Hyaluronan-Chondroitin Sulfate Anomalous Crosslinking Due to Temperature Changes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Molecular Dynamics Force Field
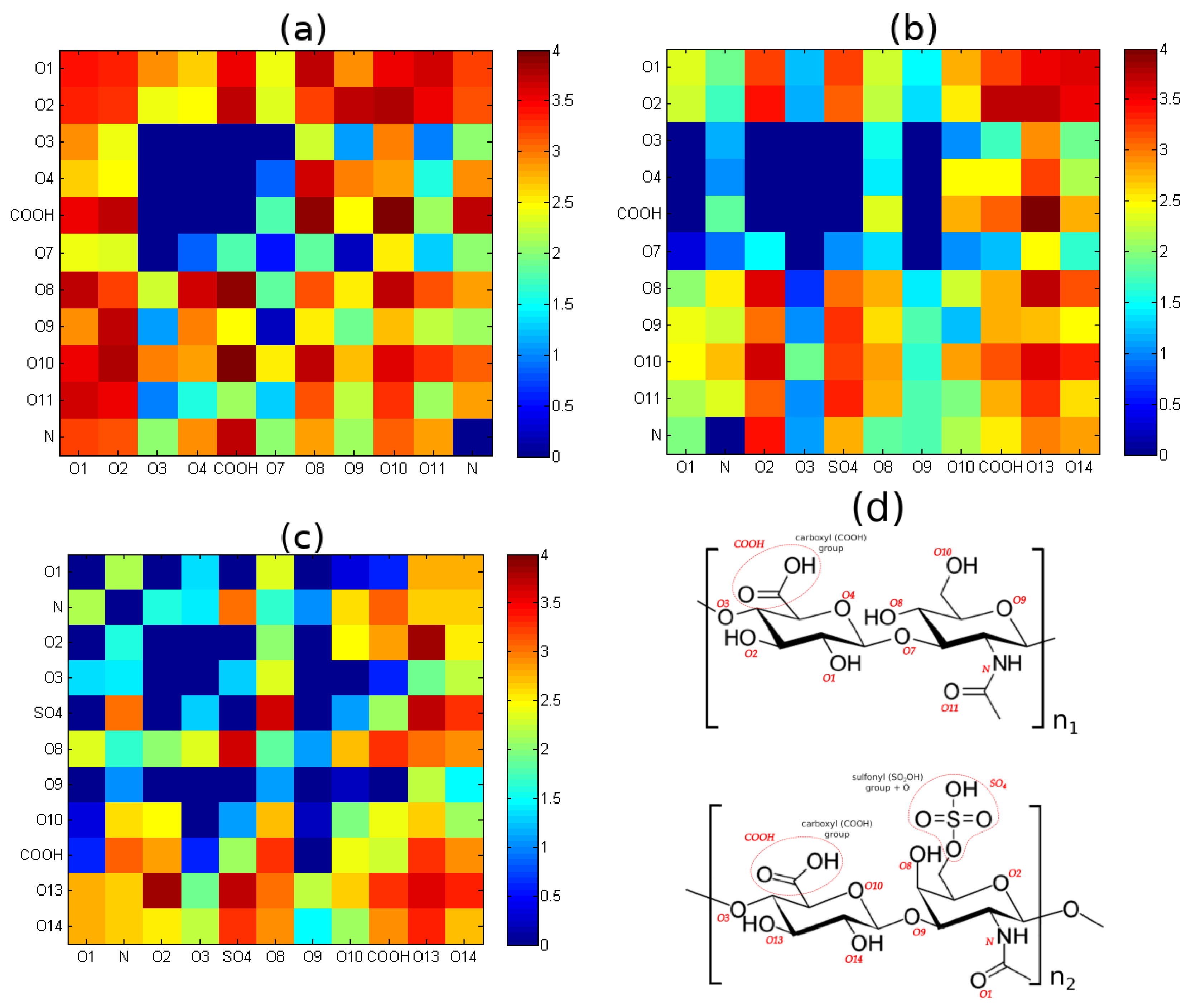
2.2.2. Hydrogen Bond Identification and Strength
2.3. Small-World Network Analysis
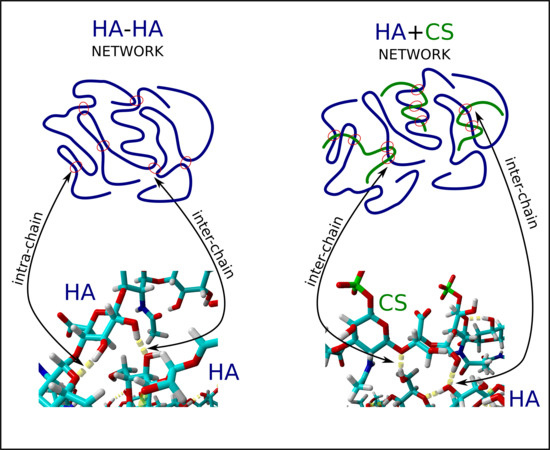
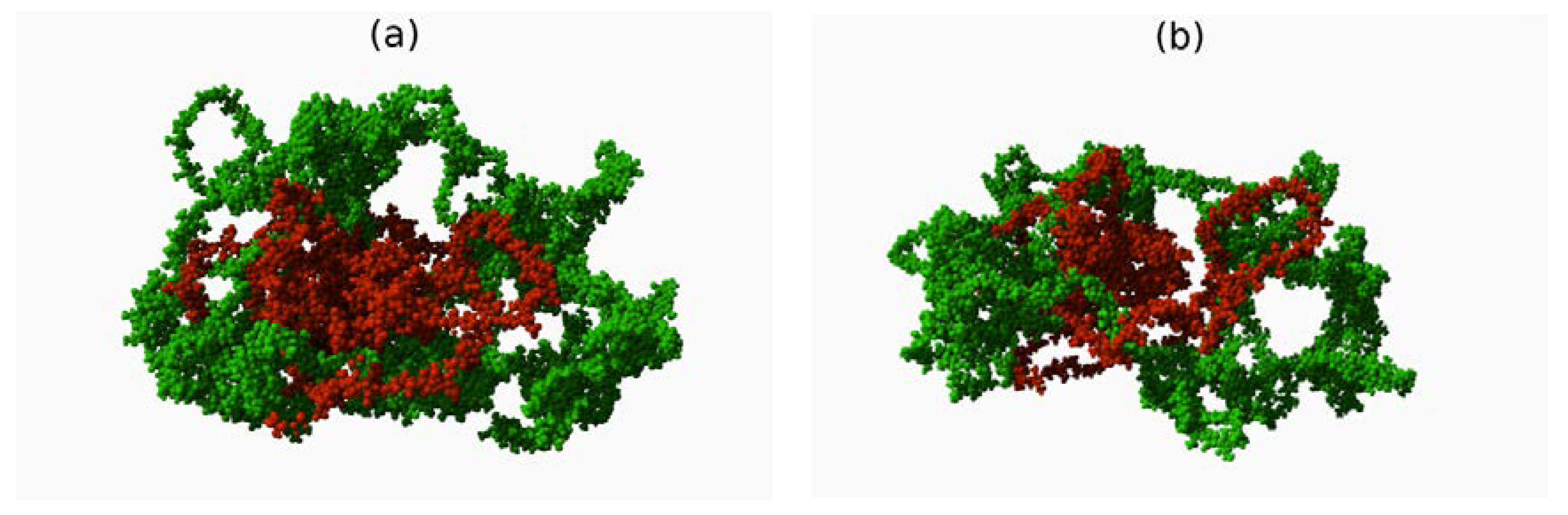
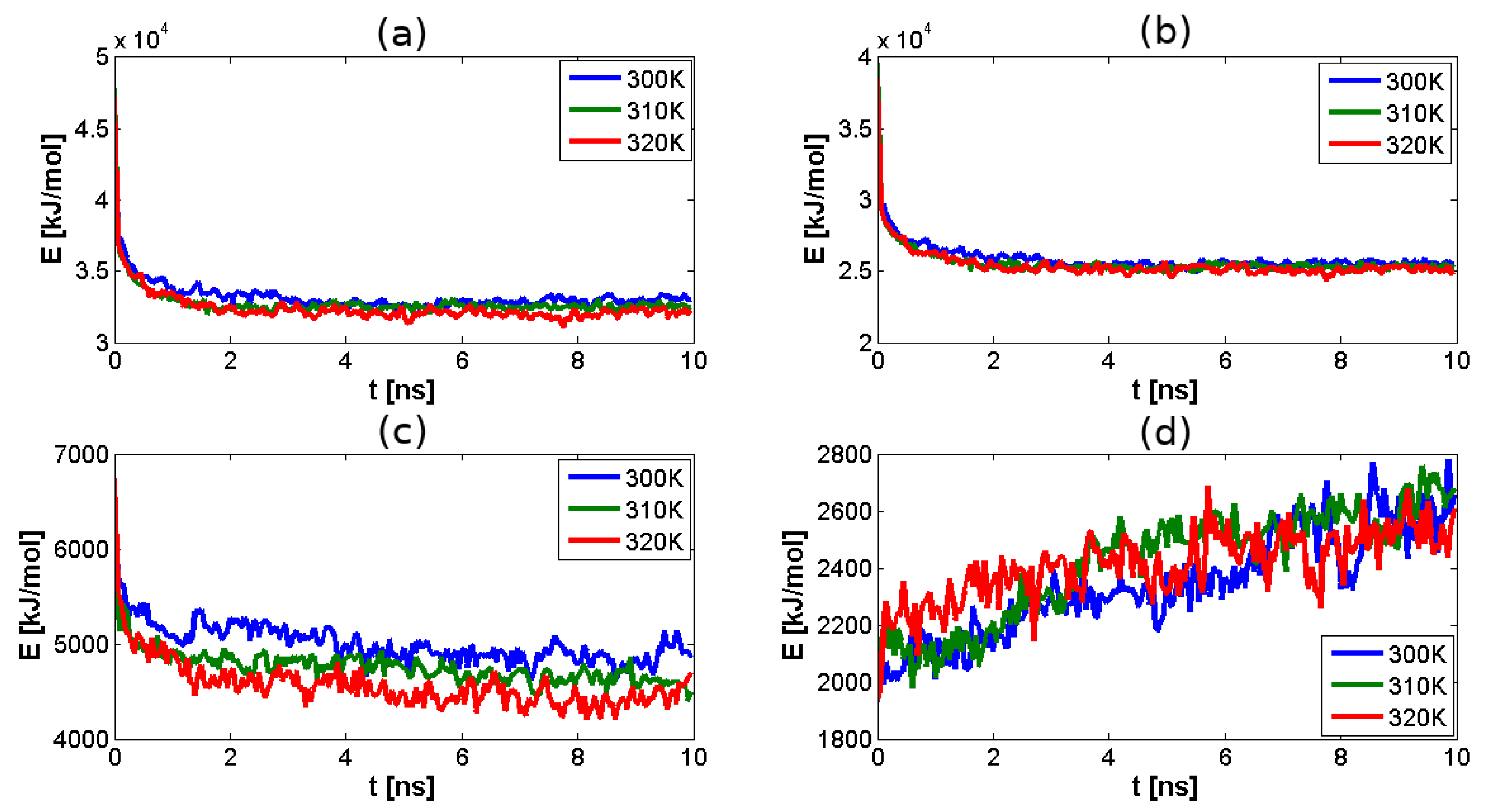
3. Results
4. Discussion
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| HA | Hyaluronic acid |
| CS | Chondroitin sulfate |
