Molecular Modeling of PEGylated Peptides, Dendrimers, and Single-Walled Carbon Nanotubes for Biomedical Applications
Abstract
:1. Introduction
2. Development of All-Atom and Coarse-Grained PEO/PEG Force Fields

3. Simulations of PEGylated Peptides
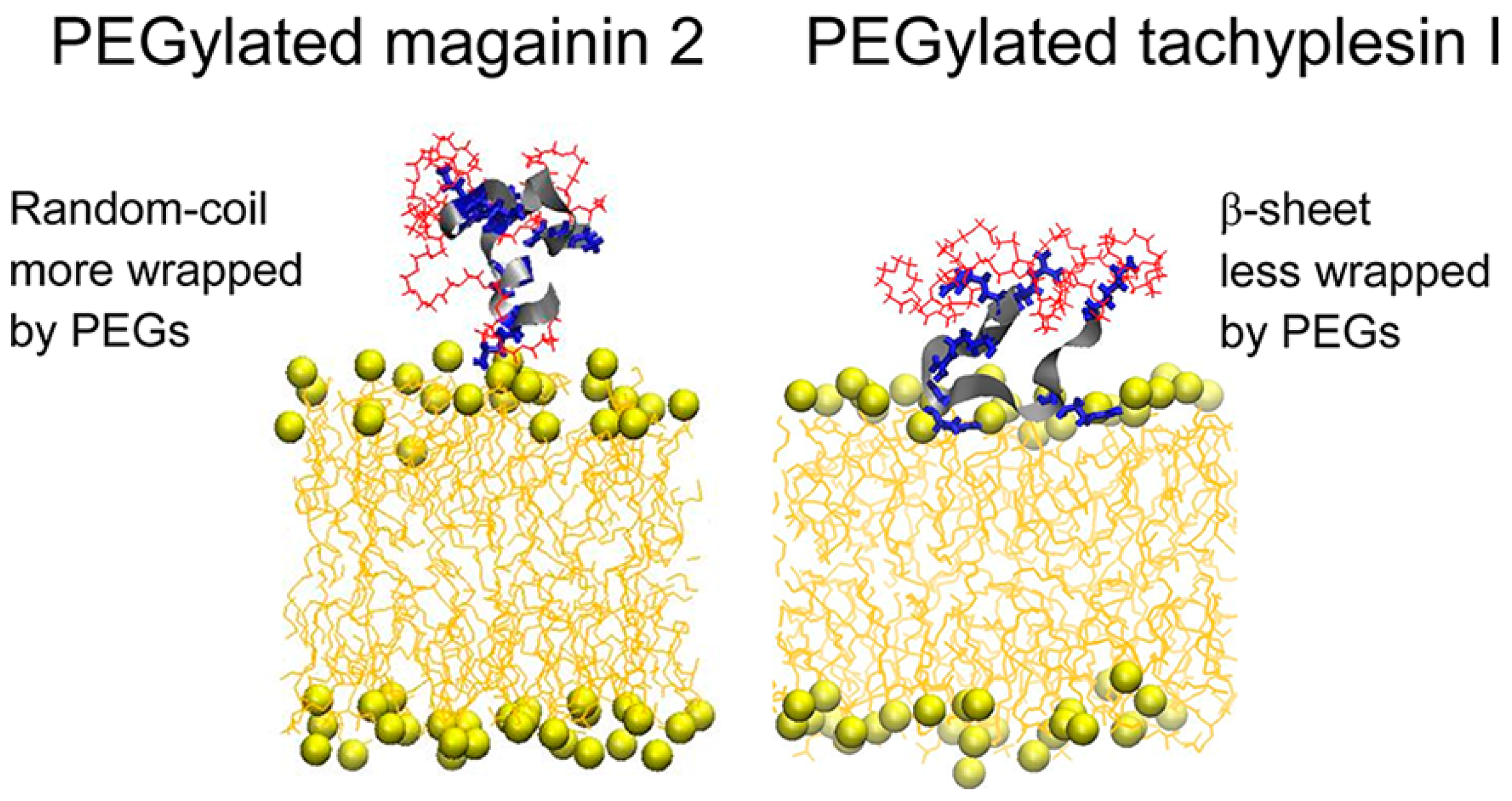
3.1. Antimicrobial Peptides
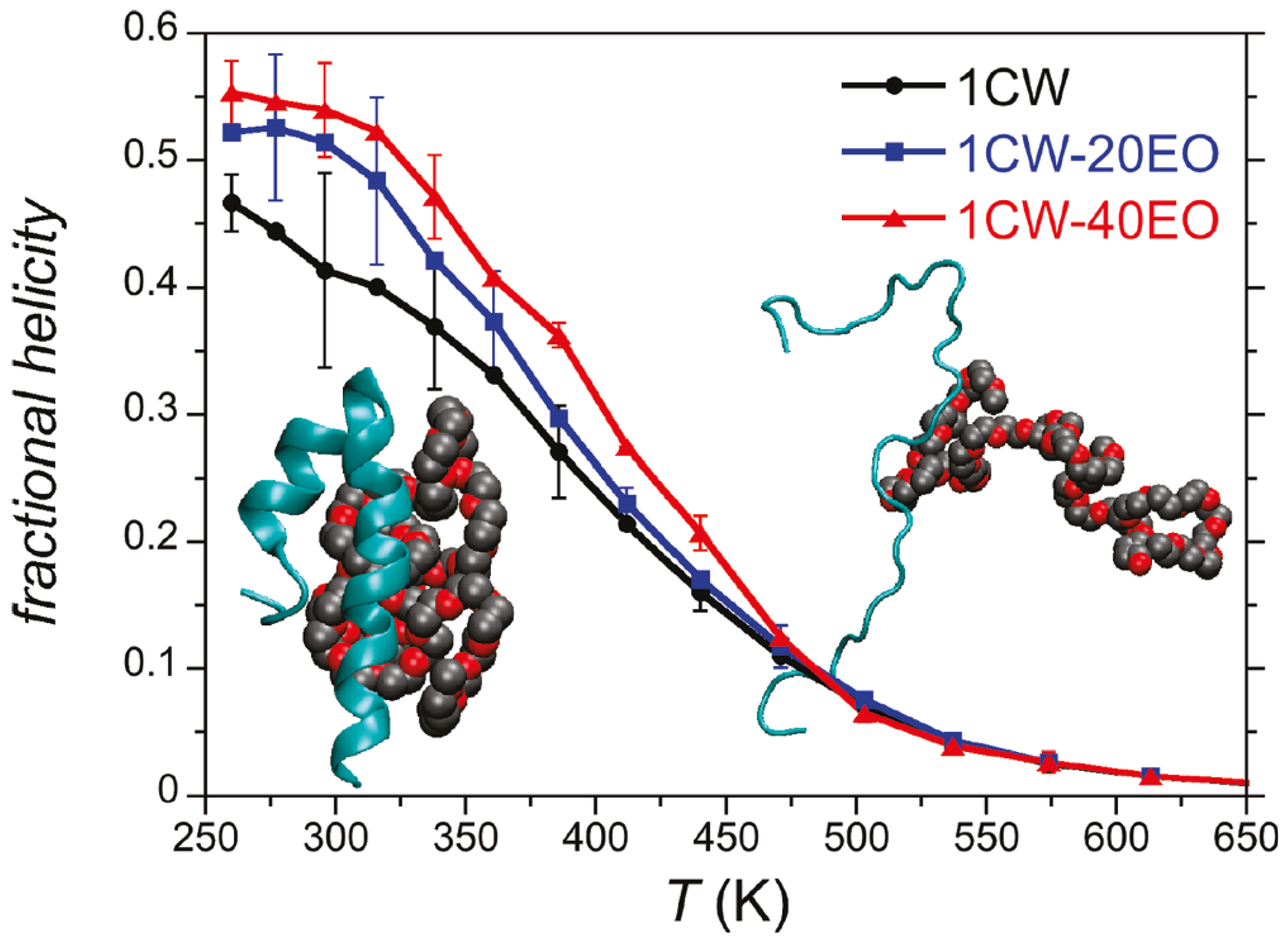
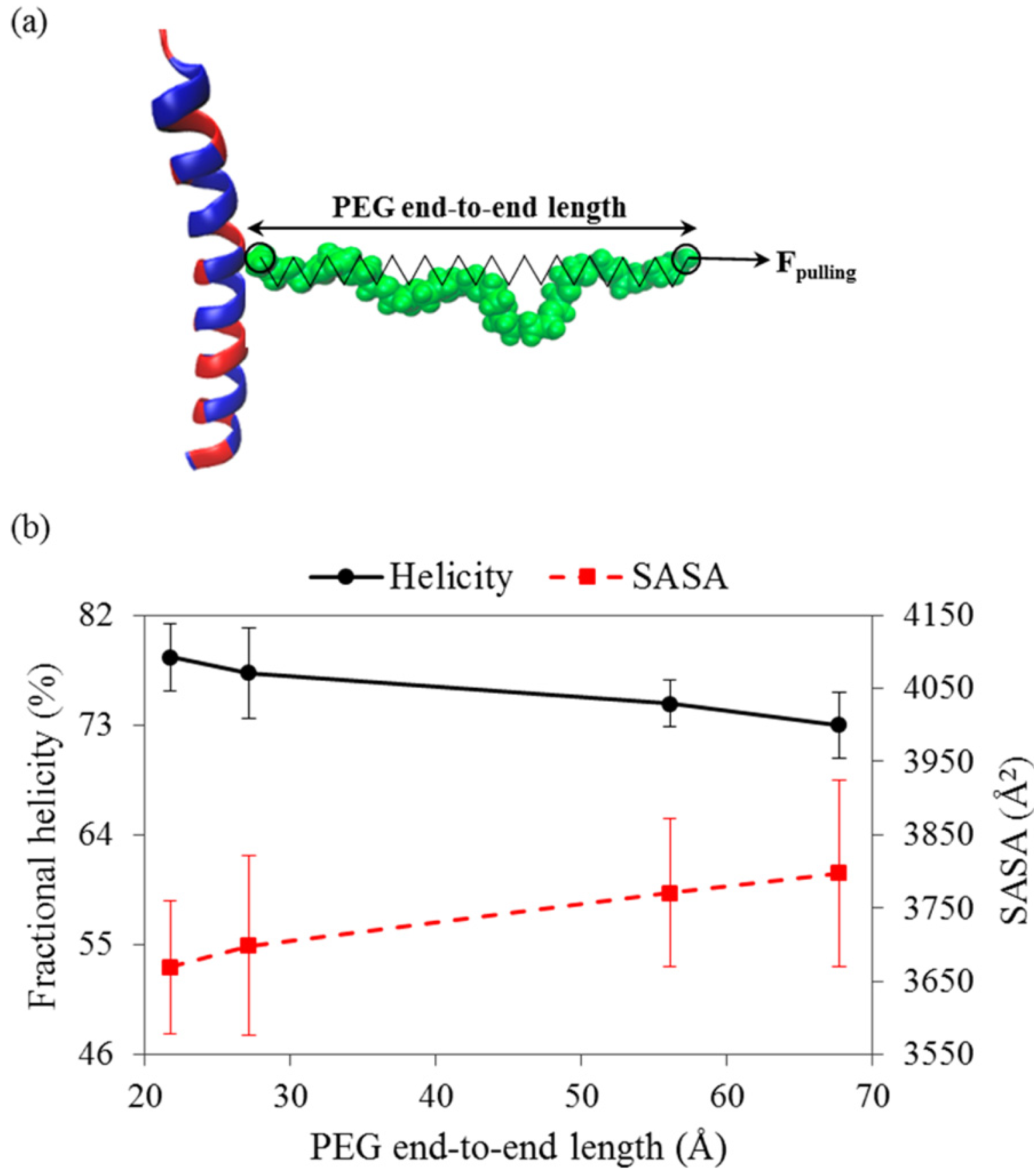
3.2. Coiled-Coil Peptides
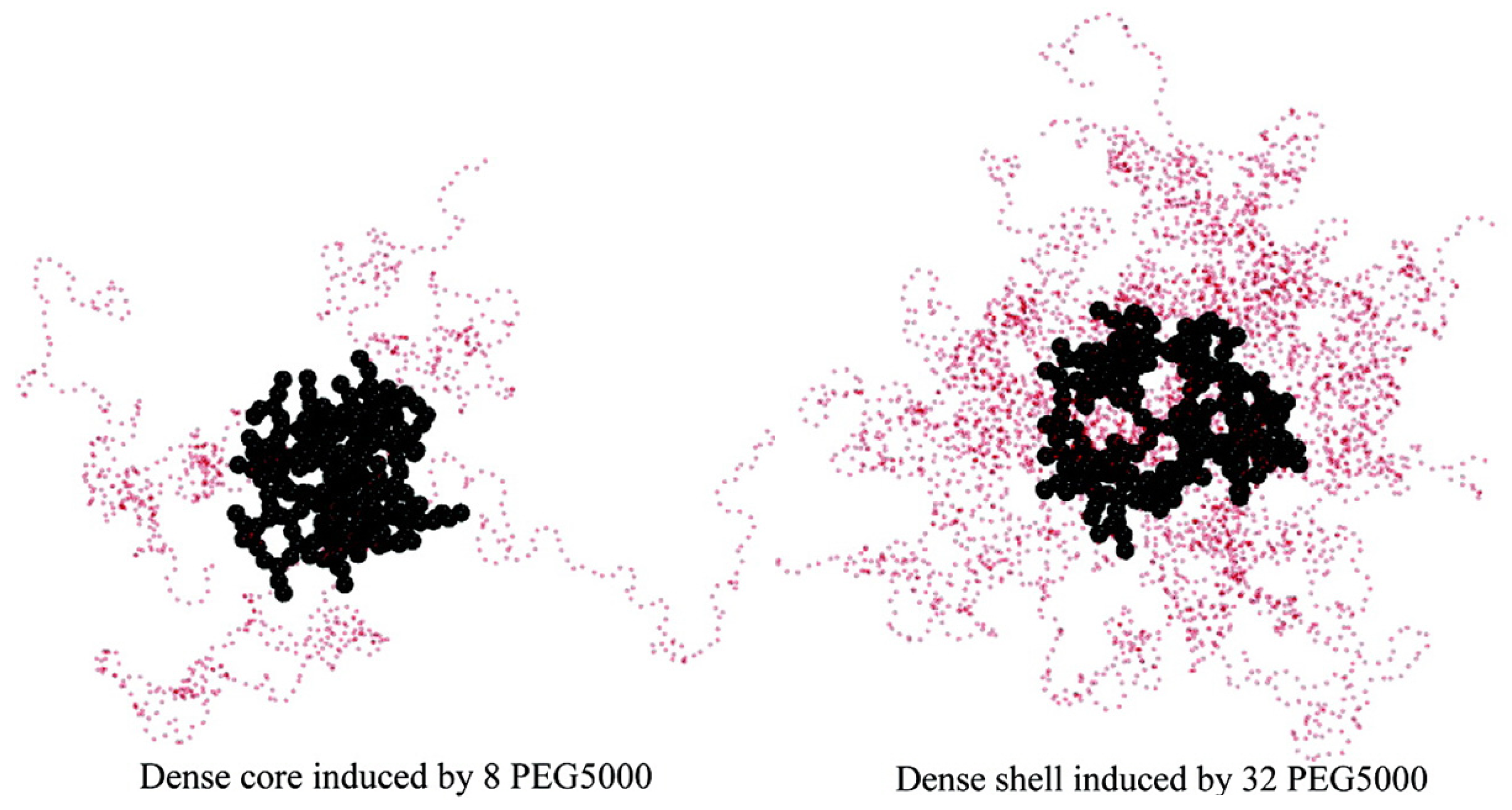
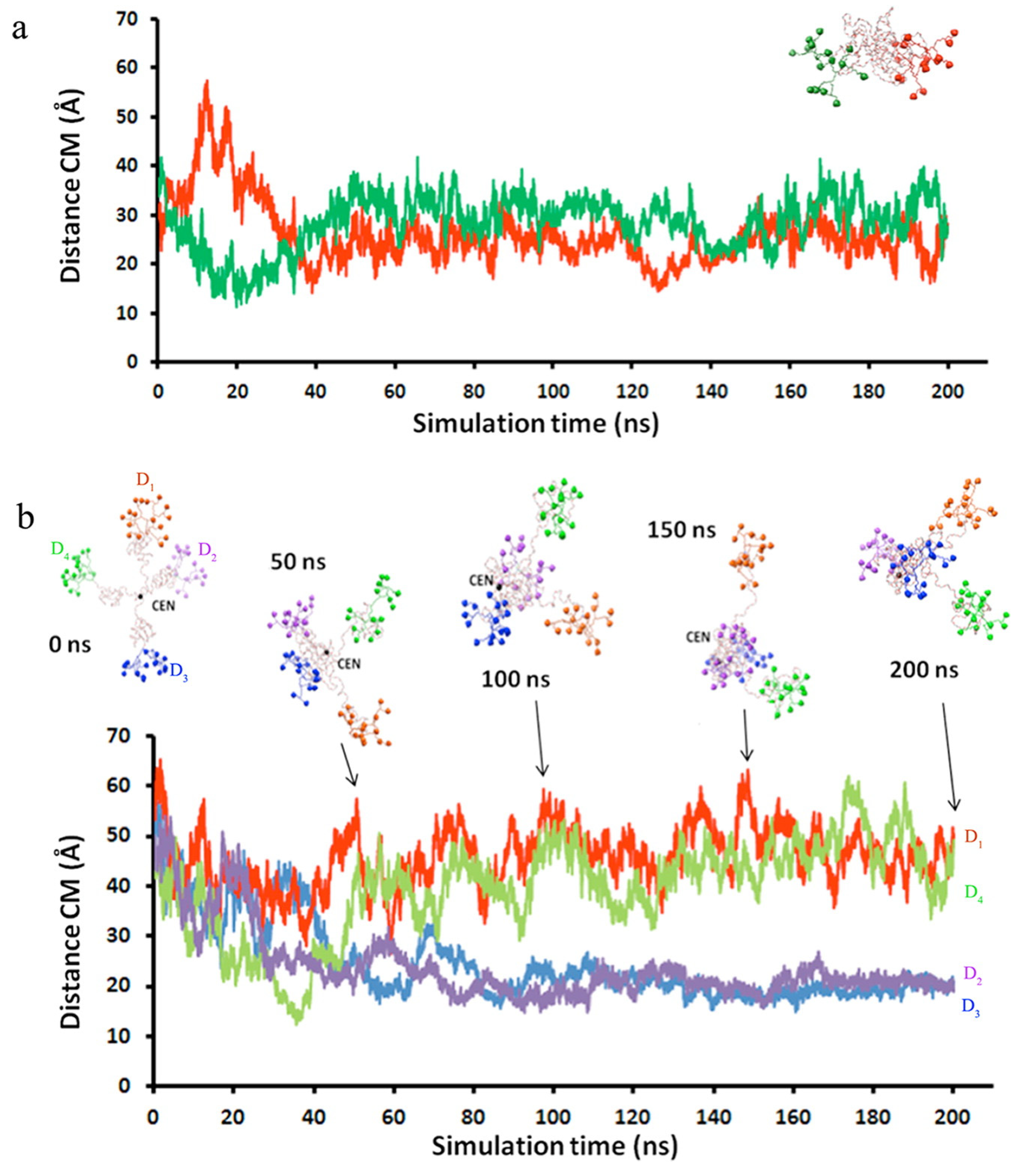
4. Simulations of PEGylated Dendrimers

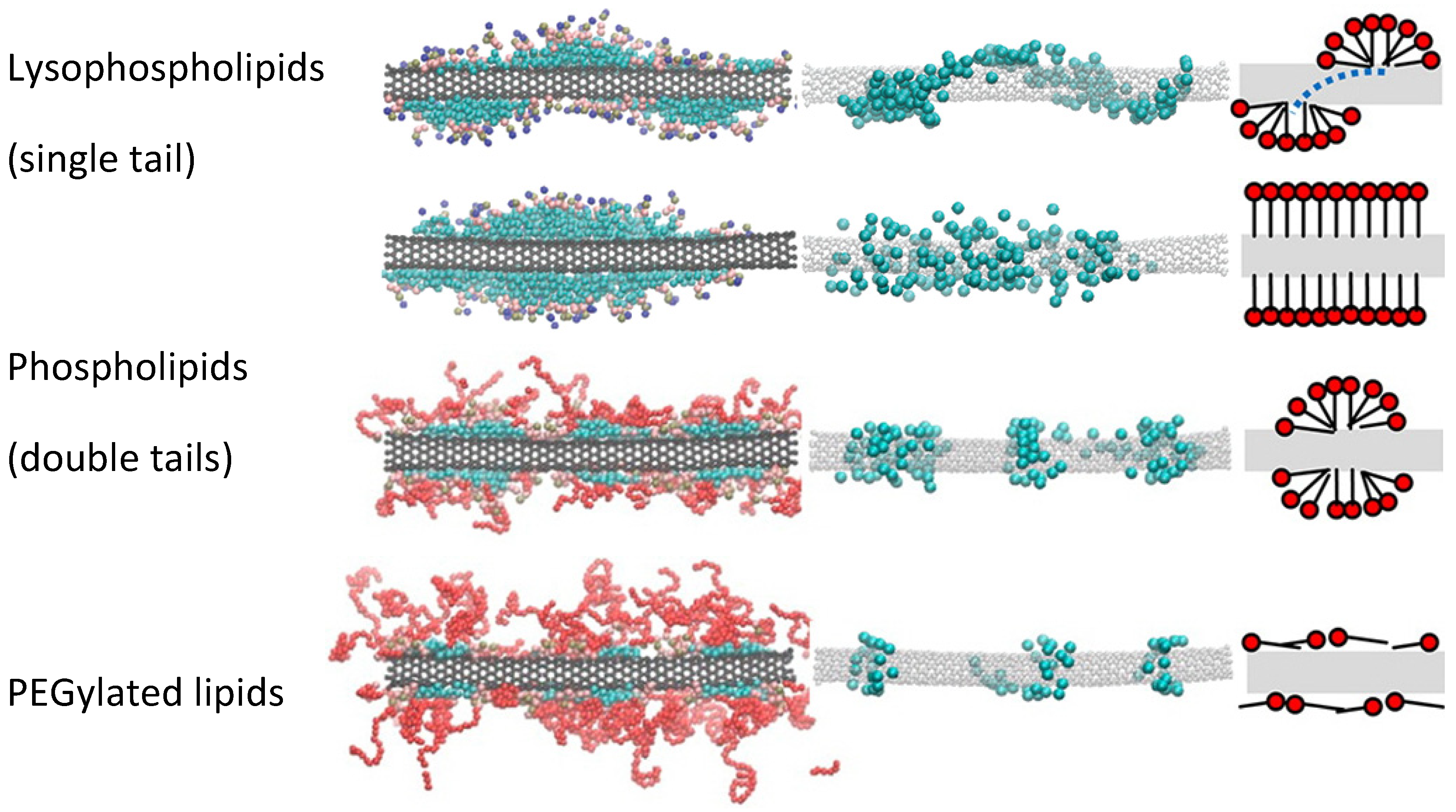
5. Simulations of PEGylated Carbon Nanotubes

