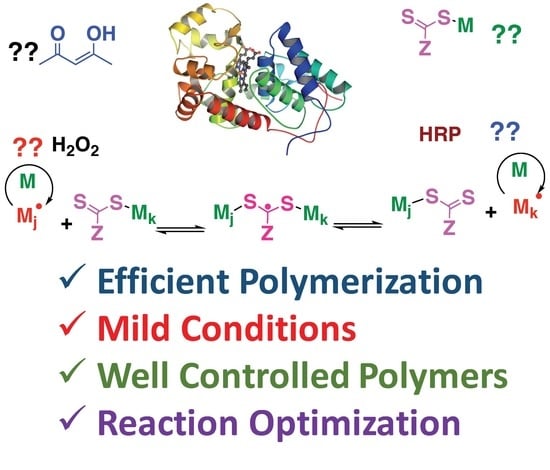
Investigating the Mechanism of Horseradish Peroxidase as a RAFT-Initiase
Abstract
:1. Introduction
2. Experimental
2.1. Materials
2.2. Typical HRP Catalyzed RAFT Polymerization of DMAm
2.3. Typical HRP Activity Assay
2.4. NMR
2.5. UV-Visible Spectroscopy
2.6. Size Exclusion Chromatography (SEC)
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Schmid, A.; Dordick, J.S.; Hauer, B.; Kiener, A.; Wubbolts, M.; Witholt, B. Industrial biocatalysis today and tomorrow. Nature 2001, 409, 258–268. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, R.A. E factors, green chemistry and catalysis: An odyssey. Chem. Commun. 2008, 3352–3365. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Kumar, J.; Tripathy, S.; Samuelson, L.A. Enzymatic Synthesis of Conducting Polyaniline in Micelle Solutions. Langmuir 2002, 18, 9696–9704. [Google Scholar] [CrossRef]
- Uyama, H.; Kobayashi, S. Enzyme-catalyzed polymerization to functional polymers. J. Mol. Catal. Enzym. 2002, 19–20, 117–127. [Google Scholar] [CrossRef]
- Gross, R.A.; Kumar, A.; Kalra, B. Polymer Synthesis by In Vitro Enzyme Catalysis. Chem. Rev. 2001, 101, 2097–2124. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.; Uyama, H.; Kimura, S. Enzymatic Polymerization. Chem. Rev. 2001, 101, 3793–3818. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.; Makino, A. Enzymatic Polymer Synthesis: An Opportunity for Green Polymer Chemistry. Chem. Rev. 2009, 109, 5288–5353. [Google Scholar] [CrossRef] [PubMed]
- Sen, S.; Puskas, E.J. Green Polymer Chemistry: Enzyme Catalysis for Polymer Functionalization. Molecules 2015, 20, 9358–9379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moad, G.; Solomon, D.H. Invited Review. Understanding and Controlling Radical Polymerization. Aust. J. Chem. 1990, 43, 215–239. [Google Scholar] [CrossRef]
- Moad, G.; Solomon, D.H. The Chemistry of Radical Polymerization; Elsevier: New York, NY, USA, 2006. [Google Scholar]
- Kalra, B.; Gross, R.A. Horseradish Peroxidase Mediated Free Radical Polymerization of Methyl Methacrylate. Biomacromolecules 2000, 1, 501–505. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Leija, R.J.; Torres-Lubian, J.R.; Resendiz-Rubio, A.; Luna-Barcenas, G.; Mota-Morales, J.D. Enzyme-mediated free radical polymerization of acrylamide in deep eutectic solvents. RSC Adv. 2016, 6, 13072–13079. [Google Scholar] [CrossRef]
- Cai, Z.-Q.I.; Wang, W.; Ruan, G.; Wen, X. Kinetic study of acrylamide radical polymerization initiated by the horseradish peroxidase–mediated system. Int. J. Chem. Kinet. 2012, 44, 475–481. [Google Scholar] [CrossRef]
- Durand, A.; Lalot, T.; Brigodiot, M.; Maréchal, E. Enzyme-mediated initiation of acrylamide polymerization: Reaction mechanism. Polymer 2000, 41, 8183–8192. [Google Scholar] [CrossRef]
- Durand, A.; Lalot, T.; Brigodiot, M.; Maréchal, E. Enzyme-mediated radical initiation of acrylamide polymerization: Main characteristics of molecular weight control. Polymer 2001, 42, 5515–5521. [Google Scholar] [CrossRef]
- Emery, O.; Lalot, T.; Brigodiot, M.; Maréchal, E. Free-radical polymerization of acrylamide by horseradish peroxidase-mediated initiation. J. Polym. Sci. Part A Polym. Chem. 1997, 35, 3331–3333. [Google Scholar] [CrossRef]
- Hollmann, F.; Arends, I.W.C.E. Enzyme Initiated Radical Polymerizations. Polymers 2012, 4, 759. [Google Scholar] [CrossRef]
- Lalot, T.; Brigodiot, M.; Maréchal, E. A kinetic approach to acrylamide radical polymerization by horse radish peroxidase-mediated initiation. Polym. Int. 1999, 48, 288–292. [Google Scholar] [CrossRef]
