An Overview of the Top Ten Detergents Used for Membrane Protein Crystallization
Abstract
:1. Introduction
2. Results
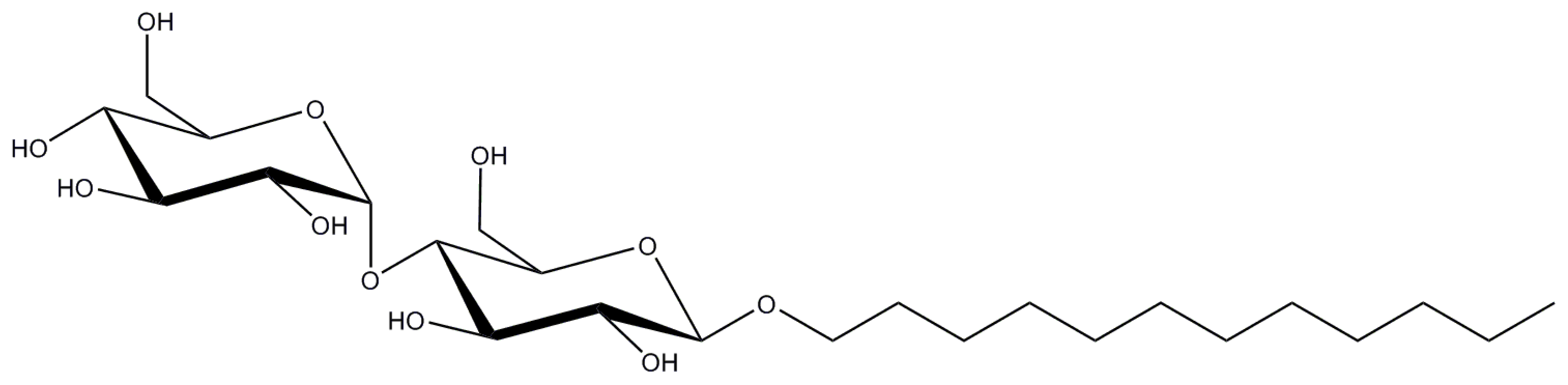
2.1. n-Dodecyl-β-d-Maltopyranoside
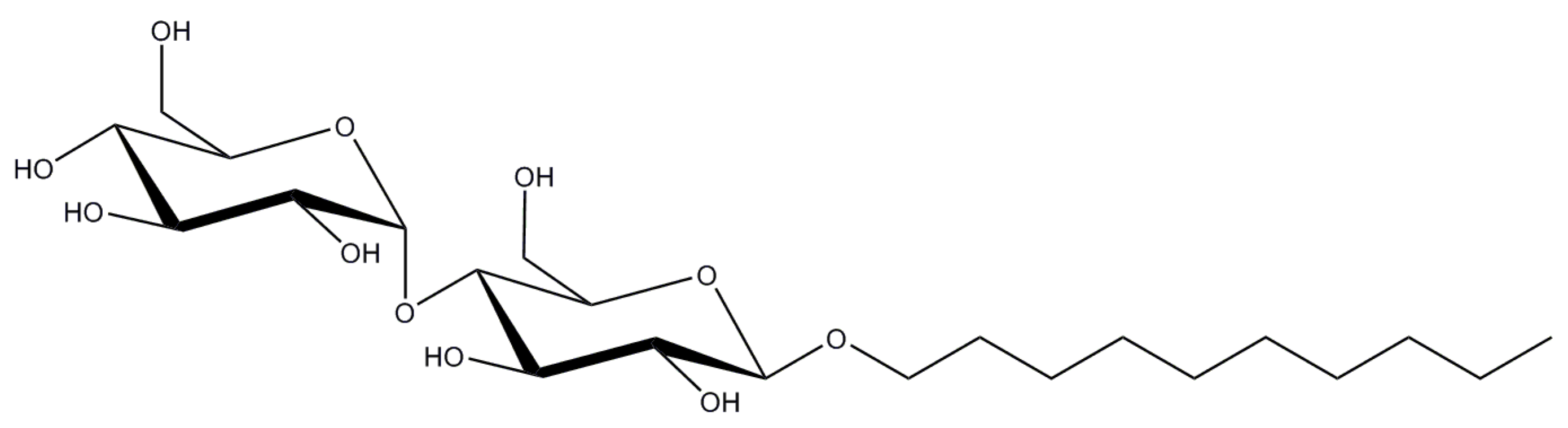
2.2. n-Decyl-β-d-Maltopyranoside
2.3. n-Octyl-β-d-Glucopyranoside
2.4. n-Nonyl β-d-Glucopyranoside
2.5. Lauryldimethylamine-N-Oxide
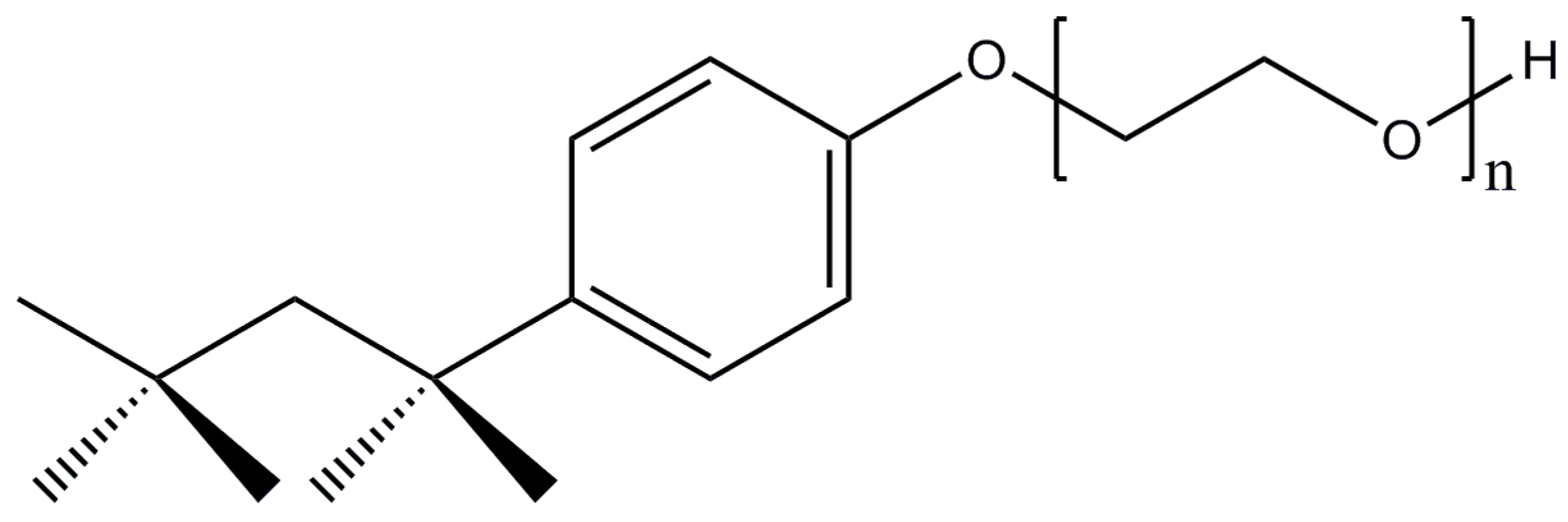
2.6. Polyoxyethylene 8 (9) dodecyl Ether
2.7. n-Undecyl-β-d-Maltopyranoside
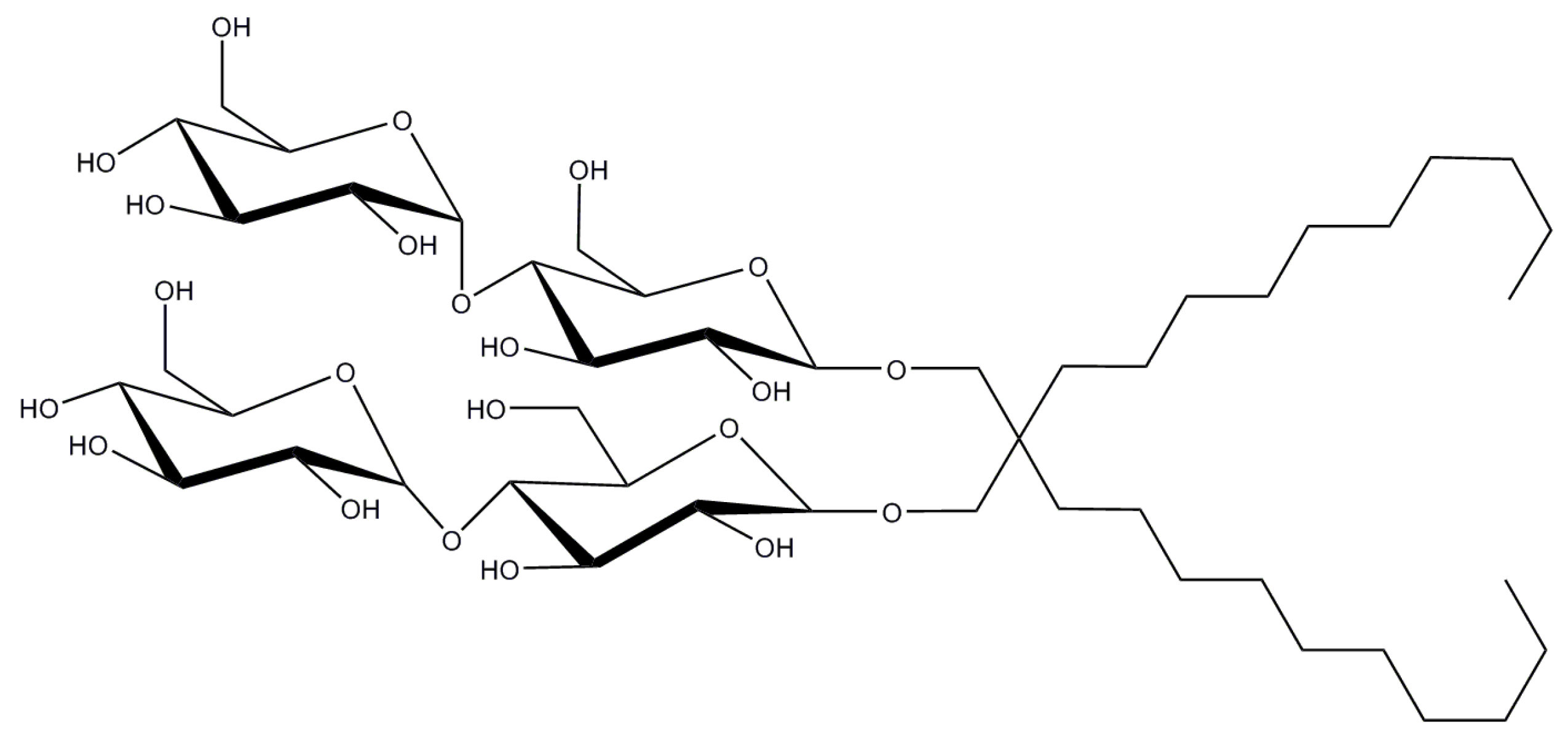
2.8. Lauryl Maltose Neopentyl Glycol
2.9. Triton X-100
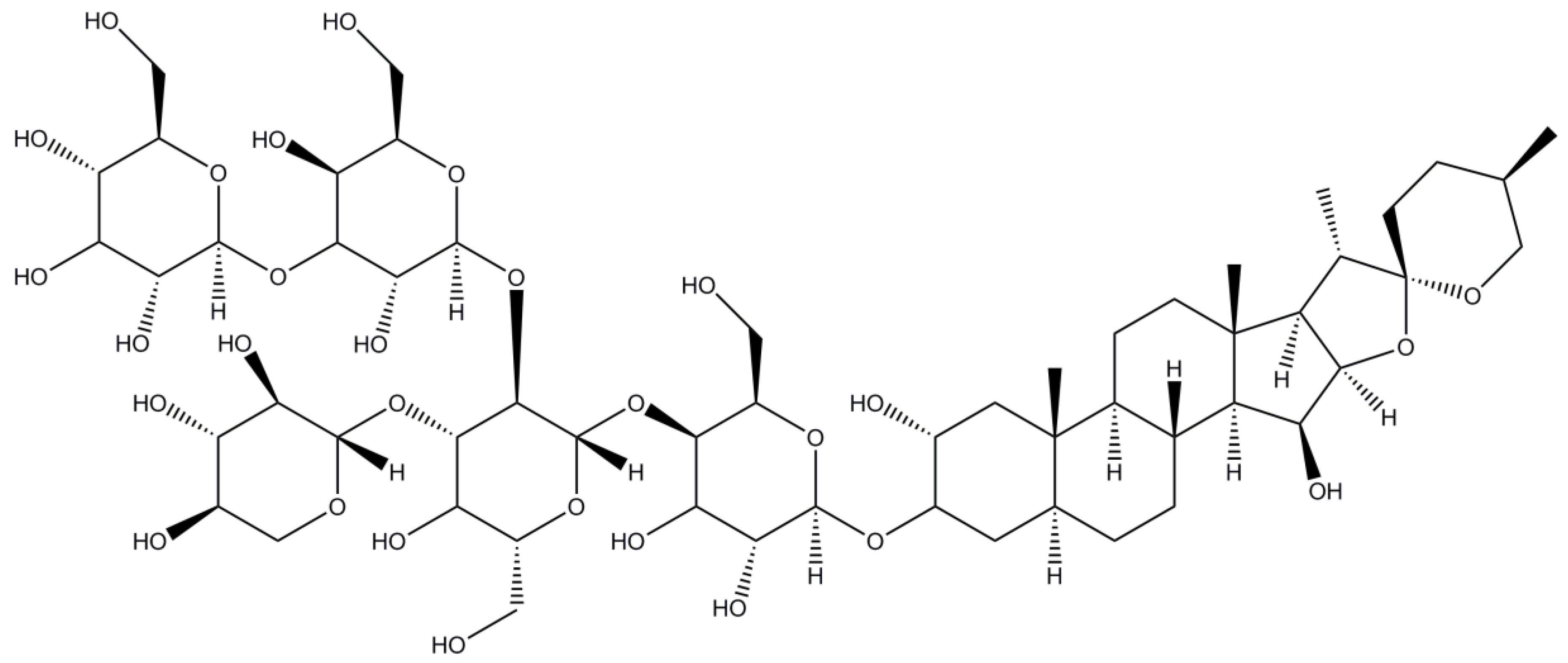
2.10. Digitonin
2.11. Cymal-5 (Cymal-6)
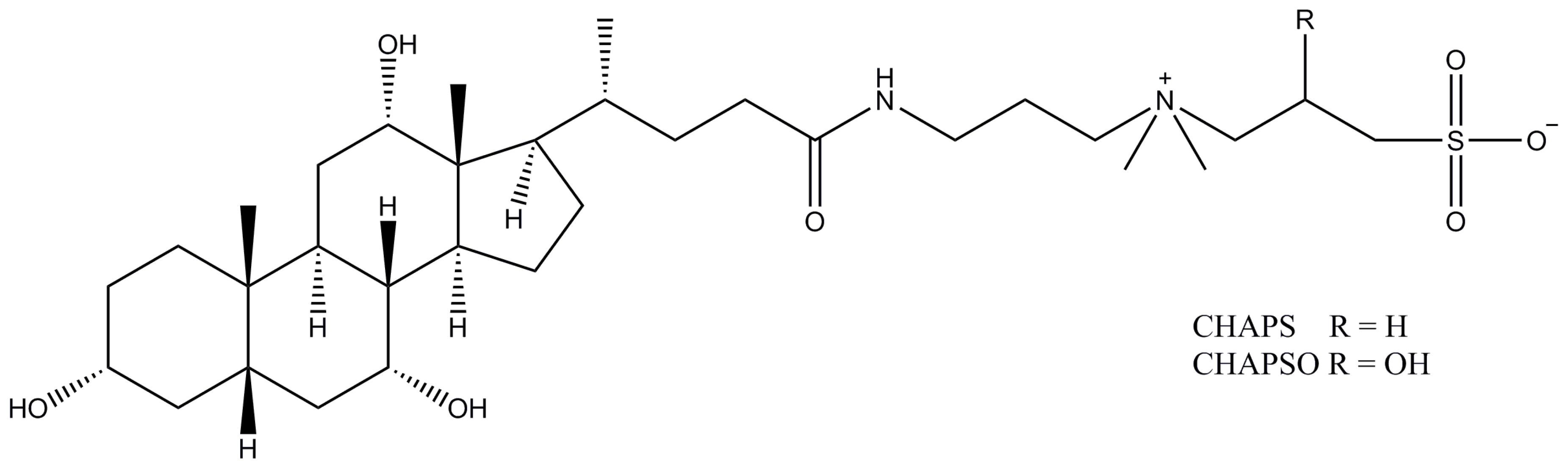
2.12. CHAPS (CHAPSO)
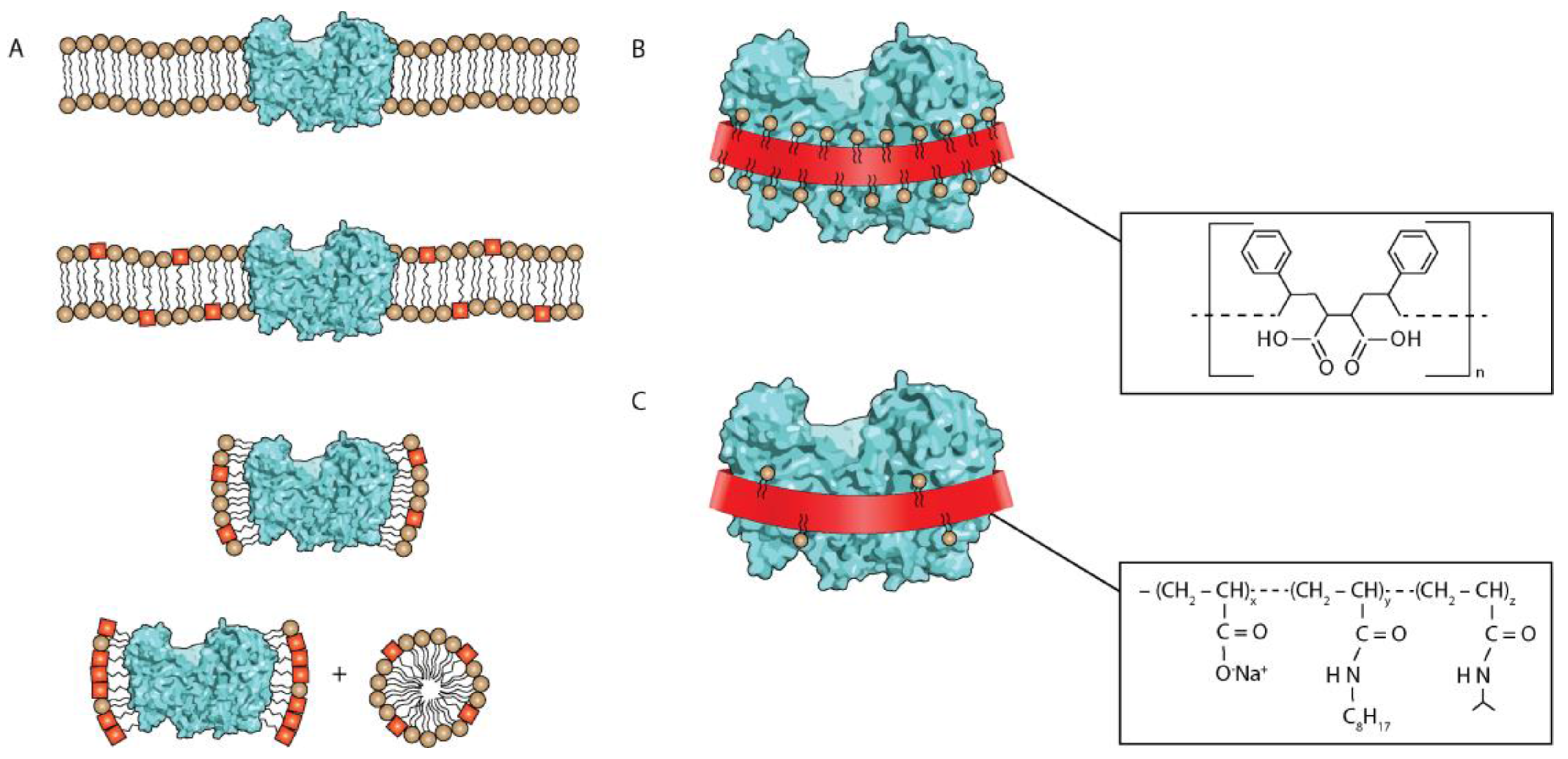
3. Discussion and Outlook
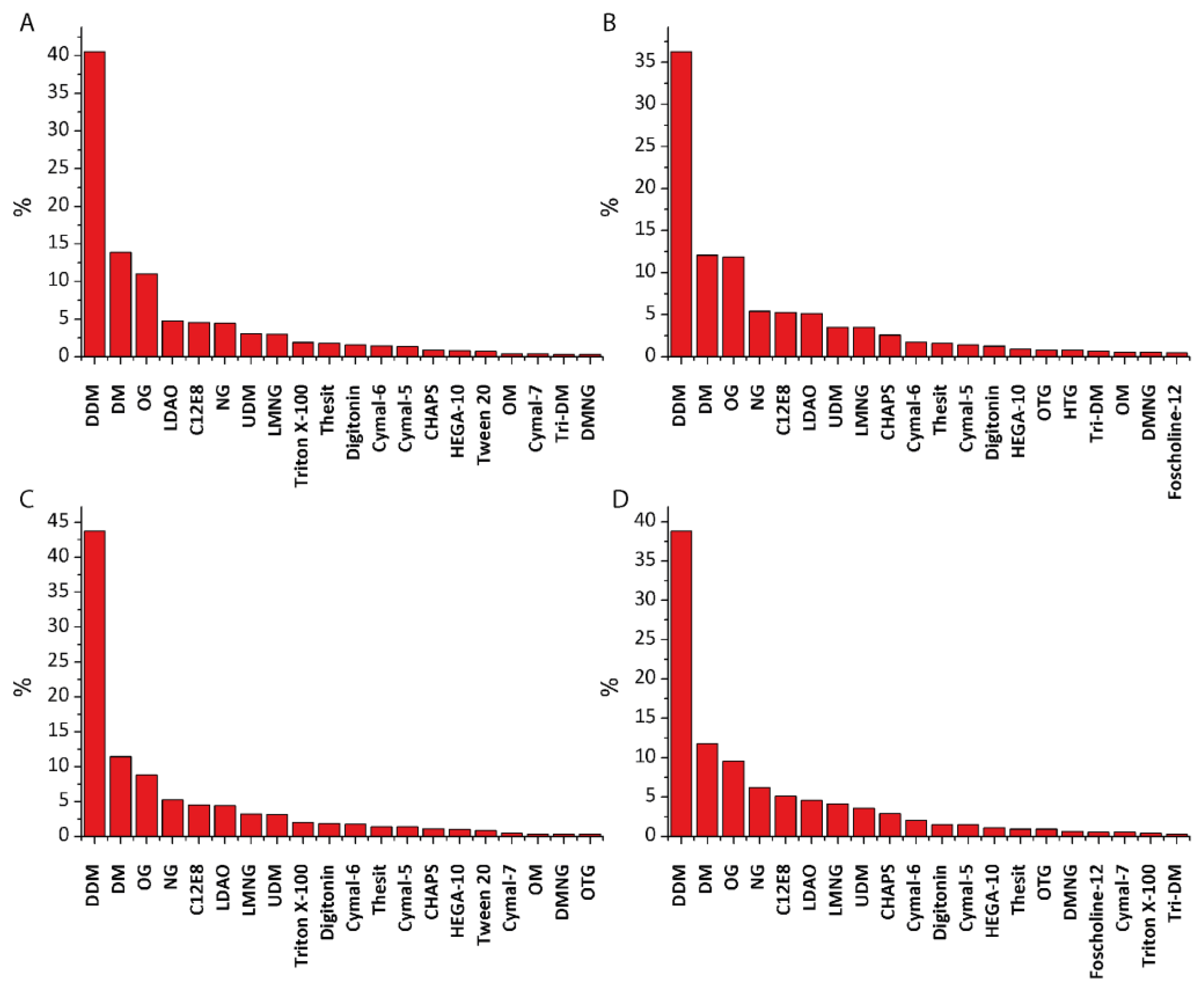
4. Materials and Methods
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Flügge, U.-I.; Westhoff, P.; Leister, D. Recent advances in understanding photosynthesis. F1000Research 2016, 5, 2890. [Google Scholar] [CrossRef] [PubMed]
- Ellisdon, A.M.; Halls, M.L. Compartmentalization of GPCR signalling controls unique cellular responses. Biochem. Soc. Trans. 2016, 44, 562–567. [Google Scholar] [CrossRef] [PubMed]
- Kern, J.; Guskov, A. Lipids in photosystem II: Multifunctional cofactors. J. Photochem. Photobiol. B Biol. 2011, 104, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Mazor, Y.; Borovikova, A.; Caspy, I.; Nelson, N. Structure of the plant photosystem I supercomplex at 2.6 Å resolution. Nat. Plants 2017, 3, 17014. [Google Scholar] [CrossRef] [PubMed]
- Hunte, C.; Richers, S. Lipids and membrane protein structures. Curr. Opin. Struct. Biol. 2008, 18, 406–411. [Google Scholar] [CrossRef] [PubMed]
- Valiyaveetil, F.I.; Zhou, Y.; MacKinnon, R. Lipids in the structure, folding, and function of the KcsA K+ channel. Biochemistry 2002, 41, 10771–10777. [Google Scholar] [CrossRef] [PubMed]
- Lemieux, M.J.; Song, J.; Kim, M.J.; Huang, Y.; Villa, A.; Auer, M.; Li, X.-D.; Wang, D.-N. Three-dimensional crystallization of the Escherichia coli glycerol-3-phosphate transporter: A member of the major facilitator superfamily. Protein Sci. 2003, 12, 2748–2756. [Google Scholar] [CrossRef] [PubMed]
- Sonoda, Y.; Newstead, S.; Hu, N.-J.; Alguel, Y.; Nji, E.; Beis, K.; Yashiro, S.; Lee, C.; Leung, J.; Cameron, A.D.; et al. Benchmarking Membrane Protein Detergent Stability for Improving Throughput of High-Resolution X-ray Structures. Structure 2011, 19, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Cherezov, V.; Rosenbaum, D.M.; Hanson, M.A.; Rasmussen, S.G.F.; Thian, F.S.; Kobilka, T.S.; Choi, H.-J.; Kuhn, P.; Weis, W.I.; Kobilka, B.K.; et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science 2007, 318, 1258–1265. [Google Scholar] [CrossRef] [PubMed]
