Molecular Weight-Dependent Activity of Aminated Poly(α)glutamates as siRNA Nanocarriers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis of PGAamine Polymers
2.2.1. Preparation of Poly-α-glutamic Acid
2.2.2. Preparation of PGAamine Polymers
2.3. Physico-Chemical Characterization of PGAamine Polymers and the Obtained Polyplexes
2.3.1. 1H-Nuclear Magnetic Resonance (NMR)
2.3.2. Multi Angle Static Light Scattering (MALS)
2.3.3. Electrophoretic Mobility Shift Assay (EMSA)
2.3.4. Zeta Potential Determination
2.3.5. Dynamic Light Scattering (DLS)
2.3.6. Transmission Electron Microscopy (TEM)
2.3.7. Isothermal Titration Calorimetry (ITC)
2.4. In Vitro Evaluation of Polyplexes
2.4.1. Cell Culture
2.4.2. In Vitro Silencing Efficacy
2.4.3. Cell Viability Assay
2.4.4. Western Blot Analysis
2.4.5. Wound Healing Assay
2.4.6. Intracellular Trafficking Study
2.5. Molecular Dynamics (MD) Simulation
2.6. Biostability and Toxicity Assays
2.6.1. Plasma Stability
2.6.2. Heparin Displacement Assay
2.6.3. Red Blood Cell Lysis
2.7. Statistical Analysis
3. Results
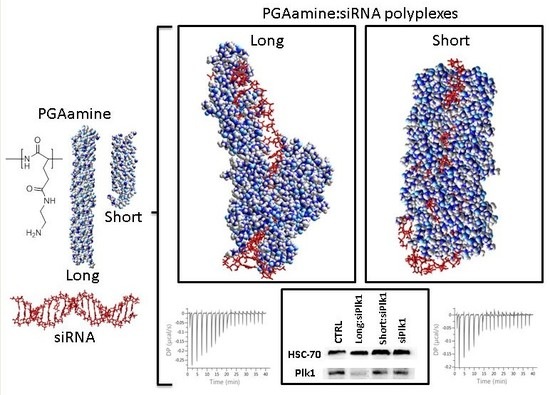
3.1. Synthesis and Characterization of Long and Short Aminated Poly(α)glutamates
3.2. Physico-Chemical Characterization of the Polyplexes Formed by Electrostatic-Based Complexation of Long or Short PGAamine with siRNA
3.3. Isothermal Titration Calorimetry (ITC)
3.4. Long PGAamine:siRNA Polyplexes Demonstrated Silencing Activity and Anticancer Efficacy in HeLa Cells
3.5. Long and Short PGAamine Polyplexes Demonstrated Similar Colocalization Ratios with Endocytic Markers
3.6. Computerized Model of Long and Short PGAamine Binding with siRNA
3.7. The Biological Stability and Toxicity of Long and Short PGAamine:siRNA Polyplexes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.Y.; Lopez-Berestein, G.; Calin, G.A.; Sood, A.K. RNAi therapies: Drugging the undruggable. Sci. Transl. Med. 2014, 6, 240ps7. [Google Scholar] [CrossRef] [PubMed]
- Scomparin, A.; Polyak, D.; Krivitsky, A.; Satchi-Fainaro, R. Achieving successful delivery of oligonucleotides—From physico-chemical characterization to in vivo evaluation. Biotechnol. Adv. 2015, 33, 1294–1309. [Google Scholar] [CrossRef] [PubMed]
- Tiram, G.; Scomparin, A.; Ofek, P.; Satchi-Fainaro, R. Interfering cancer with polymeric siRNA nanomedicines. J. Biomed. Nanotechnol. 2014, 10, 50–66. [Google Scholar] [CrossRef] [PubMed]
- Ben-Shushan, D.; Markovsky, E.; Gibori, H.; Tiram, G.; Scomparin, A.; Satchi-Fainaro, R. Overcoming obstacles in microRNA delivery towards improved cancer therapy. Drug Deliv. Transl. Res. 2014, 4, 38–49. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.; Wu, J.; Sawa, T.; Matsumura, Y.; Hori, K. Tumor vascular permeability and the EPR effect in macromolecular therapeutics: A review. J. Controll. Release 2000, 65, 271–284. [Google Scholar] [CrossRef]
- Matsumura, Y.; Maeda, H. A new concept for macromolecular therapeutics in cancer chemotherapy: Mechanism of tumoritropic accumulation of proteins and the antitumor agent smancs. Cancer Res. 1986, 46, 6387–6392. [Google Scholar] [PubMed]
- Davis, M.E.; Zuckerman, J.E.; Choi, C.H.; Seligson, D.; Tolcher, A.; Alabi, C.A.; Yen, Y.; Heidel, J.D.; Ribas, A. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature 2010, 464, 1067–1070. [Google Scholar] [CrossRef] [PubMed]
