Aggregation of Cationic Amphiphilic Block and Random Copoly(vinyl ether)s with Antimicrobial Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis of Amphiphilic Copolymers
2.3. Dye Uptake Experiment
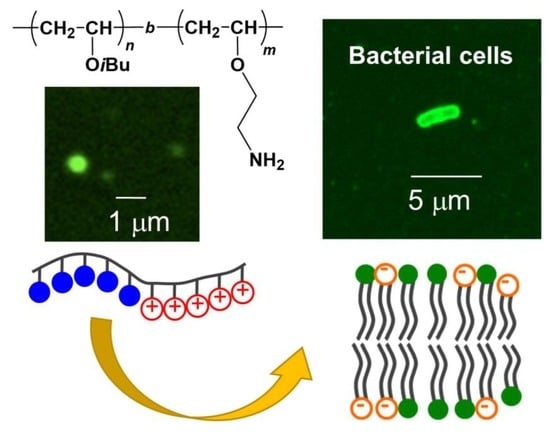
2.4. Fluorescence Microscopic Observation
2.5. Cryo-TEM Observation
3. Results and Discussion
3.1. Polymer Design, Synthesis, and Antimicrobial Activity
3.2. Dye Uptakes by Copolymers
3.3. Cryo-TEM Observations of the Block Copolymer Aggregates
3.4. Fluorescnt Study of Block Copolymer Aggregates
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Fischbach, M.A.; Walsh, C.T. Antibiotics for emerging pathogens. Science 2009, 325, 1089–1093. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, P. Antibacterial discovery and development—The failure of success? Nat. Biotechnol. 2006, 24, 1497–1503. [Google Scholar] [CrossRef] [PubMed]
- Levy, S.B. The Antibiotic Paradox. How Miracle Drugs Are Destroying the Miracle; Springer: New York, NY, USA, 1992; ISBN 978-1-4899-6042-9. [Google Scholar] [CrossRef]
- Hancock, R.E.W.; Lehrer, R. Cationic peptides: A new source of antibiotics. Trends Biotechnol. 1998, 16, 82–88. [Google Scholar] [CrossRef]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Caputo, G.A.; Vemparala, S.; Kuroda, K. Synthetic random copolymers as a molecular platform to mimic host-defense antimicrobial peptides. Bioconj. Chem. 2017, 28, 1340–1350. [Google Scholar] [CrossRef] [PubMed]
- Tew, G.N.; Scott, R.W.; Klein, M.L.; DeGrado, W.F. De novo design of antimicrobial polymers, foldamers, and small molecules: From discovery to practical applications. Acc. Chem. Res. 2010, 43, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Ilker, M.F.; Nüesslein, K.; Tew, G.N.; Coughlin, E.B. Tuning the hemolytic and antibacterial activities of amphiphilic polynorbornene derivatives. J. Am. Chem. Soc. 2004, 126, 15870–15875. [Google Scholar] [CrossRef] [PubMed]
- Mowery, B.P.; Lee, S.E.; Kissounko, D.A.; Epand, R.F.; Epand, R.M.; Weisblum, B.; Stahl, S.S.; Gellman, S.H. Mimicry of antimicrobial host-defense peptides by random copolymers. J. Am. Chem. Soc. 2007, 129, 15474–15476. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, K.; Caputo, G.A.; Degradol, W.F. The role of hydrophobicity in the antimicrobial and hemolytic activities of polymethacrylate derivatives. Chem. Eur. J. 2009, 15, 1123–1133. [Google Scholar] [CrossRef] [PubMed]
- Mowery, B.P.; Lindner, A.H.; Weisblum, B.; Stahl, S.S.; Gellman, S.H. Structure-activity relationships among random nylon-3 copolymers that mimic antibacterial host-defense peptides. J. Am. Chem. Soc. 2009, 131, 9735–9745. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, K.; DeGrado, W.F. Amphiphilic polymethacrylate derivatives as antimicrobial agents. J. Am. Chem. Soc. 2005, 127, 4128–4129. [Google Scholar] [CrossRef] [PubMed]
