NMR Studies and Molecular Dynamic Simulation of Synthetic Dendritic Antigens
Abstract
: A series of synthetic benzylpenicillinoylated dendrimers has been prepared using up to 4th generation PAMAM dendrimers. These nanoconjugates, as nanosized Dendritic Antigens, are useful in the diagnostic evaluation of drug allergy due to specific molecular recognition with the Human Immunological System (IgE). The morphology and dimensions of the conjugates coupled to the orientation of the peripheral benzylpenicillin residues in the dendrimers may play key roles in such molecular recognition processes. Herein, the characterization and conformation of these structures are studied by a detailed analysis of 1D (1H and 13C NMR) and 2D NMR (1H,1H-NOESY) spectra. These dendrimers in explicit solvent were studied by the atomistic forcefield-based molecular dynamics. Structural properties such as shape, radius-of-gyration and distribution of the monomers will be discussed in relation to the experimental observations.1. Introduction
Specific molecular recognition processes between antigens (hapten-carrier conjugate type) and Immunoglobulin E (IgE) are dependent on the morphology and dimensions of the antigens coupled to the orientation and availability of the hapten residues in the carrier macromolecule. These antigen-IgE recognition processes are the cause of IgE-mediated allergic reactions to drugs [1], the diagnosis of which is based on the determination of specific IgE antibodies. In order to detect antibodies to penicillin, we have to bear in mind that the ability for binding IgE antibodies may depend on both natures, that of the hapten and that of the carrier. Moreover, the density and the distribution of the hapten in the carrier may also influence the capacity for capturing antibodies.
In this context, we have reported the design and synthesis of precisely defined monodisperse nanoconjugates, consisting of PAMAM dendrimers up to the 2nd generation decorated with penicillin hapten units, or benzylpenicilloyl (BPO), at their periphery (GnBPO, n = 0–2) [2]. Their specific recognition by IgE antibodies has been confirmed with clinical assays using sera from patients allergic to penicillin [2,3]. It can therefore be concluded that the hapten-carrier (dendrimer) conjugates studied mimic recognition with natural hapten-carrier (protein) conjugates. The structures were characterized based on 1D NMR spectroscopy and MALDI-TOF spectrometry. However, the 3 dimensional conformation of the defined dendrimer structures also allows the study of other important parameters to understand the recognition process requisites. We extended the preparation of these Dendritic Antigens up to the 4th generation and studied the characterization and conformation of these structures by a detailed analysis of 1D (1H and 13C NMR) and 2D NMR (1H,1H-NOESY) spectra.
2. Results and Discussion
In the context of penicillin allergy, the formation of the antigenic determinant structures is based on the electrophilic properties of the β-lactam ring against nucleophilic reagents such as alcohols, thiols and amines. A simple approach to establish what happens with the hapten molecule after reaction with natural high molecular weight nucleophiles such as proteins was carried out with a low-molecular-weight nucleophile (Scheme 1(a)). After controlling the resulting structure of the antigenic determinant, synthetic dendrimers were employed to mimic carrier proteins in order to evaluate multivalent defined structures with a monodisperse structure.
2.1. Model Low Molecular Weight Antigenic Determinant
The reaction using butylamine as the nucleophile reagent and benzylpenicillin in aqueous media afforded immediately just the product corresponding to the aminolysis of the β-lactam ring (Bu-BPO). The shift of the peaks corresponding to the β-lactam protons, H5 and H6, was useful to monitor the progress of the reaction in D2O. These two protons resonate as doublets and in benzylpenicillin shift from 5.45 and 5.49 ppm (J5,6 = 3.9 Hz) to 4.86 and 4.25 ppm (J5,6 = 9.6 Hz) for H-5 and H-6 respectively, in the open β-lactam form. The residue thus derived from benzylpenicillin is called benzylpenicilloyl, the 1H NMR spectrum of which is shown in Figure 1(a). Moreover, 2D NMR techniques were employed to determine the correct stereochemistry corresponding to the protons H-5 and H-6 (Table 1 and Figure 1(b)). The NOESY experiment showed a NOE effect between H-6 and H-3 and a small NOE effect between H-5 and H-6. These data, together with the coupling constant value, point towards an important contribution of the anti conformation between H-5 and H-6 to the 3 dimensional structure of Bu-BPO in aqueous solution, as shown in Figure 1(b).
2.2. Preparation of Dendritic Antigens GnBPO (n = 0–4)
The strategy to functionalize the terminal amine groups of the PAMAM dendrimers with benzylpenicilloyl residues consisted of using the same reactivity employed for the formation of Bu-BPO. The dendritic antigens of different generations were obtained from the reaction between the corresponding PAMAM dendrimers and an excess of benzylpenicillin in an aqueous buffer at pH 10.8 (to ensure that every terminal amino group in the PAMAM dendrimer was deprotonated).
The completion of the reaction was monitored by 1H and 13C NMR, from these 13C NMR provides more information regarding the functionalization of the terminal amine groups. The control of the temperature and time stated in each case proved to be a strongly influential factor to obtain a complete peripheral substitution (Table 2). Synthetic dendritic antigens of lowest generations (0 and 1) were obtained at room temperature; whereas, the reaction at low temperature favors the functionalization of the periphery in dendrimers from 2nd to 4th generation. This could be due to a reduced mobility of the substituted branches at low temperatures, which may decrease the steric hindrance in the nucleophilic primary amines [2].