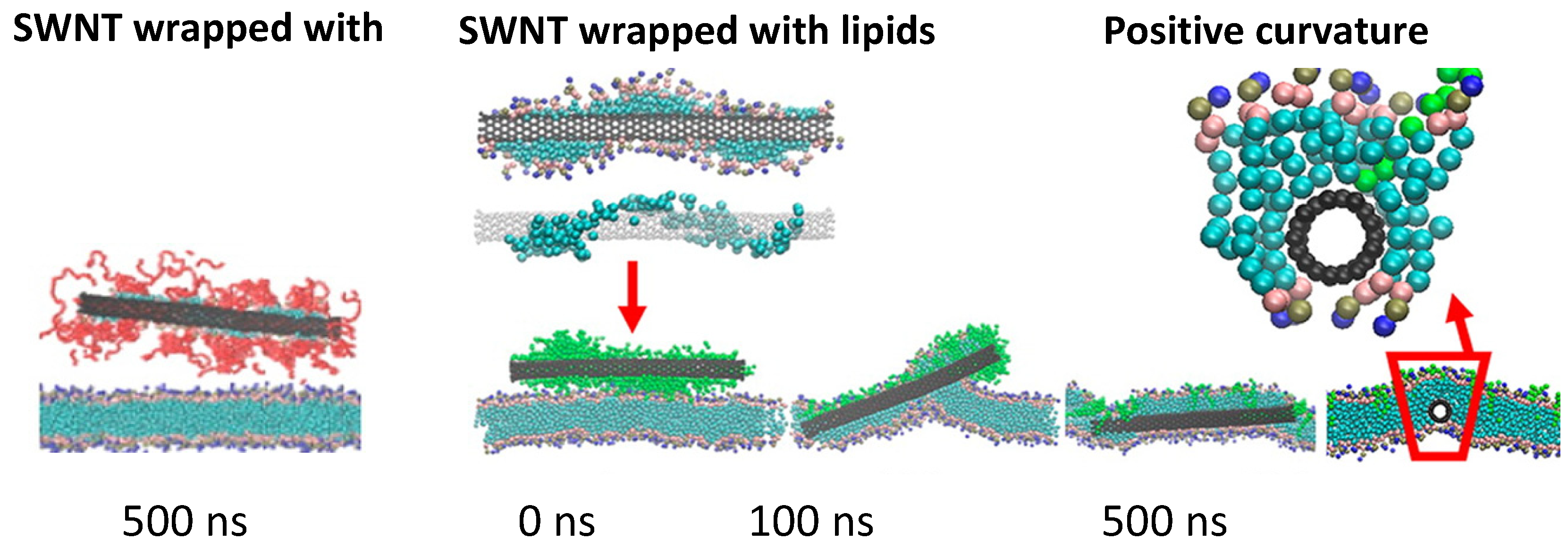
6. Conclusions
Acknowledgments
Conflicts of Interest
References
- Harris, J.M.; Martin, N.E.; Modi, M. PEGylation—A novel process for modifying pharmacokinetics. Clin. Pharmacokinet. 2001, 40, 539–551. [Google Scholar] [CrossRef]
- Harris, J.M.; Chess, R.B. Effect of PEGylation on pharmaceuticals. Nat. Rev. Drug Discov. 2003, 2, 214–221. [Google Scholar] [CrossRef]
- Imura, Y.; Nishida, M.; Ogawa, Y.; Takakura, Y.; Matsuzaki, K. Action mechanism of tachyplesin i and effects of PEGylation. Biochim. Biophys. Acta 2007, 1768, 1160–1169. [Google Scholar] [CrossRef]