- Sovadinova, I.P.; Palermo, E.F.; Urban, M.; Mpiga, P.; Caputo, G.A.; Kuroda, K. Activity and mechanism of antimicrobial peptide-mimetic amphiphilic polymethacrylate derivatives. Polymers 2011, 3, 1512–1532. [Google Scholar] [CrossRef]
- Palermo, E.F.; Lee, D.K.; Ramamoorthy, A.; Kuroda, K. Role of cationic group structure in membrane binding and disruption by amphiphilic copolymers. J. Phys. Chem. B 2011, 115, 366–375. [Google Scholar] [CrossRef] [PubMed]
- Oda, Y.; Kanaoka, S.; Sato, T.; Aoshima, S.; Kuroda, K. Block versus random amphiphilic copolymers as antibacterials agents. Biomacromolecules 2011, 12, 3581–3591. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Zhou, X.; Tan, Z.; Zhou, C. Highly efficient antibacterial diblock copolypeptides based on lysine and phenylalanine. Biopolymers 2017, 107, e23041. [Google Scholar] [CrossRef] [PubMed]
- Venkataraman, S.; Tan, J.P.K.; Ng, V.W.L.; Tan, E.W.P.; Hedrick, J.L.; Yang, Y.Y. Amphiphilic and hydrophilic block copolymers from aliphatic N-substituted 8-membered cyclic carbonates: A versatile macromolecular platform for biomedical applications. Biomacromolecules 2017, 18, 178–188. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Matsuda, Y. Macromolecular assemblies in solution: Characterization by light scattering. Polym. J. 2009, 41, 241–251. [Google Scholar] [CrossRef]
- Nakashima, K.; Bahadur, P. Aggregation of water-soluble block copolymers in aqueous solutions: Recent trends. Adv. Colloid Interface Sci. 2006, 123–126, 75–96. [Google Scholar] [CrossRef] [PubMed]
- Oda, Y.; Kanaoka, S.; Aoshima, S. Synthesis of dual pH/temperature-responsive polymers with amino groups by living cationic polymerization. J. Polym. Sci. Part A Polym. Sci. 2010, 48, 1207–1213. [Google Scholar] [CrossRef]
- Chattopadhyay, A.; London, E. Fluorimetric determination of critical micelle concentration avoiding interference from detergent charge. Anal. Biochem. 1984, 139, 408–412. [Google Scholar] [CrossRef]
- Szczubia-lka, K.; Ishikawa, K.; Morishima, Y. Associating behavior of sulfonated polyisoprene block copolymers with short polystyrene blocks at both chain ends. Langmuir 2000, 16, 2083–2092. [Google Scholar] [CrossRef]
- Sugihara, S.; Hashimoto, K.; Okabe, S.; Shibayama, M.; Kanaoka, S.; Aoshima, S. Stimuli-responsive diblock copolymers by living cationic polymerization: Precision synthesis and highly sensitive physical gelation. Macromolecules 2004, 37, 336–343. [Google Scholar] [CrossRef]
- Takahashi, R.; Sato, T.; Terao, K.; Qiu, X.-P.; Winnik, F.M. Self-association of a thermosensitive poly(alkyl-2-oxazoline) block copolymer in aqueous solution. Macromolecules 2012, 45, 6111–6119. [Google Scholar] [CrossRef]
- Sato, T.; Tanaka, K.; Toyokura, A.; Mori, R.; Takahashi, R.; Terao, K.; Yusa, S. Self-association of a thermosensitive amphiphilic block copolymer poly(N-isopropylacrylamide)-b-poly(N-vinyl-2-pyrrolidone) in aqueous solution upon heating. Macromolecules 2013, 46, 226–235. [Google Scholar] [CrossRef]