2.3. 1D NMR of Dendritic Antigens
The NMR experiments were carried out in basic D2O (pD 11) reached by addition of Na2CO3. In these conditions all amino groups in PAMAM dendrimers are unprotonated, and the resonances of the methylene protons directly bond to the terminal amino groups proved very useful for monitoring the coupling reaction since they shift from 2.70 to 3.22 ppm. The shift of the resonances of β-lactam protons indicates the opening of the ring and the signals corresponding to the rest of the benzylpenicilloyl residue are consistent with those obtained for the same residue in the Bu-BPO molecule. As a representative example, 1H NMR of G0BPO conjugate is shown in Figure 2. 13C NMR was very homogeneous and confirmed the complete functionalization of the dendrimers by the absence of carbons corresponding to the ethylene residue directly bonded to primary amino groups (carbons e and d in G0BPO4, 42.8 and 39.5 ppm), and the appearance of peaks corresponding to the condensation reaction (39.6 and 39.5 ppm). Although experiments with longer acquisition times are employed for the higher generations due to a lower concentration obtained for such higher molecular weight molecules, it is difficult to confirm the disappearance of signals, the lack of which indicates complete substitution.
A summary of NMR spectroscopic data for all these dendritic conjugates is given in Tables 3, 4 and 5. Interestingly, these nanoconjugates show homogeneous 1H and 13C NMR spectra and very similar for all generations, which is indicative of highly symmetric structures and a monodisperse nature. Proton signals corresponding to the benzylpenicilloyl residues are representative of this monodispersity (Table 3).
2.4. 2D NMR of Dendritic Antigens
The NOESY interactions between different protons provide information regarding their spatial locations relative to various parts of the dendritic chain [4]. This enables information to be inferred about the dominant orientation of the peripheral benzylpenicillin residues in the dendritic chains in aqueous solutions. 2D-NOESY spectra were carried out with the second and fourth generation of conjugates to evaluate how the dendrimer's size and the number of ligands on their periphery influence the conformation of the antigenic determinant residues in the nanoconjugate surfaces.
The spectra of both nanoconjugates show that the benzylpenicilloyl units retain the stereochemistry (in protons H-5 and H-6) from the starting benzylpenicillin. Additionally, NOE effects between H-6 and H-3 and a small NOE effect between H-5 and H-6 point towards an important contribution of the anti conformation between H-5 and H-6, as in the corresponding simpler equivalent conjugate Bu-BPO.
As shown in Figure 4, the 2D-NOESY for G2BPO16 spectra exhibit strong cross-peaks between most of the protons corresponding to the benzylpenicilloyl units and those of methylenes (l, m) in the dendritic skeleton. Nonetheless, no cross-peaks between protons of the thiazolidine ring and dendritic methylene protons were found. These results indicate that the benzylpenicilloyl residues orient the negatively charged carboxylate groups to the aqueous exterior media; whereas the rest of the benzylpenicilloyl fragments are located in a more unpolar environment surrounded by dendritic branches. Moreover, no NOE interactions were observed between protons on different antigenic ligands, indicating the chain ends are hosting the hydrophobic segment and the thiazolidine ring is primarily oriented outward from the skeleton of the dendrimers in a non-compacted way.
The 2D-NOESY for G4BPO64 (Figure 5) shows an additional NOE interaction between both methyl groups (α and β), compared to G2BPO16 spectra, and only a single cross-peak between the benzylpenicilloyl residue and the dendritic skeleton. These data suggest that G4BPO64 presents a densely packed surface on which methyl groups of different penicilloyl residues are situated in a close distance, and only the phenyl moieties are placed in the interior of the dendrimers.
This is in agreement with molecular simulations of the structure of PAMAM dendrimers which showed that lower generation dendrimers (G < 4) have an open structure, but that higher generation dendrimers (G ≥ 4) possess a densely packed surface [5]. Therefore, the functionalization of the fourth generation dendrimer with penicillins results in a dendrimer, G4BPO64, the size of which may be comparable to a fifth generation PAMAM dendrimer, with a highly packed surface and a globular structure, where the antigenic ligands could be favored to locate their phenyl groups within the interior of the dendrimer skeleton. Compared to the G4BPO64 conjugate, the antigenic dendrimer G2BPO16 may present a more open structure allowing the penetration of the more hydrophobic moieties of the antigenic determinants in the peripheral dendritic branches. These benzyl side chains are isolated from the external aqueous media with negatively charged hydrophilic fragments in the periphery, corresponding to thiazolidine moieties.
2.5. Molecular Dynamic Simulation
Equilibration
The measure of the relaxation of dendrimers was determined from the autocorrelation function of the squared radius-of-gyration [6], which is evaluated from the expression:
The resulting graph for dendrimers is presented in Figure 6 and in all examined models decays to 0 at timescales shorter than 1 ns. We can conclude that the simulation times are sufficient to sample enough independent configurations for averaging of the static properties. The details of the five systems studied are listed in Table 6.