- Van den Brink-van der Laan, E.; Chupin, V.; Killian, J.A.; de Kruijff, B. Stability of KcsA tetramer depends on membrane lateral pressure. Biochemistry 2004, 43, 4240–4250. [Google Scholar] [CrossRef] [PubMed]
- Parton, D.L.; Klingelhoefer, J.W.; Sansom, M.S.P. Aggregation of model membrane proteins, modulated by hydrophobic mismatch, membrane curvature, and protein class. Biophys. J. 2011, 101, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Newby, Z.E.R.; O’Connell, J.D.; Gruswitz, F.; Hays, F.A.; Harries, W.E.C.; Harwood, I.M.; Ho, J.D.; Lee, J.K.; Savage, D.F.; Miercke, L.J.W.; et al. A general protocol for the crystallization of membrane proteins for X-ray structural investigation. Nat. Protoc. 2009, 4, 619–637. [Google Scholar] [CrossRef] [PubMed]
- Hitscherich, C.; Aseyev, V.; Wiencek, J.; Loll, P.J. Effects of PEG on detergent micelles: Implications for the crystallization of integral membrane proteins. Acta Crystallogr. D Biol. Crystallogr. 2001, 57, 1020–1029. [Google Scholar] [CrossRef] [PubMed]
- Bordier, C. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 1981, 256, 1604–1607. [Google Scholar] [PubMed]
- Sánchez-Ferrer, A.; Bru, R.; García-Carmona, F. Phase separation of biomolecules in polyoxyethylene glycol nonionic detergents. Crit. Rev. Biochem. Mol. Biol. 1994, 29, 275–313. [Google Scholar] [CrossRef] [PubMed]
- Arnold, T.; Linke, D. Phase separation in the isolation and purification of membrane proteins. BioTechniques 2007, 43, 427–442. [Google Scholar] [CrossRef] [PubMed]
- Koszelak-Rosenblum, M.; Krol, A.; Mozumdar, N.; Wunsch, K.; Ferin, A.; Cook, E.; Veatch, C.K.; Nagel, R.; Luft, J.R.; DeTitta, G.T.; et al. Determination and application of empirically derived detergent phase boundaries to effectively crystallize membrane proteins. Protein Sci. 2009, 18, 1828–1839. [Google Scholar] [CrossRef] [PubMed]
- Laganowsky, A.; Reading, E.; Allison, T.M.; Ulmschneider, M.B.; Degiacomi, M.T.; Baldwin, A.J.; Robinson, C.V. Membrane proteins bind lipids selectively to modulate their structure and function. Nature 2014, 510, 172–175. [Google Scholar] [CrossRef] [PubMed]
- Chaptal, V.; Delolme, F.; Kilburg, A.; Magnard, S.; Montigny, C.; Picard, M.; Prier, C.; Monticelli, L.; Bornert, O.; Agez, M.; et al. Quantification of Detergents Complexed with Membrane Proteins. Sci. Rep. 2017, 7, 41751. [Google Scholar] [CrossRef] [PubMed]
- Allison, T.M.; Reading, E.; Liko, I.; Baldwin, A.J.; Laganowsky, A.; Robinson, C.V. Quantifying the stabilizing effects of protein-ligand interactions in the gas phase. Nat. Commun. 2015, 6, 8551. [Google Scholar] [CrossRef] [PubMed]
- Guskov, A.; Slotboom, D.J. Size exclusion chromatography with multi-angle laser light scattering (SEC-MALLS) to determine protein oligomeric states. In From Molecules to Living Organisms: An Interplay between Biology and Physics; Lecture Notes of the Les Houches School of Physics; Volume 102, July 2014; Oxford University Press: New York, NY, USA, 2016; pp. 169–183. [Google Scholar]