- Shogren, R.L.; Willett, J.L.; Biswas, A. HRP-mediated synthesis of starch–polyacrylamide graft copolymers. Carbohydr. Polym. 2009, 75, 189–191. [Google Scholar] [CrossRef]
- Singh, A.; Ma, D.; Kaplan, D.L. Enzyme-Mediated Free Radical Polymerization of Styrene. Biomacromolecules 2000, 1, 592–596. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, D.; Lalot, T.; Brigodiot, M.; Maréchal, E. β-Diketones as Key Compounds in Free-Radical Polymerization by Enzyme-Mediated Initiation. Macromolecules 1999, 32, 70–72. [Google Scholar] [CrossRef]
- Gormley, A.J.; Chapman, R.; Stevens, M.M. Polymerization Amplified Detection for Nanoparticle-Based Biosensing. Nano Lett. 2014, 14, 6368–6373. [Google Scholar] [CrossRef] [PubMed]
- Zavada, S.; Battsengel, T.; Scott, T. Radical-Mediated Enzymatic Polymerizations. Int. J. Mol. Sci. 2016, 17, 195. [Google Scholar] [CrossRef] [PubMed]
- Russell, G.T. The Kinetics of Free-Radical Polymerization: Fundamental Aspects. Aust. J. Chem. 2002, 55, 399–414. [Google Scholar] [CrossRef]
- Braunecker, W.A.; Matyjaszewski, K. Controlled/living radical polymerization: Features, developments, and perspectives. Prog. Polym. Sci. 2007, 32, 93–146. [Google Scholar] [CrossRef]
- Goto, A.; Fukuda, T. Kinetics of living radical polymerization. Prog. Polym. Sci. 2004, 29, 329–385. [Google Scholar] [CrossRef]
- Chiefari, J.; Chong, Y.K.; Ercole, F.; Krstina, J.; Jeffery, J.; Le, T.P.T.; Mayadunne, R.T.A.; Meijs, G.F.; Moad, C.L.; Moad, G.; et al. Living Free-Radical Polymerization by Reversible Addition-Fragmentation Chain Transfer: The RAFT Process. Macromolecules 1998, 31, 5559–5562. [Google Scholar] [CrossRef]
- Georges, M.K.; Veregin, R.P.N.; Kazmaier, P.M.; Hamer, G.K. Narrow molecular weight resins by a free-radical polymerization process. Macromolecules 1993, 26, 2987–2988. [Google Scholar] [CrossRef]
- Kato, M.; Kamigaito, M.; Sawamoto, M.; Higashimura, T. Polymerization of Methyl Methacrylate with the Carbon Tetrachloride/Dichlorotris-(triphenylphosphine)ruthenium(II)/Methylaluminum Bis(2,6-di-tert-butylphenoxide) Initiating System: Possibility of Living Radical Polymerization. Macromolecules 1995, 28, 1721–1723. [Google Scholar] [CrossRef]
- Wang, J.-S.; Matyjaszewski, K. Controlled/“living” radical polymerization. atom transfer radical polymerization in the presence of transition-metal complexes. J. Am. Chem. Soc. 1995, 117, 5614–5615. [Google Scholar] [CrossRef]
- Ng, Y.-H.; di Lena, F.; Chai, C.L.L. Metalloenzymatic radical polymerization using alkyl halides as initiators. Polym. Chem. 2011, 2, 589–594. [Google Scholar] [CrossRef]
- Ng, Y.-H.; di Lena, F.; Chai, C.L.L. PolyPEGA with predetermined molecular weights from enzyme-mediated radical polymerization in water. Chem. Commun. 2011, 47, 6464–6466. [Google Scholar] [CrossRef] [PubMed]
- Silva, T.B.; Spulber, M.; Kocik, M.K.; Seidi, F.; Charan, H.; Rother, M.; Sigg, S.J.; Renggli, K.; Kali, G.; Bruns, N. Hemoglobin and Red Blood Cells Catalyze Atom Transfer Radical Polymerization. Biomacromolecules 2013, 14, 2703–2712. [Google Scholar] [CrossRef] [PubMed]
- Simakova, A.; Mackenzie, M.; Averick, S.E.; Park, S.; Matyjaszewski, K. Bioinspired Iron-Based Catalyst for Atom Transfer Radical Polymerization. Angew. Chem. Int. Ed. 2013, 52, 12148–12151. [Google Scholar] [CrossRef] [PubMed]