| SF | Synovial fluid |
References
- Hari, G.G.; Hales, C.A. Chemistry and Biology of Hyaluronan; Elsevier Science: Amsterdam, The Netherlands, 2008; ISBN 9780080472225. [Google Scholar]
- Bełdowski, P.; Weber, P.; Andrysiak, T.; Augé, W.K., II; Ledziński, D.; De Leon, T.; Gadomski, A. Anomalous Behavior of Hyaluronan Crosslinking Due to the Presence of Excess Phospholipids in the Articular Cartilage System of Osteoarthritis. Int. J. Mol. Sci. 2017, 18, 2779. [Google Scholar] [CrossRef] [PubMed]
- Matej, D. Boundary cartilage lubrication: Review of current concepts. Wien. Med. Wochenschr. 2014, 164, 88–94. [Google Scholar]
- Fakhari, A.; Berkland, C. Applications and emerging trends of hyaluronic acid in tissue engineering, as a dermal filler and in osteoarthritis treatment. Acta Biomater. 2013, 9, 7081–7092. [Google Scholar] [CrossRef] [PubMed]
- Chondroitin Sulfate: Uses, Side Effects, Interactions, Dosage, and Warning. Available online: https://www.webmd.com/vitamins/ai/ingredientmono-744/chondroitin-sulfate (accessed on 14 May 2018).