Dendrimer Size and Shape
The dendrimer size can be quantified by considering the radius-of-gyration Rg of the molecular structure. This is especially useful to characterize polymer solutions to have a rough measure for the compactness of a structure. It is computed as follows:
Figure 7(b) shows the behavior of the Rg as a function of the dendrimer number of atoms N in logarithmic scale. The linear relation between log Rg and log N is an indication for a space-filling structure and the related fractal dimensionality dc takes account of the compactness of the dendrimer series. In this case, the size scale ,and the dc is 2.68, indicating that these dendrimers have a more polymeric-like structure than the PAMAM dendrimers, where dc is 3, showing an homogeneous and densely packed structure [8]. This compact structure requires a space filling geometry that implies back-folding of these dendrimers. This back-folding is evident from several lines of evidence, as will be discussed below.
A structure-persistent characteristic of these dendrimers can be obtained by considering the average values of the three principal moments of inertia Iz, Iy, Ix, where Iz, Iy, Ix represent the eigenvalues of the moment of inertia tensor in descending order. The ratio of these three principal moments is a measure of the eccentricity (minor-major axes ratio) of the ellipsoid shape of the dendrimer. Figure 8(a) shows the average aspect ratios Iz/Ix and Iz/Iy for the different generation dendrimers (Table 7). The values of the Iz/Iy are in the range 1.1–1.3 and Iz/Ix are in the range 1.2–2.4. This means that these dendrimers are compact ellipsoid-spheroid in shape.
The change in shape is better reflected by the relative shape anisotropy, which is defined as [9]:
We checked the position of the ethylendiamine core (EDA) to see if it stays in the center of the dendrimers. We measured the distance between the centers of mass of the whole dendrimer and of the EDA core. The average distances from G0BPO4 to G4BPO64 were 4.04, 2.56, 4.70, 2.77, 3.57 Å respectively. So it is obvious that the core deviates from the center of the dendrimers, especially in G0BPO4 and G2BPO16 generations.
Typical equilibrated conformations of the dendrimer generations are shown in Figure 9. To simplify the figure, the hydrogen atoms have been omitted, the carbon atoms are depicted cyan, nitrogen atoms blue, oxygen atoms red and sulfur atoms yellow. These snapshots are generated using VMD software [10].
Monomer density distribution
The conformation of the dendrimer can be conveniently expressed through the average radial monomer density ρ(r), that can be defined by counting the number N(r) of atoms whose centers of mass are located within the spherical shell of radius r and thickness δr. Hence, the integration over r yields the total number of atoms as: , where r is the distance from the center-of-mass of the dendrimers. This number was divided by the volume of the shell to produce ρ(r).
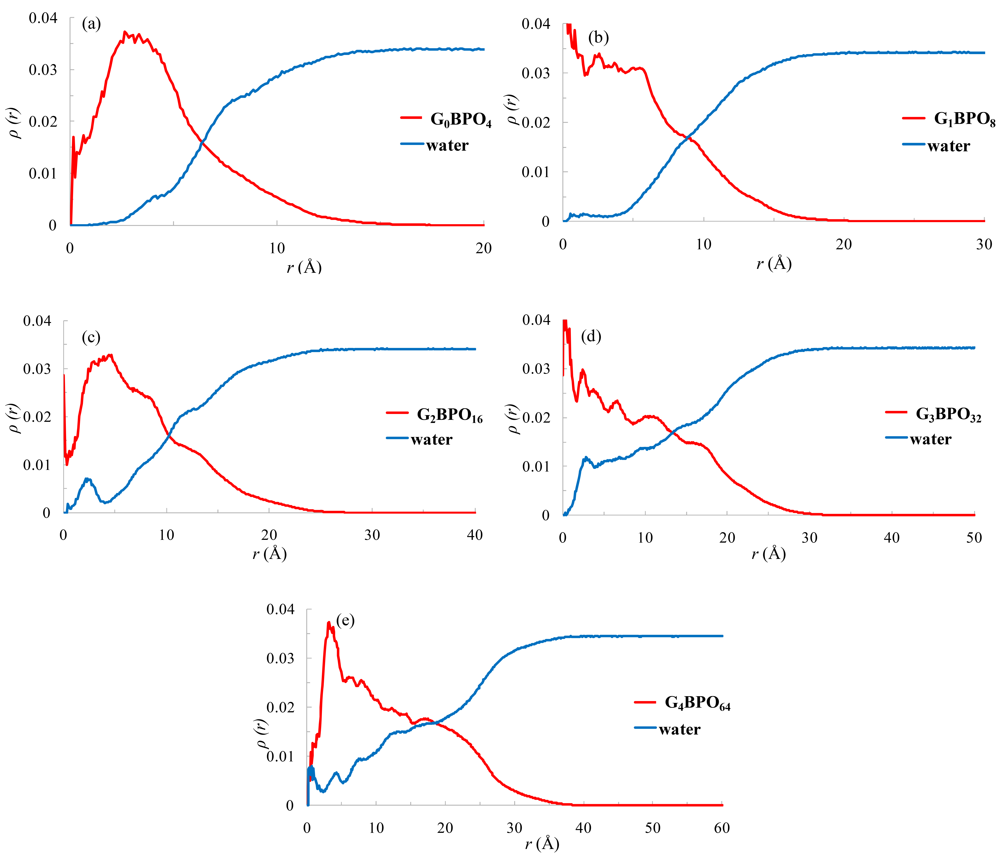
The distribution of atoms within the dendrimers is shown using radial density profiles. The curves corresponding to the five generations of dendrimers, from G0BPO4 to G4BPO64, are shown in Figure 10.