- Imura, Y.; Nishida, M.; Matsuzaki, K. Action mechanism of PEGylated magainin 2 analogue peptide. Biochim. Biophys. Acta 2007, 1768, 2578–2585. [Google Scholar] [CrossRef]
- Jaschke, A.; Furste, J.P.; Nordhoff, E.; Hillenkamp, F.; Cech, D.; Erdmann, V.A. Synthesis and properties of oligodeoxyribonucleotide polyethylene-glycol conjugates. Nucleic Acids Res. 1994, 22, 4810–4817. [Google Scholar] [CrossRef]
- Jones, D.S.; Hachmann, J.P.; Osgood, S.A.; Hayag, M.S.; Barstad, P.A.; Iverson, G.M.; Coutts, S.M. Conjugates of double-stranded oligonucleotides with poly(ethylene glycol) and keyhole limpet hemocyanin—A model for treating systemic lupus-erythematosus. Bioconj. Chem. 1994, 5, 390–399. [Google Scholar] [CrossRef]
- Kabanov, A.V.; Vinogradov, S.V.; Suzdaltseva, Y.G.; Alakhov, V.Y. Water-soluble block polycations as carriers for oligonucleotide delivery. Bioconj. Chem. 1995, 6, 639–643. [Google Scholar] [CrossRef]
- Wang, S.; Lee, R.J.; Cauchon, G.; Gorenstein, D.G.; Low, P.S. Delivery of antisense oligodeoxyribonucleotides against the human epidermal growth-factor receptor into cultured kb cells with liposomes conjugated to folate via polyethylene-glycol. Proc. Natl. Acad. Sci. USA 1995, 92, 3318–3322. [Google Scholar] [CrossRef]