- Folta-Stogniew, E. Oligomeric states of proteins determined by size-exclusion chromatography coupled with light scattering, absorbance, and refractive index detectors. Methods Mol. Biol. 2006, 328, 97–112. [Google Scholar] [PubMed]
- Linke, D. Detergents: An overview. Methods Enzymol. 2009, 463, 603–617. [Google Scholar] [PubMed]
- Smith, S.M. Strategies for the Purification of Membrane Proteins. In Protein Chromatography: Methods and Protocols; Walls, D., Loughran, S., Eds.; Springer: Berlin, Germany, 2017; pp. 389–400. [Google Scholar]
- Lin, S.-H.; Guidotti, G. Purification of membrane proteins. Methods Enzymol. 2009, 463, 619–629. [Google Scholar] [PubMed]
- Lee, S.C.; Pollock, N.L. Membrane proteins: Is the future disc shaped? Biochem. Soc. Trans. 2016, 44, 1011–1018. [Google Scholar] [CrossRef] [PubMed]
- Dörr, J.M.; Scheidelaar, S.; Koorengevel, M.C.; Dominguez, J.J.; Schäfer, M.; van Walree, C.A.; Killian, J.A. The styrene-maleic acid copolymer: A versatile tool in membrane research. Eur. Biophys. J. 2016, 45, 3–21. [Google Scholar] [CrossRef] [PubMed]
- Jamshad, M.; Grimard, V.; Idini, I.; Knowles, T.J.; Dowle, M.R.; Schofield, N.; Sridhar, P.; Lin, Y.; Finka, R.; Wheatley, M.; et al. Structural analysis of a nanoparticle containing a lipid bilayer used for detergent-free extraction of membrane proteins. Nano Res. 2015, 8, 774–789. [Google Scholar] [CrossRef]
- Lee, S.C.; Knowles, T.J.; Postis, V.L.G.; Jamshad, M.; Parslow, R.A.; Lin, Y.-P.; Goldman, A.; Sridhar, P.; Overduin, M.; Muench, S.P.; et al. A method for detergent-free isolation of membrane proteins in their local lipid environment. Nat. Protoc. 2016, 11, 1149–1162. [Google Scholar] [CrossRef] [PubMed]
- Broecker, J.; Eger, B.T.; Ernst, O.P. Crystallogenesis of Membrane Proteins Mediated by Polymer-Bounded Lipid Nanodiscs. Structure 2017, 25, 384–392. [Google Scholar] [CrossRef] [PubMed]
- Dörr, J.M.; Koorengevel, M.C.; Schäfer, M.; Prokofyev, A.V.; Scheidelaar, S.; van der Cruijsen, E.A.W.; Dafforn, T.R.; Baldus, M.; Killian, J.A. Detergent-free isolation, characterization, and functional reconstitution of a tetrameric K+ channel: The power of native nanodiscs. Proc. Natl. Acad. Sci. USA 2014, 111, 18607–18612. [Google Scholar] [CrossRef] [PubMed]
- Postis, V.; Rawson, S.; Mitchell, J.K.; Lee, S.C.; Parslow, R.A.; Dafforn, T.R.; Baldwin, S.A.; Muench, S.P. The use of SMALPs as a novel membrane protein scaffold for structure study by negative stain electron microscopy. Biochim. Biophys. Acta 2015, 1848, 496–501. [Google Scholar] [CrossRef] [PubMed]
- Craig, A.F.; Clark, E.E.; Sahu, I.D.; Zhang, R.; Frantz, N.D.; Al-Abdul-Wahid, M.S.; Dabney-Smith, C.; Konkolewicz, D.; Lorigan, G.A. Tuning the size of styrene-maleic acid copolymer-lipid nanoparticles (SMALPs) using RAFT polymerization for biophysical studies. Biochim. Biophys. Acta 2016, 1858, 2931–2939. [Google Scholar] [CrossRef] [PubMed]