For all dendrimers the maximum density is close to the center-of-mass and decays toward the edge of the molecule. The density measured in the region of the center-of-mass is not for the core part. In our model the core deviates from the center-of-mass and the dendrimers here have not perfect symmetrical structures. The smaller dendrimers G0BPO4 and G1BPO8 show a quick decrease in atom density after about 4–6 Å of the center-of-mass, while for the upper generations G2BPO16 to G4BPO64 the inner parts of the dendrimer exhibit a high atom density and at longer distances a steady decrease in atom density takes place, and it is the upper generation G4 that shows the higher density profile, suggesting that the dendrimer's atoms are densely packed. Finally, in the tail zone, the density decays monotonically toward the exterior of the molecules.
As can be readily verified in Figure 10, in all cases the water density reaches the bulk value at distances beyond the dendrimer boundary, after the dendrimer density distribution drops to zero. Voids inside the dendrimer are occupied by water molecules, as shown by the increase in the water concentration accompanying any decrease in dendrimer atom density.
Radial monomer density profiles are displayed for all the dendrimers and for each type of monomer, shown in Figure 11. These profiles allow us to determine whether or not the different residue types are present within the interior of the dendrimer. The EDA and PAA density distribution of these monomers show, for all dendrimers, a displacement from the center-of-mass of the molecule. The value of the radial density is very noisy near the center-of-mass because the error is big when the number is divided by the very small volume within the shell near the center of the mass.
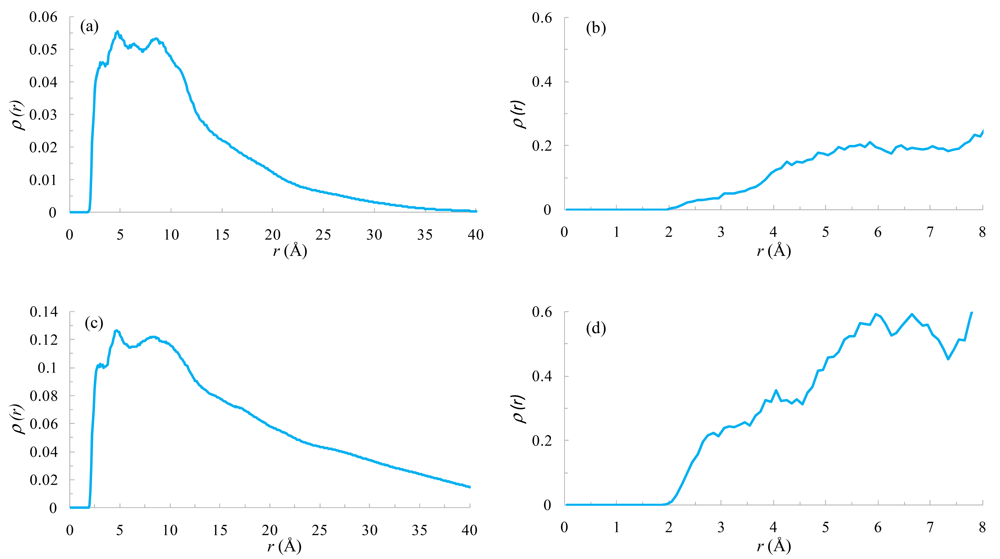
To elucidate the NOE effect found between the hydrogen atoms of the BPO-Ph subunit and the hydrogens of the methylenes (l, m) corresponding to the PAH monomer in the G2BPO16, and with the methylene (t) in the G4BPO64, we evaluated the density distribution functions between these atoms (Figure 12(a,c)). Both distributions show sharp peaks in the range between 2.5 and 5 Å. Therefore, these atoms are close enough to have a NOE effect between them.
In a similar way, Figure 12(b,d) show the density distribution functions between alpha and beta methyl protons of different BPO subunits in G2BPO16 and G4BPO64 respectively. These distributions show a higher density for G4BPO64 in the range between 2.0 and 3.0 Å.
As the dendrimer generation increases, the probability of finding a monomer, PAH or BPO, throughout the molecule also increases. The number of terminal monomers doubles with each generation, so the BPO groups are spread over an increasingly large volume. These results suggest a back-folding of the terminal monomers. The density of these groups moves generally to larger distances as the generation increases, indicating considerable flexibility in the dendritic structure.
The benzylpenicilloyl residues could be considered as negative charged thiazolidine rings (BPO-CO2−) plus the rest of the benzylpenicilloyl fragments (BPO-Ph). Figure 13 shows the radial density profiles of these subunits for all the dendrimers. In all of the dendrimers, the apolar subunit (BPO-Ph) is always oriented toward the core of the dendrimers whereas the polar BPO-CO2− is oriented toward the exterior. In higher generations this subunit shows a plateau at the periphery, suggesting a more uniform distribution on the outside of the molecule.
It is observed that sodium counterion distribution accompanies the BPO moieties. In addition, the BPO distribution increases with the generation of the dendrimers, and always on the same side of the BPO-CO2− subunit. Notice that back-folding in dendrimers is only entropically favorable, but electrostatically unfavorable when there are no counterions present.