- Zuckerman, J.E.; Gritli, I.; Tolcher, A.; Heidel, J.D.; Lim, D.; Morgan, R.; Chmielowski, B.; Ribas, A.; Davis, M.E.; Yen, Y. Correlating animal and human phase Ia/Ib clinical data with CALAA-01, a targeted, polymer-based nanoparticle containing siRNA. Proc. Natl. Acad. Sci. USA 2014, 111, 11449–11454. [Google Scholar] [CrossRef] [PubMed]
- Tabernero, J.; Shapiro, G.I.; LoRusso, P.M.; Cervantes, A.; Schwartz, G.K.; Weiss, G.J.; Paz-Ares, L.; Cho, D.C.; Infante, J.R.; Alsina, M.; et al. First-in-humans trial of an RNA interference therapeutic targeting VEGF and KSP in Cancer Patients with liver involvement. Cancer Discov. 2013, 3, 406–417. [Google Scholar] [CrossRef] [PubMed]
- Rossi, J.J. RNAi therapeutics: Snalping siRNAs in vivo. Gene Ther. 2006, 13, 583–584. [Google Scholar] [CrossRef] [PubMed]
- Tolcher, A.W.; Rodrigueza, W.V.; Rasco, D.W.; Patnaik, A.; Papadopoulos, K.P.; Amaya, A.; Moore, T.D.; Gaylor, S.K.; Bisgaier, C.L.; Sooch, M.P.; et al. A phase 1 study of the BCL2-targeted deoxyribonucleic acid inhibitor (DNAi) PNT2258 in patients with advanced solid tumors. Cancer Chemother. Pharmacol. 2014, 73, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Haag, R.; Kratz, F. Polymer therapeutics: Concepts and applications. Angew. Chem. Int. Ed. 2006, 45, 1198–1215. [Google Scholar] [CrossRef] [PubMed]
- Duncan, R. The dawning era of polymer therapeutics. Nat. Rev. Drug Discov. 2003, 2, 347–360. [Google Scholar] [CrossRef] [PubMed]
- Markovsky, E.; Baabur-Cohen, H.; Eldar-Boock, A.; Omer, L.; Tiram, G.; Ferber, S.; Ofek, P.; Polyak, D.; Scomparin, A.; Satchi-Fainaro, R. Administration, distribution, metabolism and elimination of polymer therapeutics. J. Controll. Release 2012, 161, 446–460. [Google Scholar] [CrossRef] [PubMed]
- Gragoudas, E.S.; Adamis, A.P.; Cunningham, E.T., Jr.; Feinsod, M.; Guyer, D.R. Pegaptanib for neovascular age-related macular degeneration. N. Engl. J. Med. 2004, 351, 2805–2816. [Google Scholar] [CrossRef] [PubMed]
- Marcinow, A.M.; Hall, N.; Byrum, A.; Teknos, T.N.; Old, M.O.; Agrawal, A. Use of a novel receptor-targeted (CD206) radiotracer, 99MTC-tilmanocept, and SPECT/CT for sentinel lymph node detection in oral cavity squamous cell carcinoma. JAMA Otolaryngol. Head Neck Surg. 2013, 139, 895–902. [Google Scholar] [CrossRef] [PubMed]
- Boussif, O.; Lezoualc’h, F.; Zanta, M.A.; Mergny, M.D.; Scherman, D.; Demeneix, B.; Behr, J.P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: Polyethylenimine. Proc. Natl. Acad. Sci. USA 1995, 92, 7297–7301. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, C.; Mennesson, E.; Fuchs, R.; Gorvel, J.P.; Midoux, P.; Pichon, C. Macropinocytosis of polyplexes and recycling of plasmid via the clathrin-dependent pathway impair the transfection efficiency of human hepatocarcinoma cells. Mol. Ther. 2004, 10, 373–385. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Cheng, J. Hexamethyldisilazane-mediated controlled polymerization of α-amino acid n-carboxyanhydrides. J. Am. Chem. Soc. 2007, 129, 14114–14115. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Deming, T.J. Synthesis of polypeptides by ring-opening polymerization of α-amino acid n-carboxyanhydrides. In Peptide-Based Materials; Springer: Berlin/Heidelberg, Germany, 2011; pp. 1–26. [Google Scholar]