- Zoonens, M.; Popot, J.-L. Amphipols for Each Season. J. Membr. Biol. 2014, 247, 759–796. [Google Scholar] [CrossRef] [PubMed]
- Popot, J.L.; Althoff, T.; Bagnard, D.; Banéres, J.L.; Bazzacco, P.; Billon-Denis, E.; Catoire, L.J.; Champeil, P.; Charvolin, D.; Cocco, M.J.; et al. Amphipols from a to Z. Annu. Rev. Biophys. 2011, 40, 379–408. [Google Scholar] [CrossRef] [PubMed]
- Giusti, F.; Popot, J.-L.; Tribet, C. Well-defined critical association concentration and rapid adsorption at the air/water interface of a short amphiphilic polymer, amphipol A8–35: A study by Förster resonance energy transfer and dynamic surface tension measurements. Langmuir 2012, 28, 10372–10380. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Clarke, O.B.; Kim, J.; Stowe, S.; Kim, Y.-K.; Assur, Z.; Cavalier, M.; Godoy-Ruiz, R.; Desiree, C.; Manzini, C.; et al. Structure of the STRA6 receptor for retinol uptake. Science 2016, 353, aad8266. [Google Scholar] [CrossRef] [PubMed]
- Zubcevic, L.; Herzik, M.A.; Chung, B.C.; Liu, Z.; Lander, G.C.; Lee, S.-Y. Cryo-electron microscopy structure of the TRPV2 ion channel. Nat. Struct. Mol. Biol. 2016, 23, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.-C.; Rajendra, E.; Yang, G.; Shi, Y.; Scheres, S.H.W. Sampling the conformational space of the catalytic subunit of human γ-secretase. Elife 2015, 4, e11182. [Google Scholar] [CrossRef] [PubMed]
- Elter, S.; Raschle, T.; Arens, S.; Viegas, A.; Gelev, V.; Etzkorn, M.; Wagner, G. The use of amphipols for NMR structural characterization of 7-TM proteins. J. Membr. Biol. 2014, 247, 957–964. [Google Scholar] [CrossRef] [PubMed]
- Planchard, N.; Point, É.; Dahmane, T.; Giusti, F.; Renault, M.; Le Bon, C.; Durand, G.; Milon, A.; Guittet, É.; Zoonens, M.; et al. The use of amphipols for solution NMR studies of membrane proteins: Advantages and constraints as compared to other solubilizing media. J. Membr. Biol. 2014, 247, 827–842. [Google Scholar] [CrossRef] [PubMed]
- Polovinkin, V.; Gushchin, I.; Sintsov, M.; Round, E.; Balandin, T.; Chervakov, P.; Schevchenko, V.; Utrobin, P.; Popov, A.; Borshchevskiy, V.; et al. High-resolution structure of a membrane protein transferred from amphipol to a lipidic mesophase. J. Membr. Biol. 2014, 247, 997–1004. [Google Scholar] [CrossRef] [PubMed]
- Caffrey, M. A comprehensive review of the lipid cubic phase or in meso method for crystallizing membrane and soluble proteins and complexes. Acta Crystallogr. F Struct. Biol. Commun. 2015, 71, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Denisov, I.G.; Sligar, S.G. Nanodiscs for structural and functional studies of membrane proteins. Nat. Struct. Mol. Biol. 2016, 23, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Matar-Merheb, R.; Rhimi, M.; Leydier, A.; Huché, F.; Galián, C.; Desuzinges-Mandon, E.; Ficheux, D.; Flot, D.; Aghajari, N.; Kahn, R.; et al. Structuring detergents for extracting and stabilizing functional membrane proteins. PLoS ONE 2011, 6, e18036. [Google Scholar] [CrossRef] [PubMed]