3. Experimental Section
3.1. Synthesis and NMR Experiments
Materials
Poly(amidoamine) dendrimers (ethylenediamine core) with amino surface groups (PAMAM generations 0–4) were purchased from Dendritic Nanotechnologies, Inc. (dnt). Benzylpenicillin sodium salt was supplied by CEPA, and S.L. Standard chemicals were obtained from Aldrich or Merck.
Synthesis
The conjugates Bu-BPO and GnBPO were prepared using procedures previously described and adapted for each generation (Table 2) [2].
NMR Experiments
1H NMR, 13C NMR and 1H,1H-NOESY experiments were performed on a Bruker Avance III 400 MHz NMR instrument and chemical shifts (δ) are given in ppm relative to the residual solvent peak. All NMR spectra were recorded using D2O as solvent (with Na2CO3 to ensure a basic pD = 11) at room temperature. 1H NMR spectra were acquired with a spectral window of 14 ppm, an acquisition time of 3 s and a relaxation delay of 1 s. 13C NMR spectra were acquired with a spectral window of 240 ppm, an acquisition time of 0.7 s and a relaxation delay of 2 s. NOESY 2D-NMR spectra of conjugates were collected with 3,605 Hz spectral windows in f1 and f2, a 0.5 s mixing time and 9.28 μs 1H 90° pulse width. The experiments were done with a 1 s relaxation delay and a 0.284 s acquisition time. Eight transients were averaged for each 256 × 2,048 complex t1 increments.
3.2. Computational Details
Dendrimer Construction
Dendrimers were built using the program Starmaker which is part of Silico [11]. This is a flexible program which can be used to construct a wide variety of dendrimers, including heterogeneously functionalized (variegated) dendrimers. Starmaker builds dendrimers from a core by repeated addition of generations of monomers. As each monomer unit is added, a stochastic search is used to attain an extended conformation for the monomer, while leaving the remainder of the dendrimer intact. This extended conformation is reached through application of a penalty varying inversely with distance, coupled with a significantly larger penalty for each close contact.
Each dendron structure was built using four different residue types: a central ethylendiamine (EDA), a repetitive polyamidoamine (PAA), a terminal polyamidoamine (PAH), and a terminal benzylpenicilloyl group (BPO). The missing bonds, angle torsions, or van der Waals parameters not included in the parm99 force field were transferred from the general AMBER force field (GAFF) [12]. These residue types were optimized and subjected to a conformational analysis using TINKER-Software Tools for Molecular Design [13]. The conformation of minimum energy was submitted to MP2/6-31G* calculation to obtain the electrostatic potentials using G09 [14]. Next, the restrained potential (RESP) method was used for charge fitting [15]. Finally, the residue was created using the Antechamber module of AmberTools (version 1.5) [16].
Molecular Dynamics
Molecular dynamics simulations were performed with the program NAMD2 (version 2.6b1) [17,18], using the AMBER force field which has previously been successfully applied in past computational studies for the description of PAMAM dendrimers [19-21]. All simulations were run using a timestep of 2 fs, a cutoff of 10 Å with a switching function 8 Å, and the Langevin thermostat. Full periodic boundary conditions were used and to treat the long-range electrostatic interactions the particle-mesh Ewald (PME) algorithm was employed [22]. The PME algorithm used a tolerance of 1 × 10−6 Å and an interpolation order of 4. Unit cell sizes were chosen to ensure that the number of points along any dimension in the PME grid was composed of factors of 2, 3 and 5. Bond lengths involving bonds to hydrogen atoms were constrained using the SHAKE algorithm [23]. Water molecules were modeled using the TIP3P model [24].
At the first step, the dendrimers were subjected to a conjugate gradient energy minimization (100,000 steps) in vacuum using a relative dielectric constant of 1. In the second stage, to preserve the overall charge neutrality and to represent more realistic conditions, an appropriate number of Na+ counterions were added, and the molecules were then hydrated in cubic cells the dimension of which was chosen to provide at least a 10 Å solvation shell around the dendrimer structure. These systems were minimized (100,000 steps) and followed by four cycles of constant volume, constant temperature ensemble (NVT) simulated annealing, each approx. 2 ns long (100 ps of heating to 600 K, followed by 1.9 ns of cooling to 300 K). The third stage of the simulation protocol involved an equilibration period of at least 2 ns using a constant temperature and constant pressure ensemble (NPT, p = 1 atm, T = 300 K) [25]. Finally, starting from the configurations produced by the above procedure, production runs of 14–15 ns trajectories were performed under an NPT ensemble. All of the molecular dynamics simulations were carried out working in parallel on 32 processors of the IBM calculation cluster of the Supercomputing and Bioinformatic Center of the University of Malaga.
4. Conclusions
The shape that a target epitope adopts on the surface of a carrier macromolecule is a key aspect in the process of molecular recognition by specific ligands. The preparation of the GnBPO dendrimers described here, as artificial dendritic antigens recognized by specific IgEs from sera of patients allergic to Benzylpenicillin, enables us to study how these penicilloyl residues (BPO) become exposed to interact with IgEs at the surface of dendrimers. The evidence from NMR is well supported by fully atomistic Molecular Dynamic Simulations. From the NOESY data, the thiazolidine moiety of the BPO residue is projected to the outer space of the dendritic domain, whereas the benzyl acyl side chain mainly interacts with the branches of the dendrimer, in an inner location. These dendrimers have a more polymeric-like structure than the PAMAM dendrimers, and the aspect ratio of the dendrimers is best defined as oblate ellipsoid, except for the higher generation. These macromolecules are flexible entities, where the water and the counterions can significantly penetrate the dendrimer.