- Bertin, A. Polyelectrolyte complexes of DNA and polycations as gene delivery vectors. In Polyelectrolyte Complexes in the Disperesed and Solid State II; Müller, M., Ed.; Springer: Berlin/Heidelberg, Germany, 2014; Volume 256, pp. 103–195. [Google Scholar]
- Honary, S.; Zahir, F. Effect of zeta potential on the properties of nano-drug delivery systems—A review (part 2). Trop. J. Pharm. Res. 2013, 12, 265–273. [Google Scholar]
- Krivitsky, A.; Polyak, D.; Scomparin, A.; Eliyahu, S.; Ofek, P.; Tiram, G.; Kalinski, H.; Avkin-Nachum, S.; Feiner Gracia, N.; Albertazzi, L.; et al. Amphiphilic poly(alpha)glutamate polymeric micelles for systemic administration of siRNA to tumors. Nanomedicine 2018, 14, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Gibori, H.; Eliyahu, S.; Krivitsky, A.; Ben-Shushan, D.; Epshtein, Y.; Tiram, G.; Blau, R.; Ofek, P.; Lee, J.S.; Ruppin, E.; et al. Amphiphilic nanocarrier-induced modulation of PLK1 and miR-34a leads to improved therapeutic response in pancreatic cancer. Nat. Commun. 2018, 9, 16. [Google Scholar] [CrossRef] [PubMed]
- Krivitsky, A.; Polyak, D.; Scomparin, A.; Eliyahu, S.; Ori, A.; Avkin-Nachum, S.; Krivitsky, V.; Satchi-Fainaro, R. Structure-function correlation of aminated poly(α)glutamate as siRNA nanocarriers. Biomacromolecules 2016, 17, 2787–2800. [Google Scholar] [CrossRef] [PubMed]
- Polyak, D.; Krivitsky, A.; Scomparin, A.; Eliyahu, S.; Kalinski, H.; Avkin-Nachum, S.; Satchi-Fainaro, R. Systemic delivery of siRNA by aminated poly(α)glutamate for the treatment of solid tumors. J. Controll. Release 2017, 257, 132–143. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, S.M.; Raines, R.T. Internalization of cationic peptides: The road less (or more?) traveled. Cell. Mol. Life Sci. CMLS 2006, 63, 1819–1822. [Google Scholar] [CrossRef] [PubMed]
- McLendon, P.M.; Buckwalter, D.J.; Davis, E.M.; Reineke, T.M. Interaction of poly(glycoamidoamine) DNA delivery vehicles with cell-surface glycosaminoglycans leads to polyplex internalization in a manner not solely dependent on charge. Mol. Pharm. 2010, 7, 1757–1768. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, A.; Seelig, J. High affinity of the cell-penetrating peptide HIV-1 TAT-PTD for DNA. Biochemistry 2007, 46, 8138–8145. [Google Scholar] [CrossRef] [PubMed]
- Chou, S.T.; Hom, K.; Zhang, D.; Leng, Q.; Tricoli, L.J.; Hustedt, J.M.; Lee, A.; Shapiro, M.J.; Seog, J.; Kahn, J.D.; et al. Enhanced silencing and stabilization of siRNA polyplexes by histidine-mediated hydrogen bonds. Biomaterials 2014, 35, 846–855. [Google Scholar] [CrossRef] [PubMed]
- Conejos-Sanchez, I.; Duro-Castano, A.; Birke, A.; Barz, M.; Vicent, M.J. A controlled and versatile nca polymerization method for the synthesis of polypeptides. Polym. Chem. 2013, 4, 3182–3186. [Google Scholar] [CrossRef]