- Popot, J.-L. Amphipols, nanodiscs, and fluorinated surfactants: Three nonconventional approaches to studying membrane proteins in aqueous solutions. Annu. Rev. Biochem. 2010, 79, 737–775. [Google Scholar] [CrossRef] [PubMed]
- Deisenhofer, J.; Epp, O.; Miki, K.; Huber, R.; Michel, H. Structure of the protein subunits in the photosynthetic reaction centre of Rhodopseudomonas viridis at 3Å resolution. Nature 1985, 318, 618–624. [Google Scholar] [CrossRef] [PubMed]
- Warschawski, D.E.; Arnold, A.A.; Beaugrand, M.; Gravel, A.; Chartrand, É.; Marcotte, I. Choosing membrane mimetics for NMR structural studies of transmembrane proteins. Biochim. Biophys. Acta 2011, 1808, 1957–1974. [Google Scholar] [CrossRef] [PubMed]
- Rosevear, P.; VanAken, T.; Baxter, J.; Ferguson-Miller, S. Alkyl glycoside detergents: A simpler synthesis and their effects on kinetic and physical properties of cytochrome c oxidase. Biochemistry 1980, 19, 4108–4115. [Google Scholar] [CrossRef] [PubMed]
- Chae, P.S.; Rasmussen, S.G.F.; Rana, R.R.; Gotfryd, K.; Chandra, R.; Goren, M.A.; Kruse, A.C.; Nurva, S.; Loland, C.J.; Pierre, Y.; et al. Maltose–neopentyl glycol (MNG) amphiphiles for solubilization, stabilization and crystallization of membrane proteins. Nat. Methods 2010, 7, 1003–1008. [Google Scholar] [CrossRef] [PubMed]
- Slotboom, D.J.; Duurkens, R.H.; Olieman, K.; Erkens, G.B. Static light scattering to characterize membrane proteins in detergent solution. Methods 2008, 46, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Strop, P.; Brunger, A.T. Refractive index-based determination of detergent concentration and its application to the study of membrane proteins. Protein Sci. 2005, 14, 2207–2211. [Google Scholar] [CrossRef] [PubMed]
- Lorber, B.; Bishop, J.B.; DeLucas, L.J. Purification of octyl beta-D-glucopyranoside and re-estimation of its micellar size. Biochim. Biophys. Acta 1990, 1023, 254–265. [Google Scholar] [CrossRef]
- Sonoda, Y.; Cameron, A.; Newstead, S.; Omote, H.; Moriyama, Y.; Kasahara, M.; Iwata, S.; Drew, D. Tricks of the trade used to accelerate high-resolution structure determination of membrane proteins. FEBS Lett. 2010, 584, 2539–2547. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.L.; Newstead, S. Current trends in α-helical membrane protein crystallization: An update. Protein Sci. 2012, 21, 1358–1365. [Google Scholar] [CrossRef] [PubMed]
- Thiyagarajan, P.; Tiede, D.M. Detergent micelle structure and micelle-micelle interactions determined by small-angle neutron scattering under solution conditions used for membrane protein crystallization. J. Phys. Chem. 1994, 98, 10343–10351. [Google Scholar] [CrossRef]