These studies are of importance to understand further how the Human Immunological System can interact with dendritic structures.












| Position | 1H NMR δ (ppm) | 13C NMR δ (ppm) | Correlations HMBC (J2, J3) | Correlations H2BC (J2) |
|---|---|---|---|---|
| 2-CH3 (α) | 1.21 (s, 3H) | 26.7 | C-2, C-3 | C-2 |
| 2-CH3 (β) | 1.56 (s, 3H) | 27.1 | C-2, C-3 | C-2 |
| 3 | 3.44 (s, 1H) | 74.4 | CH3(α/β), C-2, C-5 | C-2 |
| 5 | 4.86 (d, 1H, J = 9.6 Hz) | 64.6 | C-3, C-6, C-7 | C-6 |
| 6 | 4.25 (d, 1H, J = 9.6 Hz) | 59.3 | C-5, C-7, C-13 | C-5 |
| 9 | 2.75 (m, 2H) | 39.0 | C-3, C-6, C-7 | C-10 |
| 10 | 1.026 (m, 2H) | 30.3 | C-9, C-11, C-12 | C-11 |
| 11 | 0.86 (m, 2H) | 19.3 | C-9, C-10, C-12 | C-10, C-12 |
| 12 | 0.48 (t, 3H, J = 7.4 Hz) | 13.0 | C-10, C-11 | C-11 |
| 14 | 3.62 (d, J = 20 Hz) | 42.0 | C-13, Ph |
| Dendritic-Antigens | Yield | Temperature Reaction | Reaction time |
|---|---|---|---|
| G0BPO4 | 73% | 20–23 °C | 1 day |
| G1BPO8 | 70% | 20–23 °C | 2 days |
| G2BPO16 | 80% | 4 °C | 4 days |
| G3BPO32 | 70% | 4 °C | 5 days |
| G4BPO64 | 82% | 4 °C | 6 days |
| Kind of H | Ph | H-5 | H-6 |  | H-3 | CH3 | CH3 | |
|---|---|---|---|---|---|---|---|---|
| δ (ppm) | 7.28 | 4.88 | 4.26 | 3.61 | 3.43 | 1.54 | 1.20 | |
| description | m | d (J = 8.5 Hz) | d (J = 8.5 Hz) | s | s | s | s | |
| G0BPO4 | N° of hydrogens | 20 H | 4 H | 4 H | 8 H | 4 H | 12 H | 12 H |
| G1BPO8 | 40 H | 8 H | 8 H | 16 H | 8 H | 24 H | 24 H | |
| G2BPO16 | 80 H | 16 H | 16 H | 32 H | 16 H | 48 H | 48 H | |
| G3BPO32 | 160 H | 32 H | 32 H | 64 H | 32 H | 96 H | 96 H | |
| G4BPO64 | 320 H | 64 H | 64 H | 128 H | 64 H | 192 H | 192 H | |
| Assignment/number of H | ||||
|---|---|---|---|---|
| Kind of H |  |  | 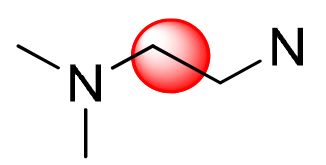 | 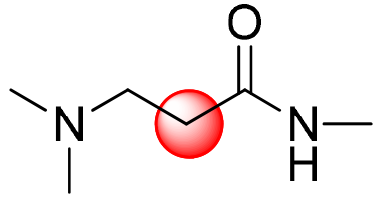 |
| Range of δ (ppm)/description | 3.30–3.08 (m) | 2.79–2.57 (m) | 2.55–2.40 (m) | 2.32–2.17 (m) |
| G0BPO4 | d, e | b | a | c |
| 16 H | 8 H | 4 H | 8 H | |
| G1BPO8 | d, h, i | b, f | a, e | c, g |
| 40 H | 24 H | 12 H | 24 H | |
| G2BPO16 | d, h, l, m | b, f, j | a, e, i | c, g, k |
| 88 H | 56 H | 28 H | 56 H | |
| G3BPO32 | d, h, l, p, q | b, f, j, n | a, e, i, m | c, g, k, o |
| 184 H | 120 H | 60 H | 120 H | |
| G4BPO64 | d, h, l, p, t, u | b, f, j, n, r | a, e, i, m, q | c, g, k, o, s |
| 376 H | 248 H | 124 H | 148 H | |
| δ range (ppm) | Kind of Carbon |
|---|---|
| 176.0–173.5 |  |
| 173.0–171.0 | C3-CO2−Na+ |
| 136.0–135.0 | |
| 130.2–129.5 | |
| 129.9–128.5 | Aromatic |
| 127.6–128.3 | |
| 75.7–74.4 | C-3 |
| 66.9–64.6 | C-5 |
| 60.3–59.6 | C-6 |
| 60.2–59.5 | C-2 |
| 52.2–50.8 | N-CH2-CH2-N |
| 50.0–49.3 | NHCO-CH2-CH2-N |
| 43.0–42.4 | Ph-CH2 |
| 39.6–38.5 | CH2-NH-CO |
| 33.7–32.8 | CH2-CO-NH |
| 28.3–26.6 | (CH3)2-C2 |
| Dendrimer | Nden | NNa+ | Nwater | Ntotal | box (Å) | V(Å3) | Length of the simulation (ns) |
|---|---|---|---|---|---|---|---|
| G0BPO4 | 244 | 4 | 12,276 | 12,524 | 59 × 51 × 55 | 165,495 | 15.0 |
| G1BPO8 | 548 | 8 | 20,550 | 21,106 | 60 ×71 ×64 | 272,640 | 15.0 |
| G2BPO16 | 1,156 | 16 | 45,294 | 46,466 | 87 × 81 × 82 | 577,854 | 15.0 |
| G3BPO32 | 1,372 | 32 | 79,317 | 80,721 | 96 × 98 ×105 | 987,840 | 15.0 |
| G4BPO64 | 4,804 | 64 | 123,630 | 128,498 | 124 × 110 × 113 | 1,541,320 | 14.2 |