- Kim, D.H.; Rossi, J.J. Strategies for silencing human disease using RNA interference. Nat. Rev. Genet. 2007, 8, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Dykxhoorn, D.M.; Palliser, D.; Lieberman, J. The silent treatment: SiRNAs as small molecule drugs. Gene Ther. 2006, 13, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Tang, C.; Yin, C. Effect of binding affinity for siRNA on the in vivo antitumor efficacy of polyplexes. Biomaterials 2013, 34, 5317–5327. [Google Scholar] [CrossRef] [PubMed]
- Salmaso, S.; Bersani, S.; Scomparin, A.; Balasso, A.; Brazzale, C.; Barattin, M.; Caliceti, P. A novel soluble supramolecular system for sustained rh-Gh delivery. J. Controll. Release 2014, 194, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Holzerny, P.; Ajdini, B.; Heusermann, W.; Bruno, K.; Schuleit, M.; Meinel, L.; Keller, M. Biophysical properties of chitosan/siRNA polyplexes: Profiling the polymer/siRNA interactions and bioactivity. J. Controll. Release 2012, 157, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Mertins, O.; Dimova, R. Binding of chitosan to phospholipid vesicles studied with isothermal titration calorimetry. Langmuir 2011, 27, 5506–5515. [Google Scholar] [CrossRef] [PubMed]
- Prevette, L.E.; Kodger, T.E.; Reineke, T.M.; Lynch, M.L. Deciphering the role of hydrogen bonding in enhancing pDNA-polycation interactions. Langmuir 2007, 23, 9773–9784. [Google Scholar] [CrossRef] [PubMed]
- Utsuno, K.; Uludag, H. Thermodynamics of polyethylenimine-DNA binding and DNA condensation. Biophys. J. 2010, 99, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Sheikhi Mehrabadi, F.; Hirsch, O.; Zeisig, R.; Posocco, P.; Laurini, E.; Pricl, S.; Haag, R.; Kemmner, W.; Calderón, M. Structure-activity relationship study of dendritic polyglycerolamines for efficient siRNA transfection. RSC Adv. 2015, 5, 78760–78770. [Google Scholar] [CrossRef]
- Karatasos, K.; Posocco, P.; Laurini, E.; Pricl, S. Poly(amidoamine)-based dendrimer/siRNA complexation studied by computer simulations: Effects of pH and generation on dendrimer structure and siRNA binding. Macromol. Biosci. 2012, 12, 225–240. [Google Scholar] [CrossRef] [PubMed]
- Ofek, P.; Fischer, W.; Calderon, M.; Haag, R.; Satchi-Fainaro, R. In vivo delivery of small interfering RNA to tumors and their vasculature by novel dendritic nanocarriers. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2010, 24, 3122–3134. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Crawford, M.; Yu, B.; Mao, Y.; Nana-Sinkam, S.P.; Lee, L.J. MicroRNA delivery by cationic lipoplexes for lung cancer therapy. Mol. Pharm. 2011, 8, 1381–1389. [Google Scholar] [CrossRef] [PubMed]
- McCaskill, J.; Singhania, R.; Burgess, M.; Allavena, R.; Wu, S.; Blumenthal, A.; McMillan, N.A. Efficient biodistribution and gene silencing in the lung epithelium via intravenous liposomal delivery of siRNA. Mol. Ther. Nucl. Acids 2013, 2, e96. [Google Scholar] [CrossRef] [PubMed]
- Fehring, V.; Schaeper, U.; Ahrens, K.; Santel, A.; Keil, O.; Eisermann, M.; Giese, K.; Kaufmann, J. Delivery of therapeutic siRNA to the lung endothelium via novel lipoplex formulation DACC. Mol. Ther. 2014, 22, 811–820. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Gong, L.; Li, W.; Chen, L. Overexpression of Plk1 promotes malignant progress in human esophageal squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 2010, 136, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Han, D.P.; Cui, J.T.; Lu, A.G.; Chen, X.H.; Feng, B.; Zong, Y.P.; Qu, S.; Cao, Q.F.; Zheng, M.H. [Influence of silencing polo-like kinase 1 on migration and invasion of colorectal cancer cells]. Zhonghua Wei Chang Wai Ke Za Zhi 2011, 14, 61–64. [Google Scholar] [PubMed]