- Psachoulia, E.; Bond, P.J.; Sansom, M.S.P. MD Simulations of Mistic: Conformational Stability in Detergent Micelles and Water. Biochemistry 2006, 45, 9053–9058. [Google Scholar] [CrossRef] [PubMed]
- Le Maire, M.; Champeil, P.; Moller, J.V. Interaction of membrane proteins and lipids with solubilizing detergents. Biochim. Biophys. Acta 2000, 1508, 86–111. [Google Scholar] [CrossRef]
- Alonso, H.; Roujeinikova, A. Characterization and two-dimensional crystallization of membrane component AlkB of the medium-chain alkane hydroxylase system from Pseudomonas putida GPo1. Appl. Environ. Microbiol. 2012, 78, 7946–7953. [Google Scholar] [CrossRef] [PubMed]
- Mast, R.C.; Haynes, L.V. The use of the fluorescent probes perylene and magnesium 8-anilinonaphthalene-1-sulfonate to determine the critical micelle concentration of surfactants in aqueous solution. J. Colloid Interface Sci. 1975, 53, 35–41. [Google Scholar] [CrossRef]
- Kang, Y.; Zhou, X.E.; Gao, X.; He, Y.; Liu, W.; Ishchenko, A.; Barty, A.; White, T.A.; Yefanov, O.; Han, G.W.; et al. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature 2015, 523, 561–567. [Google Scholar] [CrossRef] [PubMed]
- White, J.F.; Noinaj, N.; Shibata, Y.; Love, J.; Kloss, B.; Xu, F.; Gvozdenovic-Jeremic, J.; Shah, P.; Shiloach, J.; Tate, C.G.; et al. Structure of the agonist-bound neurotensin receptor. Nature 2012, 490, 508–513. [Google Scholar] [CrossRef] [PubMed]
- Hasko Paradies, H. Shape and size of a nonionic surfactant micelle. Triton X-100 in aqueous solution. J. Phys. Chem. 1980, 84, 599–607. [Google Scholar] [CrossRef]
- Streletzky, K.; Phillies, G.D.J. Temperature dependence of triton X-100 micelle size and hydration. Langmuir 1995, 11, 42–47. [Google Scholar] [CrossRef]
- Ashani, Y.; Catravas, G.N. Highly reactive impurities in Triton X-100 and Brij 35: Partial characterization and removal. Anal. Biochem. 1980, 109, 55–62. [Google Scholar] [CrossRef]
- McPherson, A.; Gavira, J.A. Introduction to protein crystallization. Acta Crystallogr. F Struct. Biol. Commun. 2014, 70, 2–20. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.R.; Fan, G.; Serysheva, I.I. Single-particle cryo-EM of the ryanodine receptor channel in an aqueous environment. Eur. J. Transl. Myol. 2015, 25, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Abeyrathne, P.D.; Arheit, M.; Kebbel, F.; Castano-Diez, D.; Goldie, K.N.; Chami, M.; Stahlberg, H.; Renault, L.; Kühlbrandt, W. Chapter 119–1.15 Analysis of 2-D Crystals of Membrane Proteins by Electron Microscopy. In Comprehensive Biophysics; Elsevier: Amsterdam, The Netherlands, 2012; pp. 277–310. [Google Scholar]
- Smith, E.L.; Pickels, E.G. Micelle Formation in Aqueous Solutions of Digitonin. Proc. Natl. Acad. Sci. USA 1940, 26, 272–277. [Google Scholar] [CrossRef] [PubMed]