- Allen, T.M.; Hansen, C.; Martin, F.; Redemann, C.; Yauyoung, A. Liposomes containing synthetic lipid derivatives of poly(ethylene glycol) show prolonged circulation half-lives in vivo. Biochim. Biophys. Acta 1991, 1066, 29–36. [Google Scholar] [CrossRef]
- Allen, T.M.; Hansen, C. Pharmacokinetics of stealth versus conventional liposomes—Effect of dose. Biochim. Biophys. Acta 1991, 1068, 133–141. [Google Scholar] [CrossRef]
- Papahadjopoulos, D.; Allen, T.M.; Gabizon, A.; Mayhew, E.; Matthay, K.; Huang, S.K.; Lee, K.D.; Woodle, M.C.; Lasic, D.D.; Redemann, C.; et al. Sterically stabilized liposomes—Improvements in pharmacokinetics and antitumor therapeutic efficacy. Proc. Natl. Acad. Sci. USA 1991, 88, 11460–11464. [Google Scholar] [CrossRef]
- Klibanov, A.L.; Maruyama, K.; Torchilin, V.P.; Huang, L. Amphipathic polyethyleneglycols effectively prolong the circulation time of liposomes. FEBS Lett. 1990, 268, 235–237. [Google Scholar] [CrossRef]
- Torchilin, V.P.; Klibanov, A.L.; Huang, L.; Odonnell, S.; Nossiff, N.D.; Khaw, B.A. Targeted accumulation of polyethylene glycol-coated immunoliposomes in infarcted rabbit myocardium. FASEB J. 1992, 6, 2716–2719. [Google Scholar]