- Sigg, S.J.; Seidi, F.; Renggli, K.; Silva, T.B.; Kali, G.; Bruns, N. Horseradish Peroxidase as a Catalyst for Atom Transfer Radical Polymerization. Macromol. Rapid Commun. 2011, 32, 1710–1715. [Google Scholar] [CrossRef] [PubMed]
- Chapman, R.; Gormley, A.J.; Herpoldt, K.-L.; Stevens, M.M. Highly Controlled Open Vessel RAFT Polymerizations by Enzyme Degassing. Macromolecules 2014, 47, 8541–8547. [Google Scholar] [CrossRef]
- Enciso, A.E.; Fu, L.; Russell, A.J.; Matyjaszewski, K. A Breathing Atom-Transfer Radical Polymerization: Fully Oxygen-Tolerant Polymerization Inspired by Aerobic Respiration of Cells. Angew. Chem. Int. Ed. 2018, 57, 933–936. [Google Scholar] [CrossRef] [PubMed]
- Yeow, J.; Chapman, R.; Gormley, A.J.; Boyer, C. Up in the air: Oxygen tolerance in controlled/living radical polymerisation. Chem. Soc. Rev. 2018, 47, 4357–4387. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Lv, Y.; An, Z. Enzymatic Cascade Catalysis for the Synthesis of Multiblock and Ultrahigh-Molecular-Weight Polymers with Oxygen Tolerance. Angew. Chem. 2017, 129, 14040–14044. [Google Scholar] [CrossRef]
- Hill, M.R.; Carmean, R.N.; Sumerlin, B.S. Expanding the Scope of RAFT Polymerization: Recent Advances and New Horizons. Macromolecules 2015, 48, 5459–5469. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, X.; Zhu, A.; Ma, K.; Lv, Y.; Wang, X.; An, Z. Enzyme-Initiated Reversible Addition–Fragmentation Chain Transfer Polymerization. Macromolecules 2015, 48, 7792–7802. [Google Scholar] [CrossRef]
- Liu, Z.; Lv, Y.; Zhu, A.; An, Z. One-Enzyme Triple Catalysis: Employing the Promiscuity of Horseradish Peroxidase for Synthesis and Functionalization of Well-Defined Polymers. ACS Macro Lett. 2018, 7, 1–6. [Google Scholar] [CrossRef]
- Tan, J.; Xu, Q.; Li, X.; He, J.; Zhang, Y.; Dai, X.; Yu, L.; Zeng, R.; Zhang, L. Enzyme-PISA: An Efficient Method for Preparing Well-Defined Polymer Nano-Objects under Mild Conditions. Macromol. Rapid Commun. 2018, 39, 1700871. [Google Scholar] [CrossRef] [PubMed]
- Danielson, A.P.; Van-Kuren, D.B.; Lucius, M.E.; Makaroff, K.; Williams, C.; Page, R.C.; Berberich, J.A.; Konkolewicz, D. Well-Defined Macromolecules Using Horseradish Peroxidase as a RAFT Initiase. Macromol. Rapid Commun. 2016, 37, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-López, J.N.; Lowe, D.J.; Hernández-Ruiz, J.; Hiner, A.N.P.; García-Cánovas, F.; Thorneley, R.N.F. Mechanism of Reaction of Hydrogen Peroxide with Horseradish Peroxidase: Identification of Intermediates in the Catalytic Cycle. J. Am. Chem. Soc. 2001, 123, 11838–11847. [Google Scholar] [CrossRef] [PubMed]
- Goto, A.; Sato, K.; Tsujii, Y.; Fukuda, T.; Moad, G.; Rizzardo, E.; Thang, S.H. Mechanism and Kinetics of RAFT-Based Living Radical Polymerizations of Styrene and Methyl Methacrylate. Macromolecules 2001, 34, 402–408. [Google Scholar] [CrossRef]
- Konkolewicz, D.; Siauw, M.; Gray-Weale, A.; Hawkett, B.S.; Perrier, S. Obtaining Kinetic Information from the Chain-Length Distribution of Polymers Produced by RAFT. J. Phys. Chem. B 2009, 113, 7086–7094. [Google Scholar] [CrossRef] [PubMed]
- Nicell, J.A.; Wright, H. A model of peroxidase activity with inhibition by hydrogen peroxide. Enzym. Microb. Technol. 1997, 21, 302–310. [Google Scholar] [CrossRef]
- Falatach, R.; McGlone, C.; Al-Abdul-Wahid, M.S.; Averick, S.; Page, R.C.; Berberich, J.A.; Konkolewicz, D. The best of both worlds: Active enzymes by grafting-to followed by grafting-from a protein. Chem. Commun. 2015, 51, 5343–5346. [Google Scholar] [CrossRef] [PubMed]