| Dendrimer | Rg (Å) | Iz/Ix | Iz/Iy | δ |
|---|---|---|---|---|
| G0BPO4 | 8.2 (4.00) | 2.2 | 1.3 | 0.052 |
| G1BPO8 | 10.4 (7.90) | 1.9 | 1.2 | 0.032 |
| G2BPO16 | 14.2 (11.80) | 2.4 | 1.2 | 0.054 |
| G3BPO32 | 17.8 (15.09) | 1.8 | 1.1 | 0.025 |
| G4BPO64 | 21.9 (18.60) | 1.2 | 1.2 | 0.005 |
Acknowledgments
We gratefully acknowledge the computer resources, technical expertise and assistance provided by the SCBI (Supercomputing and Bioinformatics) center of the University of Malaga. Grants from the Ministerio de Ciencia e Innovacion-Spain (CTQ2010-20303) and the Junta de Andalucía (PI-0551/2009) are greatly acknowledged. The authors also thank to the EU Framework Program COST Action TD0802 “Dendrimers in Biomedical Applications”. M. I. M. thanks ISCIII for Sara Borrell Post-Doc grant. The authors thank Ian Johnstone for help with the English language.
References
- Mammen, M.; Chio, S.-K.; Whitesides, G.M. Polyvalent interactions in biological systems: implications for design and use of multivalent ligands and inhibitors. Angew. Chem. Int. Ed. 1998, 37, 2755–2794. [Google Scholar]
- Sánchez-Sancho, F.; Pérez-Inestrosa, E.; Suau, R.; Mayorga, C.; Torres, M.J.; Blanca, M. Dendrimers as carrier protein mimetics for IgE antibody recognition. Synthesis and characterization of densely penicilloylated dendrimers. Bioconjugate Chem. 2002, 13, 647–653. [Google Scholar]
- Montañez, M.I.; Perez-Inestrosa, E.; Suau, R.; Mayorga, C.; Torres, M.J.; Blanca, M. Dendrimerized cellulose as a scaffold for artificial antigens with applications in drug allergy diagnosis. Biomacromolecules 2008, 9, 1461–1466. [Google Scholar]
- Chai, M.; Niu, Y.; Youngs, W.J.; Rinaldi, P.L. Structure and conformation of DAB dendrimers in solution via multidimensional NMR techniques. J. Am. Chem. Soc. 2001, 123, 4670–4678. [Google Scholar]
- Naylor, A.M.; Goddard, W.A.; Kiefer, G.E.; Tomalia, D.A. Starburst dendrimers. 5. Molecular shape control. J. Am. Chem. Soc. 1989, 111, 2339–2341. [Google Scholar]
- Dalakoglou, G.K.; Karatasos, K.; Lyulin, S.V.; Lyulin, A.V. Effects of topology and size on statics and dynamics of complexes of hyperbranched polymers with linear polyelectrolytes. J. Chem. Phys. 2007, 127, 214903. [Google Scholar]
- Rathgeber, S.; Monkenbusch, M.; Kreitschmann, M.; Urban, V.; Brulet, A. Dynamics of star-burst dendrimers in solution in relation to their structural properties. J. Chem. Phys. 2002, 117, 4047–4062. [Google Scholar]
- Muratt, M.; Grest, G.S. Molecular dynamics study of dendrimer molecules in solvents of varying quality. Macromolecules 1996, 29, 1278–1285. [Google Scholar]
- Rudnick, J.; Gaspari, G. The asphericity of random walks. J. Phys. A: Math. Gen. 1986, 19, L191–L193. [Google Scholar]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD—Visual molecular dynamics. J. Mol. Graphics 1996, 14, 33–38. [Google Scholar]
- Chalmers, D.K.; Roberts, B.P. Silico: A Perl Molecular Toolkit. Available online: http://silico.sourceforge.net (accessed on 28 June 2011).
- Wang, J.; Wang, W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2005, 25, 1157–1174. [Google Scholar]
- Tinker Molecular Modeling Package, v. 4.2. Available online: http://dasher.wustl.edu/tinker/ (accessed on 28 June 2011).
