zannis, V.I.; Atkinson, D. Structure and stability of apolipoprotein AI in solution and in discoidal high-density lipoprotein probed by double charge ablation and deletion mutation. Biochemistry 2006, 45, 1242–1254. [Google Scholar] [CrossRef] [PubMed]
- Boldog, T.; Grimme, S.; Li, M.; Sligar, S.G.; Hazelbauer, G.L. Nanodiscs separate chemoreceptor oligomeric states and reveal their signaling properties. Proc. Natl. Acad. Sci. USA 2006, 103, 11509–11514. [Google Scholar] [CrossRef] [PubMed]
- Whorton, M.R.; Bokoch, M.P.; Rasmussen, S.G.; Huang, B.; Zare, R.N.; Kobilka, B.; Sunahara, R.K. A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc. Natl. Acad. Sci. USA 2007, 104, 7682–7687. [Google Scholar] [CrossRef] [PubMed]
- Joseph, B.; Sikora, A.; Bordignon, E.; Jeschke, G.; Cafiso, D.S.; Prisner, T.F. Distance measurement on an endogenous membrane transporter in E. Coli cells and native membranes using EPR spectroscopy. Angew. Chem. Int. Ed. 2015, 54, 6196–6199. [Google Scholar] [CrossRef] [PubMed]
- Joseph, B.; Tormyshev, V.M.; Rogozhnikova, O.Y.; Akhmetzyanov, D.; Bagryanskaya, E.G.; Prisner, T.F. Selective high-resolution detection of membrane protein–ligand interaction in native membranes using trityl–nitroxide peldor. Angew. Chem. Int. Ed. 2016, 55, 11538–11542. [Google Scholar] [CrossRef] [PubMed]
- Chimento, D.P.; Mohanty, A.K.; Kadner, R.J.; Wiener, M.C. Substrate-induced transmembrane signaling in the cobalamin transporter BtuB. Nat. Struct. Mol. Biol. 2003, 10, 394–401. [Google Scholar] [CrossRef] [PubMed]
- Shultis, D.D.; Purdy, M.D.; Banchs, C.N.; Wiener, M.C. Outer membrane active transport: Structure of the BtuB:Tonb complex. Science 2006, 312, 1396–1399. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Payne, M.A.; Cao, Z.; Foster, S.B.; Feix, J.B.; Newton, S.M.C.; Klebba, P.E. Ligand-specific opening of a gated-porin channel in the outer membrane of living bacteria. Science 1997, 276, 1261–1264. [Google Scholar] [CrossRef] [PubMed]
- Merianos, H.J.; Cadieux, N.; Lin, C.H.; Kadner, R.J.; Cafiso, D.S. Substrate-induced exposure of an energy-coupling motif of a membrane transporter. Nat. Struct. Mol. Biol. 2000, 7, 205–209. [Google Scholar]
- Agard, N.J.; Prescher, J.A.; Bertozzi, C.R. A strain-promoted [3 + 2] azide–alkyne cycloaddition for covalent modification of biomolecules in living systems. J. Am. Chem. Soc. 2004, 126, 15046–15047. [Google Scholar] [CrossRef] [PubMed]
- Kucher, S.; Korneev, S.; Tyagi, S.; Apfelbaum, R.; Grohmann, D.; Lemke, E.A.; Klare, J.P.; Steinhoff, H.-J.; Klose, D. Orthogonal spin labeling using click chemistry for in vitro and in vivo applications. J. Magn. Reson. 2017, 275, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Spindler, P.E.; Glaser, S.J.; Skinner, T.E.; Prisner, T.F. Broadband inversion peldor spectroscopy with partially adiabatic shaped pulses. Angew. Chem. Int. Ed. 2013, 52, 3425–3429. [Google Scholar] [CrossRef] [PubMed]
- Spindler, P.E.; Waclawska, I.; Endeward, B.; Plackmeyer, J.; Ziegler, C.; Prisner, T.F. Carr–purcell pulsed electron double resonance with shaped inversion pulses. J. Phys. Chem. Lett. 2015, 6, 4331–4335. [Google Scholar] [CrossRef] [PubMed]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wunnicke, D.; Hänelt, I. The Synergetic Effects of Combining Structural Biology and EPR Spectroscopy on Membrane Proteins. Crystals 2017, 7, 117. https://doi.org/10.3390/cryst7040117
Wunnicke D, Hänelt I. The Synergetic Effects of Combining Structural Biology and EPR Spectroscopy on Membrane Proteins. Crystals. 2017; 7(4):117. https://doi.org/10.3390/cryst7040117
Chicago/Turabian StyleWunnicke, Dorith, and Inga Hänelt. 2017. "The Synergetic Effects of Combining Structural Biology and EPR Spectroscopy on Membrane Proteins" Crystals 7, no. 4: 117. https://doi.org/10.3390/cryst7040117