- Paeth, M.; Stapleton, J.; Dougherty, M.L.; Fischesser, H.; Shepherd, J.; McCauley, M.; Falatach, R.; Page, R.C.; Berberich, J.A.; Konkolewicz, D. Chapter Nine—Approaches for Conjugating Tailor-Made Polymers to Proteins. In Methods in Enzymology; Kumar, C.V., Ed.; Academic Press: Cambridge, MA, USA, 2017; Volume 590, pp. 193–224. [Google Scholar]
- Hiner, A.N.P.; Hernández-Ruiz, J.; Williams, G.A.; Arnao, M.B.; García-Cánovas, F.; Acosta, M. Catalase-like Oxygen Production by Horseradish Peroxidase Must Predominantly Be an Enzyme-Catalyzed Reaction. Arch. Biochem. Biophys. 2001, 392, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Chang, B.S.; Kendrick, B.S.; Carpenter, J.F. Surface-Induced Denaturation of Proteins during Freezing and its Inhibition by Surfactants. J. Pharm. Sci. 1996, 85, 1325–1330. [Google Scholar] [CrossRef] [PubMed]
- Franks, F. Biophysics and Biochemistry at Low Temperatures; Cambridge University Press: Cambridge, UK, 1985. [Google Scholar]
- Kurek, P.N.; Kloster, A.J.; Weaver, K.A.; Manahan, R.; Allegrezza, M.L.; De Alwis Watuthanthrige, N.; Boyer, C.; Reeves, J.A.; Konkolewicz, D. How Do Reaction and Reactor Conditions Affect Photoinduced Electron/Energy Transfer Reversible Addition–Fragmentation Transfer Polymerization? Ind. Eng. Chem. Res. 2018, 57, 4203–4213. [Google Scholar] [CrossRef]
- Sariri, R.; Sajedi, R.H.; Jafarian, V. Inhibition of horseradish peroxidase activity by thiol type inhibitors. J. Mol. Liquids 2006, 123, 20–23. [Google Scholar] [CrossRef]
- Konkolewicz, D.; Krys, P.; Matyjaszewski, K. Explaining Unexpected Data via Competitive Equilibria and Processes in Radical Reactions with Reversible Deactivation. Acc. Chem. Res. 2014, 47, 3028–3036. [Google Scholar] [CrossRef] [PubMed]
- Vana, P.; Davis, T.P.; Barner-Kowollik, C. Kinetic Analysis of Reversible Addition Fragmentation Chain Transfer (RAFT) Polymerizations: Conditions for Inhibition, Retardation, and Optimum Living Polymerization. Macromol. Theory Simul. 2002, 11, 823–835. [Google Scholar] [CrossRef]
- Mueller, A.H.; Zhuang, R.; Yan, D.; Litvinenko, G. Kinetic analysis of “living” polymerization processes exhibiting slow equilibria. 1. Degenerative transfer (direct activity exchange between active and “dormant” species). Application to group transfer polymerization. Macromolecules 1995, 28, 4326–4333. [Google Scholar] [CrossRef]
- Jesson, C.P.; Pearce, C.M.; Simon, H.; Werner, A.; Cunningham, V.J.; Lovett, J.R.; Smallridge, M.J.; Warren, N.J.; Armes, S.P. H2O2 Enables Convenient Removal of RAFT End-Groups from Block Copolymer Nano-Objects Prepared via Polymerization-Induced Self-Assembly in Water. Macromolecules 2017, 50, 182–191. [Google Scholar] [CrossRef]










© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Danielson, A.P.; Van-Kuren, D.B.; Bornstein, J.P.; Kozuszek, C.T.; Berberich, J.A.; Page, R.C.; Konkolewicz, D. Investigating the Mechanism of Horseradish Peroxidase as a RAFT-Initiase. Polymers 2018, 10, 741. https://doi.org/10.3390/polym10070741
Danielson AP, Van-Kuren DB, Bornstein JP, Kozuszek CT, Berberich JA, Page RC, Konkolewicz D. Investigating the Mechanism of Horseradish Peroxidase as a RAFT-Initiase. Polymers. 2018; 10(7):741. https://doi.org/10.3390/polym10070741
Chicago/Turabian StyleDanielson, Alex P., Dylan Bailey Van-Kuren, Joshua P. Bornstein, Caleb T. Kozuszek, Jason A. Berberich, Richard C. Page, and Dominik Konkolewicz. 2018. "Investigating the Mechanism of Horseradish Peroxidase as a RAFT-Initiase" Polymers 10, no. 7: 741. https://doi.org/10.3390/polym10070741