- Chae, P.S.; Rasmussen, S.G.F.; Rana, R.R.; Gotfryd, K.; Kruse, A.C.; Manglik, A.; Cho, K.H.; Nurva, S.; Gether, U.; Guan, L.; et al. A new class of amphiphiles bearing rigid hydrophobic groups for solubilization and stabilization of membrane proteins. Chemistry 2012, 18, 9485–9490. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chen, J. Atomic Structure of the Cystic Fibrosis Transmembrane Conductance Regulator. Cell 2016, 167, 1586–1597. [Google Scholar] [CrossRef] [PubMed]
- Letts, J.A.; Fiedorczuk, K.; Sazanov, L.A. The architecture of respiratory supercomplexes. Nature 2016, 537, 644–648. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, T.C.; Engel, A.; Rémigy, H.-W. A novel method for detergent concentration determination. Biophys. J. 2006, 90, 310–317. [Google Scholar] [CrossRef] [PubMed]
- D’Avanzo, N.; McCusker, E.C.; Powl, A.M.; Miles, A.J.; Nichols, C.G.; Wallace, B.A. Differential lipid dependence of the function of bacterial sodium channels. PLoS ONE 2013, 8, e61216. [Google Scholar] [CrossRef] [PubMed]
- Zana, R.; Weill, C. Effect of temperature on the aggregation behaviour of nonionic surfactants in aqueous solutions. J. Phys. Lett. 1985, 46, 953–960. [Google Scholar] [CrossRef]
- Poulos, S.; Morgan, J.L.W.; Zimmer, J.; Faham, S. Bicelles coming of age: An empirical approach to bicelle crystallization. Methods Enzymol. 2015, 557, 393–416. [Google Scholar] [PubMed]
- Breyton, C.; Chabaud, E.; Chaudier, Y.; Pucci, B.; Popot, J.-L. Hemifluorinated surfactants: A non-dissociating environment for handling membrane proteins in aqueous solutions? FEBS Lett. 2004, 564, 312–318. [Google Scholar] [CrossRef]
- Efremov, R.G.; Leitner, A.; Aebersold, R.; Raunser, S. Architecture and conformational switch mechanism of the ryanodine receptor. Nature 2015, 517, 39–43. [Google Scholar] [CrossRef] [PubMed]
- Nasr, M.L.; Baptista, D.; Strauss, M.; Sun, Z.-Y.J.; Grigoriu, S.; Huser, S.; Plückthun, A.; Hagn, F.; Walz, T.; Hogle, J.M.; et al. Covalently circularized nanodiscs for studying membrane proteins and viral entry. Nat. Methods 2017, 14, 49–52. [Google Scholar] [CrossRef] [PubMed]
- Bae, H.E.; Mortensen, J.S.; Ribeiro, O.; Du, Y.; Ehsan, M.; Kobilka, B.K.; Loland, C.J.; Byrne, B.; Chae, P.S. Tandem neopentyl glycol maltosides (TNMs) for membrane protein stabilisation. Chem. Commun. 2016, 52, 12104–12107. [Google Scholar] [CrossRef] [PubMed]
- Von Heijne, G. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 1992, 225, 487–494. [Google Scholar] [CrossRef]
- Zhang, Q.; Ma, X.; Ward, A.; Hong, W.-X.; Jaakola, V.-P.; Stevens, R.C.; Finn, M.G.; Chang, G. Designing facial amphiphiles for the stabilization of integral membrane proteins. Angew. Chem. Int. Ed. Engl. 2007, 46, 7023–7025. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Bennett, B.C.; Hong, W.-X.; Fu, Y.; Baker, K.A.; Marcoux, J.; Robinson, C.V.; Ward, A.B.; Halpert, J.R.; Stevens, R.C.; et al. Steroid-based facial amphiphiles for stabilization and crystallization of membrane proteins. Proc. Natl. Acad. Sci. USA 2013, 110, E1203–E1211. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Szewczyk, P.; Karyakin, A.; Evin, M.; Hong, W.-X.; Zhang, Q.; Chang, G. Structure of a cation-bound multidrug and toxic compound extrusion transporter. Nature 2010, 467, 991–994. [Google Scholar] [CrossRef] [PubMed]