- Noh, I.; Kim, G.-W.; Choi, Y.-J.; Kim, M.-S.; Park, Y.; Lee, K.-B.; Kim, I.-S.; Hwang, S.-J.; Tae, G. Effects of cross-linking molecular weights in a hyaluronic acid-poly(ethylene oxide) hydrogel network on its properties. Biomed. Mater. 2006, 1, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Kosińska, M.K.; Ludwig, T.E.; Liebisch, G.; Zhang, R.; Siebert, H.C.; Wilhelm, J.; Kaesser, U.; Dettmeyer, R.B.; Klein, H.; Ishaque, B.; et al. Articular joint lubricants during osteoarthritis and rheumatoid arthritis display altered levels and molecular species. PLoS ONE 2015, 10, e0125192. [Google Scholar] [CrossRef] [PubMed]
- Bełdowski, P.; Andrysiak, T.; Mreła, A.; Pawlak, Z.; Augé, W.K., II; Gadomski, A. The Anomalies of Hyaluronan Structures in Presence of Surface Active Phospholipids—Molecular Mass Dependence. Polymers 2018, 10, 273. [Google Scholar] [CrossRef]
- Snelling, S.; Rout, R.; Davidson, R.; Clark, I.; Carr, A.; Hulley, P.A.; Price, A.J. A gene expression study of normal and damaged cartilage in anteromedial gonarthrosis, a phenotype of osteoarthritis. Osteoarthr. Cartil. 2014, 22, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, M.; Yan, W.; Mukudai, Y.; Nakamura, S.; Nakamasu, K.; Kawata, M.; Kawamoto, T.; Noshiro, M.; Hamada, T.; Kato, Y. Role of chondroitin sulfate–hyaluronan interactions in the viscoelastic properties of extracellular matrices and fluids. Biochim. Biophys. Acta Gen. Subj. 1998, 1380, 1–9. [Google Scholar] [CrossRef]
- Moghadam, M.N.; Abdel-Sayed, P.; Camine, V.M.; Pioletti, D.P. Impact of synovial fluid flow on temperature regulation in knee cartilage. J. Biomech. 2015, 48, 370–374. [Google Scholar] [CrossRef] [PubMed]
- Band, P.A.; Heeter, J.; Wiśniewski, H.G.; Liublinska, V.; Pattanayak, C.W.; Karia, R.J.; Stabler, T.; Balazs, E.A.; Kraus, V.B. Hyaluronan molecular weight distribution is associated with the risk of knee osteoarthritis progression. Osteoarthr. Cartil. 2015, 23, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Temple-Wong, M.M.; Ren, S.; Quach, P.; Hansen, B.C.; Chen, A.C.; Hasegawa, A.; D’Lima, D.D.; Koziol, J.; Masuda, K.; Lotz, M.K.; et al. Hyaluronan concentration and size distribution in human knee synovial fluid: Variations with age and cartilage degeneration. Arthritis Res. Ther. 2016, 18, 18. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.W.; Banquy, X.; Das, S.; Cadirov, N.; Jay, G.; Israelachvili, J. Effects of molecular weight of grafted hyaluronic acid on wear initiation. J. Acta Biomater. 2014, 10, 1817–1823. [Google Scholar] [CrossRef] [PubMed]