- Zhang, Z.; Zhang, G.; Gao, Z.; Li, S.; Li, Z.; Bi, J.; Liu, X.; Kong, C. Comprehensive analysis of differentially expressed genes associated with Plk1 in bladder cancer. BMC Cancer 2017, 17, 861. [Google Scholar] [CrossRef] [PubMed]
- Strebhardt, K.; Ullrich, A. Targeting polo-like kinase 1 for cancer therapy. Nat. Rev. Cancer 2006, 6, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Sun, Q.; Wang, X. Plk1, a potential target for cancer therapy. Transl. Oncol. 2017, 10, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Satchi-Fainaro, R.; Duncan, R.; Barnes, C.M. Polymer therapeutics for cancer: Current status and future challenges. In Polymer Therapeutics Ii: Polymers as Drugs, Conjugates and Gene Delivery Systems; Satchi-Fainaro, R., Duncan, R., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; Volume 193, pp. 1–65. [Google Scholar]
- Lazebnik, M.; Keswani, R.K.; Pack, D.W. Endocytic transport of polyplex and lipoplex siRNA vectors in HeLa cells. Pharm. Res. 2016, 33, 2999–3011. [Google Scholar] [CrossRef] [PubMed]
- Xiang, S.; Tong, H.; Shi, Q.; Fernandes, J.C.; Jin, T.; Dai, K.; Zhang, X. Uptake mechanisms of non-viral gene delivery. J. Controll. Release 2012, 158, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Von Gersdorff, K.; Sanders, N.N.; Vandenbroucke, R.; De Smedt, S.C.; Wagner, E.; Ogris, M. The internalization route resulting in successful gene expression depends on both cell line and polyethylenimine polyplex type. Mol. Ther. 2006, 14, 745–753. [Google Scholar] [CrossRef] [PubMed]
- Rejman, J.; Bragonzi, A.; Conese, M. Role of clathrin- and caveolae-mediated endocytosis in gene transfer mediated by lipo- and polyplexes. Mol. Ther. 2005, 12, 468–474. [Google Scholar] [CrossRef] [PubMed]
- MacKerell, A.D., Jr.; Banavali, N.; Foloppe, N. Development and current status of the charmm force field for nucleic acids. Biopolymers 2000, 56, 257–265. [Google Scholar] [CrossRef]








© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krivitsky, A.; Krivitsky, V.; Polyak, D.; Scomparin, A.; Eliyahu, S.; Gibori, H.; Yeini, E.; Pisarevsky, E.; Blau, R.; Satchi-Fainaro, R. Molecular Weight-Dependent Activity of Aminated Poly(α)glutamates as siRNA Nanocarriers. Polymers 2018, 10, 548. https://doi.org/10.3390/polym10050548
Krivitsky A, Krivitsky V, Polyak D, Scomparin A, Eliyahu S, Gibori H, Yeini E, Pisarevsky E, Blau R, Satchi-Fainaro R. Molecular Weight-Dependent Activity of Aminated Poly(α)glutamates as siRNA Nanocarriers. Polymers. 2018; 10(5):548. https://doi.org/10.3390/polym10050548
Chicago/Turabian StyleKrivitsky, Adva, Vadim Krivitsky, Dina Polyak, Anna Scomparin, Shay Eliyahu, Hadas Gibori, Eilam Yeini, Evgeni Pisarevsky, Rachel Blau, and Ronit Satchi-Fainaro. 2018. "Molecular Weight-Dependent Activity of Aminated Poly(α)glutamates as siRNA Nanocarriers" Polymers 10, no. 5: 548. https://doi.org/10.3390/polym10050548