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; Nakatsuji, H.; Caricato, M.; Li, X.; Hratchian, H.P.; Izmaylov, A.F.; Bloino, J.; Zheng, G.; Sonnenberg, J.L.; Hada, M.; Ehara, M.; Toyota, K.; Fukuda, R.; Hasegawa, J.; Ishida, M.; Nakajima, T.; Honda, Y.; Kitao, O.; Nakai, H.; Vreven, T.; Montgomery, J.A., Jr.; Peralta, J.E.; Ogliaro, F.; Bearpark, M.; Heyd, J.J.; Brothers, E.; Kudin, K.N.; Staroverov, V.N.; Kobayashi, R.; Normand, J.; Raghavachari, K.; Rendell, A.; Burant, J.C.; Iyengar, S.S.; Tomasi, J.; Cossi, M.; Rega, N.; Millam, J.M.; Klene, M.; Knox, J.E.; Cross, J.B.; Bakken, V.; Adamo, C.; Jaramillo, J.; Gomperts, R.; Stratmann, R.E.; Yazyev, O.; Austin, A.J.; Cammi, R.; Pomelli, C.; Ochterski, J.W.; Martin, R.L.; Morokuma, K.; Zakrzewski, V.G.; Voth, G.A.; Salvador, P.; Dannenberg, J.J.; Dapprich, S.; Daniels, A.D.; Farkas, O.; Foresman, J.B.; Ortiz, J.V.; Cioslowski, J.; Fox, D.J. Gaussian 09, Revision, A. L.; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Bayly, C.I.; Cleplak, P.; Cornell, W.D.; Kollman, P.A. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: The RESP model. J. Phys. Chem. 1993, 97, 10269–10280. [Google Scholar]
- Case, D.A.; Darden, T.A.; Cheatham, T.E., III; Simmerling, C.L.; Wang, J.; Duke, R.E.; Luo, R.; Walker, R.C.; Zhang, W.; Merz, K.M.; Roberts, B.; Wang, B.; Hayik, S.; Roitberg, A.; Seabra, G.; Kolossvai, I.; Wong, K.F.; Paesani, F.; Vanicek, J.; Liu, J.; Wu, X.; Brozell, S.R.; Steinbrecher, T.; Gohlke, H.; Cai, Q.; Ye, X.; Wang, J.; Hsieh, M.-J.; Cui, G.; Roe, D.R.; Mathews, D.H.; Seetin, M.G.; Sagui, C.; Babin, V.; Luchko, T.; Gusarov, S.; Kovalenko, A.; Kollman, P.A. AMBER 11; University of California: San Francisco, CA, USA, 2010. [Google Scholar]
- Kale, L.; Skeel, R.; Bhandarkar, M.; Brunner, R.; Gursoy, A.; Krawetz, N.; Phillips, J.; Shinozaki, A.; Varadarajan, K.; Schulten, K. NAMD2: Greater scalability for parallel molecular dynamics. J. Comput. Phys. 1999, 151, 283–312. [Google Scholar]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kale, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar]
- Tanis, I.; Karatasos, K. Association of a weakly acidic anti-inflammatory drug (Ibuprofen) with a poly(amidoamine) dendrimer as studied by molecular dynamics simulations. J. Phys. Chem. B 2009, 113, 10984–10993. [Google Scholar]
- Posocco, P.; Ferrone, M.; Fermeglia, M.; Pricl, S. Binding at the core. Computational study of structural and ligand binding properties of naphthyridine-based dendrimers. Macromolecules 2007, 40, 2257–2266. [Google Scholar]
- Lee, H.; Baker, J.R.; Larson, R.G. Molecular dynamics studies of the size, shape, and internal structure of 0% and 90% Acetylated fifth-generation polyamidoamine dendrimers in water and methanol. J. Phys. Chem. B 2006, 110, 4014–4019. [Google Scholar]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N -log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar]
- Ryckaert, J.-P.; Ciccotti, G.; Berendsen, H.J.C. Numerical integration of the cartesian equations of motion of a system with Constraints: Molecular dynamics of n-Alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar]
- Roberts, B.P.; Scanlon, M.J.; Krippner, G.Y.; Chalmers, D.K. Molecular dynamics of poly(L-lysine) dendrimers with naphthalene disulfonate caps. Macromolecules 2009, 42, 2775–2783. [Google Scholar]
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Montañez, M.I.; Najera, F.; Perez-Inestrosa, E. NMR Studies and Molecular Dynamic Simulation of Synthetic Dendritic Antigens. Polymers 2011, 3, 1533-1553. https://doi.org/10.3390/polym3031533
Montañez MI, Najera F, Perez-Inestrosa E. NMR Studies and Molecular Dynamic Simulation of Synthetic Dendritic Antigens. Polymers. 2011; 3(3):1533-1553. https://doi.org/10.3390/polym3031533
Chicago/Turabian StyleMontañez, Maria Isabel, Francisco Najera, and Ezequiel Perez-Inestrosa. 2011. "NMR Studies and Molecular Dynamic Simulation of Synthetic Dendritic Antigens" Polymers 3, no. 3: 1533-1553. https://doi.org/10.3390/polym3031533