- Takahashi, R.; Qiu, X.-P.; Xue, N.; Sato, T.; Terao, K.; Winnik, F.M. Self-association of the thermosensitive block copolymer poly(2-isopropyl-2-oxazoline)-b-poly(N-isopropylacrylamide) in water-methanol mixtures. Macromolecules 2014, 47, 6900–6910. [Google Scholar] [CrossRef]
- Sato, T.; Takahashi, R. Competition between the micellization and the liquid-liquid phase separation in amphiphilic block copolymer solutions. Polym. J. 2017, 49, 273–277. [Google Scholar] [CrossRef]
- Krishan, A. Rapid flow cytofluorometric analysis of mammalian cell cycle by propidium iodide staining. J. Cell Biol. 1975, 66, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Xu, K.; Wang, H.; Tan, P.K.J.; Fan, W.; Venkatraman, S.S.; Li, L.; Yang, Y.Y. Self-assembled cationic peptide nanoparticles as an efficient antimicrobial agent. Nat. Nanotechnol. 2009, 4, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, I.; Vemparala, S.; Pophristic, V.; Kuroda, K.; DeGrado, W.F.; McCammon, J.A.; Klein, M.L. Characterization of nonbiological antimicrobial polymers in aqueous solution and at water-lipid interfaces from all-atom molecular dynamics. J. Am. Chem. Soc. 2006, 128, 1778–1779. [Google Scholar] [CrossRef] [PubMed]
- Song, A.; Walker, S.G.; Parker, K.A.; Sampson, N.S. Antibacterial studies of cationic polymers with alternating, random, and uniform backbones. ACS Chem. Biol. 2011, 6, 590–599. [Google Scholar] [CrossRef] [PubMed]
- Totani, M.; Ando, T.; Terada, K.; Terashima, T.; Kim, I.Y.; Ohtsuki, C.; Xi, C.; Kuroda, K.; Tanihara, M. Utilization of star-shaped polymer architecture in the creation of high-density polymer brush coatings for the prevention of platelet and bacteria adhesion. Biomater. Sci. 2014, 2, 1172–1185. [Google Scholar] [CrossRef] [PubMed]





| Polymer | Copolymer Structure | DP 1 | MPIBVE 1 (mol %) | BC99.9 2 (μg/mL) | HC50 (μg/mL) | CDPH 4 (μg/mL) | CAC 5 (μg/mL) | RH 6, Rg 7 (nm) |
|---|---|---|---|---|---|---|---|---|
| H44 | Homopolymer | 44 | 0 | 1.6 ± 0.0 | >1000 (42.5 ± 6.3%) 3 | 90 | N.D. | N.D. |
| B3826 | Block copolymer | 38 | 26 | 2.4 ± 0.91 | >1000 (37.7 ± 2.8%) 3 | 124 | 36 | 250 6 |
| R4025 | Random copolymer | 40 | 25 | 1.6 ± 0.0 | 0.49 ± 0.17 | 125 | 380 | 27 7 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oda, Y.; Yasuhara, K.; Kanaoka, S.; Sato, T.; Aoshima, S.; Kuroda, K. Aggregation of Cationic Amphiphilic Block and Random Copoly(vinyl ether)s with Antimicrobial Activity. Polymers 2018, 10, 93. https://doi.org/10.3390/polym10010093
Oda Y, Yasuhara K, Kanaoka S, Sato T, Aoshima S, Kuroda K. Aggregation of Cationic Amphiphilic Block and Random Copoly(vinyl ether)s with Antimicrobial Activity. Polymers. 2018; 10(1):93. https://doi.org/10.3390/polym10010093
Chicago/Turabian StyleOda, Yukari, Kazuma Yasuhara, Shokyoku Kanaoka, Takahiro Sato, Sadahito Aoshima, and Kenichi Kuroda. 2018. "Aggregation of Cationic Amphiphilic Block and Random Copoly(vinyl ether)s with Antimicrobial Activity" Polymers 10, no. 1: 93. https://doi.org/10.3390/polym10010093