- Kwieciński, J.J.; Dorosz, S.G.; Ludwig, T.E.; Abubacker, S.; Cowman, M.K.; Schmidt, T.A. The effect of molecular weight on hyaluronan’s cartilage boundary lubricating ability—Alone and in combination with proteoglycan 4. Osteoarthr. Cartil. 2011, 19, 1356–1362. [Google Scholar] [CrossRef] [PubMed]
- Seror, J.; Zhu, L.; Goldberg, R.; Day, A.J.; Klein, J. Supramolecular synergy in the boundary lubrication of synovial joints. Nat. Commun. 2015, 6, 6497. [Google Scholar] [CrossRef] [PubMed]
- Katta, J.; Jin, Z.; Ingham, E.; Fisher, J. Chondroitin sulphate: An effective joint lubricant? Osteoarthr. Cartil. 2009, 17, 1001–1008. [Google Scholar] [CrossRef] [PubMed]
- Weininger, D. Smiles 1. Introduction and encoding rules. J. Chem. Inf. Comput. Sci. 1988, 28, 31–36. [Google Scholar] [CrossRef]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A.; et al. PubChem Substance and Compound databases. Nucleic Acids Res. 2015, 44, D1202–D1213. [Google Scholar] [CrossRef] [PubMed]
- Krieger, E.; Vriend, G. New ways to boost molecular dynamics simulations. J. Comput. Chem. 2015, 36, 996–1007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mark, P.; Nilsson, L. Structure and Dynamics of the TIP3P, SPC, and SPC/E Water Models at 298 K. J. Phys. Chem. A 2001, 105, 9954–9960. [Google Scholar] [CrossRef]
- Duan, Y.; Wu, C.; Chowdhury, S.; Lee, M.C.; Xiong, G.; Zhang, W.; Yang, R.; Cieplak, P.; Luo, R.; Lee, T.; et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 2003, 24, 1999–2012. [Google Scholar] [CrossRef] [PubMed]
- Rapaport, D.C. The Art of Molecular Dynamics Simulation; Cambridge Univ. Press: Cambridge, UK, 2010. [Google Scholar]
- Kmiecik, S.; Gront, D.; Kolinski, M.; Wieteska, L.; Dawid, A.E.; Kolinski, A. Coarse-grained protein models and their applications. Chem. Rev. 2016, 116, 7898–7936. [Google Scholar] [CrossRef] [PubMed]
- Ingólfsson, H.I.; Lopez, C.A.; Uusitalo, J.J.; Jong, D.H.D.; Gopal, S.M.; Periole, X.; Marrink, S.J. The power of coarse graining in biomolecular simulations. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2013, 4, 225–248. [Google Scholar] [CrossRef] [PubMed]
- Baaden, M.; Marrink, S.J. Coarse-grain modelling of protein-protein interactions. Curr. Opin. Strut. Biol. 2013, 23, 878–886. [Google Scholar] [CrossRef] [PubMed]
- Atilgan, A.R.; Akan, P.; Baysal, C. Small-World Communication of Residues and Significance for Protein Dynamics. Biophys. J. 2004, 86, 85–91. [Google Scholar] [CrossRef]