- Chae, P.S.; Gotfryd, K.; Pacyna, J.; Miercke, L.J.W.; Rasmussen, S.G.F.; Robbins, R.A.; Rana, R.R.; Loland, C.J.; Kobilka, B.; Stroud, R.; et al. Tandem facial amphiphiles for membrane protein stabilization. J. Am. Chem. Soc. 2010, 132, 16750–16752. [Google Scholar] [CrossRef] [PubMed]
- Drew, D.; Lerch, M.; Kunji, E.; Slotboom, D.J.; de Gier, J.-W. Optimization of membrane protein overexpression and purification using GFP fusions. Nat. Methods 2006, 3, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Kawate, T.; Gouaux, E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 2006, 14, 673–681. [Google Scholar] [CrossRef] [PubMed]
- Desuzinges-Mandon, E.; Agez, M.; Pellegrin, R.; Igonet, S.; Jawhari, A. Novel systematic detergent screening method for membrane proteins solubilization. Anal. Biochem. 2017, 517, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Lantez, V.; Nikolaidis, I.; Rechenmann, M.; Vernet, T.; Noirclerc-Savoye, M. Rapid automated detergent screening for the solubilization and purification of membrane proteins and complexes. Eng. Life Sci. 2015, 15, 39–50. [Google Scholar] [CrossRef]

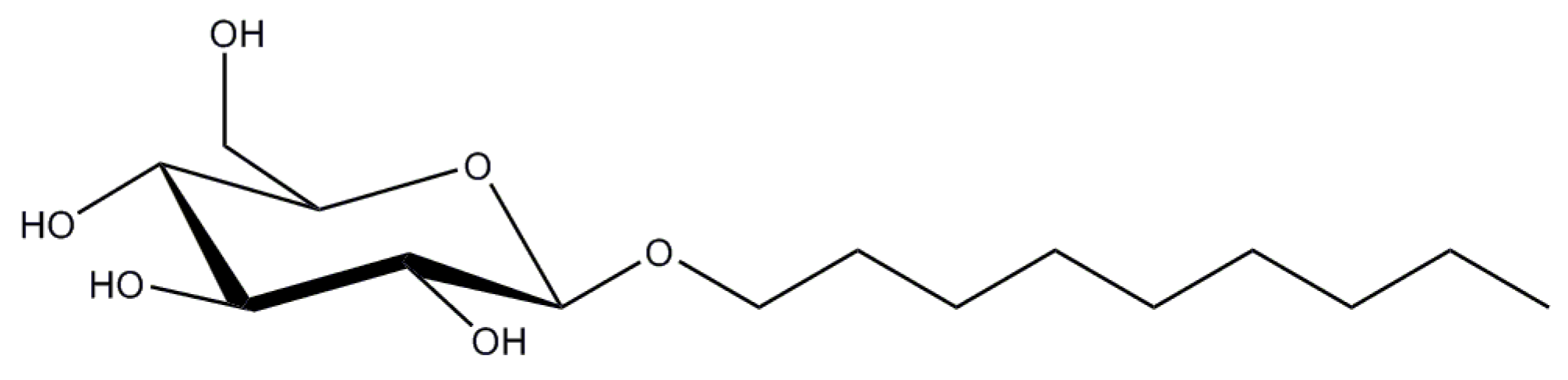
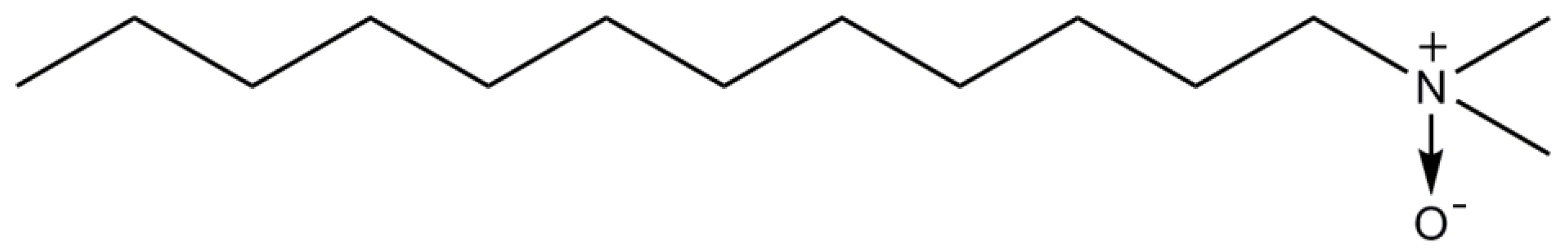
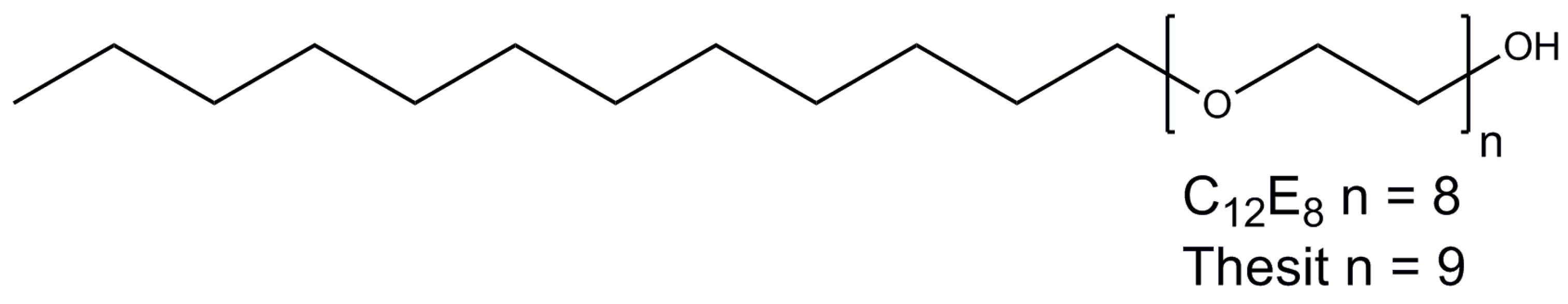
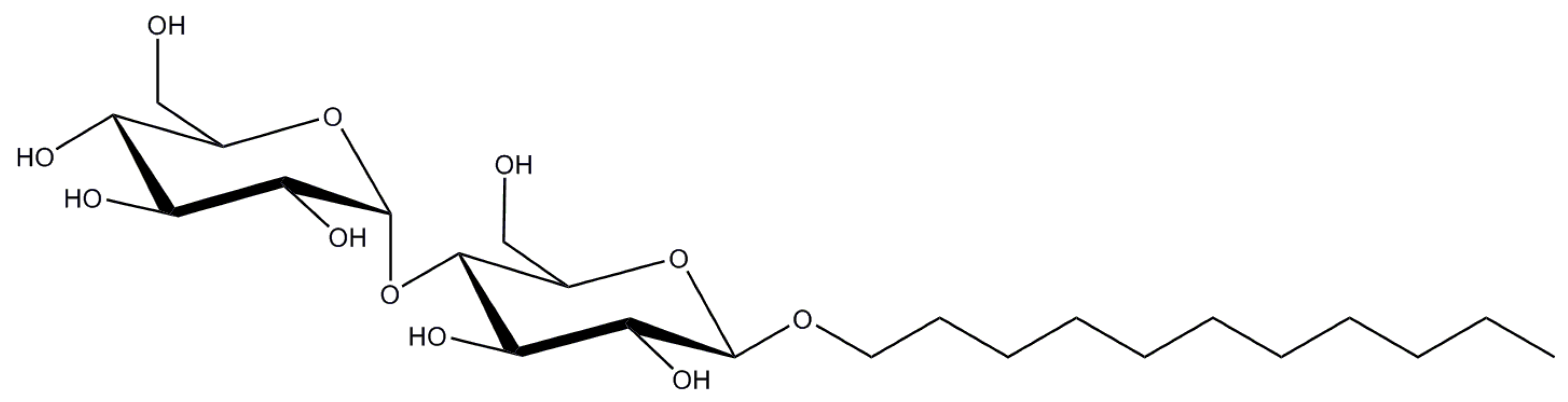

| Detergent | CMC (%/mM) | Aggregation Number, N 1 | Micelle Size Mw (kDa) | Mw (Da) |
|---|---|---|---|---|
| DDM | 0.0087/0.17 | 80–150 | 65–70 | 510.6 |
| DM | 0.087/1.8 | 69 | 40 | 482.6 |
| OG | 0.53/20 | 30–100 | 25 | 292.4 |
| NG | 0.20/6.5 | 133 | 85 | 306.4 |
| LDAO | 0.023/1-2 | 76 | 21.5 | 229.4 |
| C12E8 | 0.005/0.09 | 90–120 | 66 | 538.7 |
| C12E9 | 0.003/0.05 | 90 2 | 83 | 582.8 |
| UDM | 0.029/0.59 | 71 | 50 | 496.6 |
| LMNG | 0.001/0.01 | ~400 3 | 91–393 | 1069.2 |
| Triton X-100 | 0.01/0.2 | 75–165 | 60–90 | 624.8 (av.) |
| Digitonin | 0.002/0.5 | 60 4 | 70 | 1229.3 |
| Cymal-5 | 0.12/2.5 | 47 | 23 | 494.6 |
| Cymal-6 | 0.028/0.56 | 91 | 32 | 508.6 |
| CHAPS | 0.5/8–10 | 10 | 6 | 614.9 |
| CHAPSO | 0.5/8–10 | 11 | 7 | 630.9 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stetsenko, A.; Guskov, A. An Overview of the Top Ten Detergents Used for Membrane Protein Crystallization. Crystals 2017, 7, 197. https://doi.org/10.3390/cryst7070197
Stetsenko A, Guskov A. An Overview of the Top Ten Detergents Used for Membrane Protein Crystallization. Crystals. 2017; 7(7):197. https://doi.org/10.3390/cryst7070197
Chicago/Turabian StyleStetsenko, Artem, and Albert Guskov. 2017. "An Overview of the Top Ten Detergents Used for Membrane Protein Crystallization" Crystals 7, no. 7: 197. https://doi.org/10.3390/cryst7070197