- Watts, D.J.; Strogatz, S.H. Collective dynamics of ‘small-world’ networks. Nature 1998, 393, 440–442. [Google Scholar] [CrossRef] [PubMed]
- Newman, M.E.J. Models of the Small World. J. Stat. Phys. 2000, 101, 819–841. [Google Scholar] [CrossRef]
- Wieland, D.C.F.; Degen, P.; Zander, T.; Gayer, S.; Raj, A.; An, J.; Dédinaité, A.; Claesson, P.; Willumeit-Römer, R. Structure of DPPC-hyaluronan interfacial layers—Effects of molecular weight and ion composition. Soft Matter 2016, 12, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Banquy, X.; Zappone, B.; Israelachvili, J. Synergistic interactions between grafted hyaluronic acid and lubricin provide enhanced wear protection and lubrication. Biomacromolecules 2013, 14, 1669–1677. [Google Scholar] [CrossRef] [PubMed]
- Oates, K.M.N.; Krause, W.E.; Jones, R.L.; Colby, R.H. Rheopexy of synovial fluid and protein aggregation. J. R. Soc. Interface 2006, 3, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Green, P.F.; Kramer, E.J. Matrix effects on the diffusion of long polymer chains. Macromolecules 1986, 19, 1108–1114. [Google Scholar] [CrossRef]
- Doi, M.; Edwards, S.F. The Theory of Polymer Dynamics, 2nd ed.; Clarendon Press: Oxford, UK, 1988. [Google Scholar]



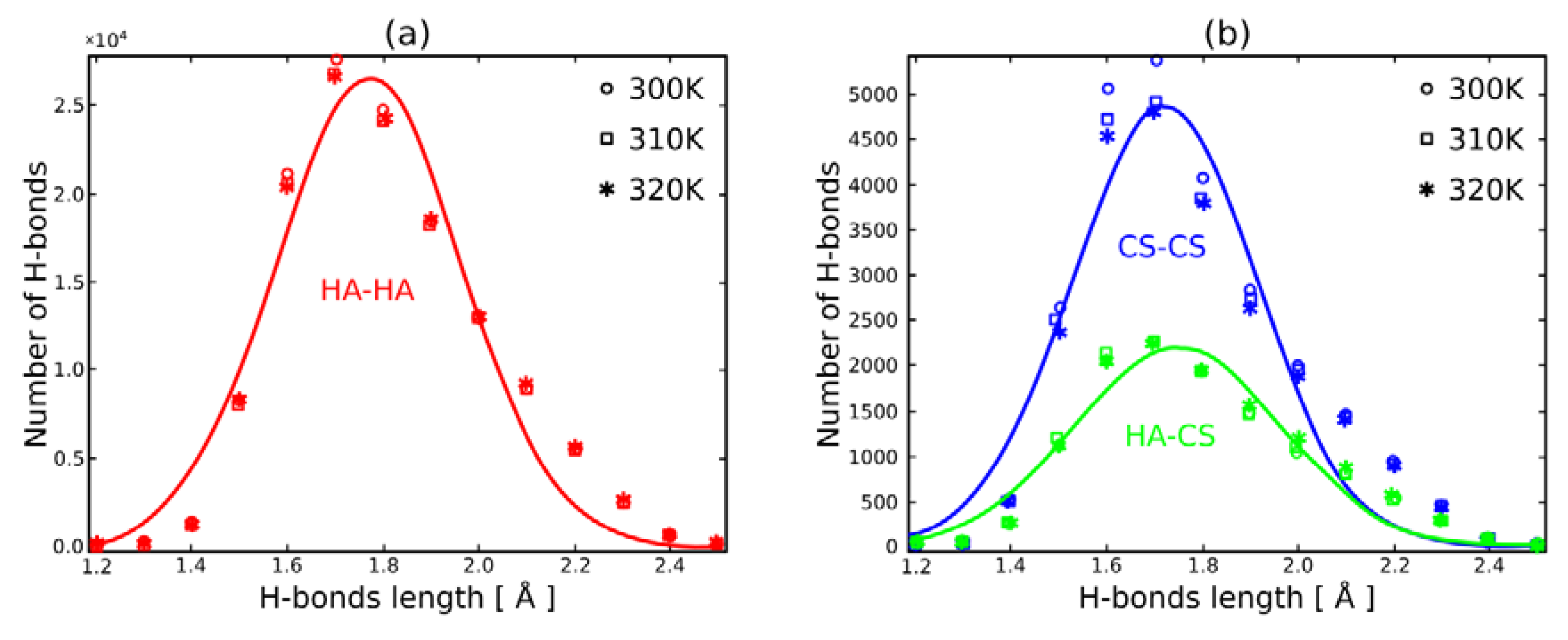
| Temperature | Length Distance | Length Distance | Length Distance |
|---|---|---|---|
| HA | HA–CS | CS | |
| 300 K | 1.20 | 1.27 | 1.42 |
| 310 K | 1.20 | 1.35 | 1.45 |
| 320 K | 1.16 | 1.29 | 1.44 |
| Temperature | Number Distance | Number Distance | Number Distance |
|---|---|---|---|
| HA | HA–CS | CS | |
| 300 K | 8.62 | 9.89 | 8.87 |
| 310 K | 8.44 | 9.51 | 8.91 |
| 320 K | 8.66 | 9.93 | 8.96 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andrysiak, T.; Bełdowski, P.; Siódmiak, J.; Weber, P.; Ledziński, D. Hyaluronan-Chondroitin Sulfate Anomalous Crosslinking Due to Temperature Changes. Polymers 2018, 10, 560. https://doi.org/10.3390/polym10050560
Andrysiak T, Bełdowski P, Siódmiak J, Weber P, Ledziński D. Hyaluronan-Chondroitin Sulfate Anomalous Crosslinking Due to Temperature Changes. Polymers. 2018; 10(5):560. https://doi.org/10.3390/polym10050560
Chicago/Turabian StyleAndrysiak, Tomasz, Piotr Bełdowski, Jacek Siódmiak, Piotr Weber, and Damian Ledziński. 2018. "Hyaluronan-Chondroitin Sulfate Anomalous Crosslinking Due to Temperature Changes" Polymers 10, no. 5: 560. https://doi.org/10.3390/polym10050560