- Sawhney, A.S.; Pathak, C.P.; Hubbell, J.A. Bioerodible hydrogels based on photopolymerized poly(ethylene glycol)-co-poly(α-hydroxy acid) diacrylate macromers. Macromolecules 1993, 26, 581–587. [Google Scholar] [CrossRef]
- Burdick, J.A.; Anseth, K.S. Photoencapsulation of osteoblasts in injectable rgd-modified PEG hydrogels for bone tissue engineering. Biomaterials 2002, 23, 4315–4323. [Google Scholar] [CrossRef]
- Kim, Y.; Klutz, A.M.; Jacobson, K.A. Systematic investigation of polyamidoamine dendrimers surface-modified with poly(ethylene glycol) for drug delivery applications: Synthesis, characterization, and evaluation of cytotoxicity. Bioconj. Chem. 2008, 19, 1660–1672. [Google Scholar] [CrossRef]
- Kojima, C.; Kono, K.; Maruyama, K.; Takagishi, T. Synthesis of polyamidoamine dendrimers having poly(ethylene glycol) grafts and their ability to encapsulate anticancer drugs. Bioconj. Chem. 2000, 11, 910–917. [Google Scholar] [CrossRef]
- Luo, D.; Haverstick, K.; Belcheva, N.; Han, E.; Saltzman, W.M. Poly(ethylene glycol)-conjugated pamam dendrimer for biocompatible, high-efficiency DNA delivery. Macromolecules 2002, 35, 3456–3462. [Google Scholar] [CrossRef]
- Chun, D.; Wudl, F.; Nelson, A. Supramacromolecular assembly driven by complementary molecular recognition. Macromolecules 2007, 40, 1782–1785. [Google Scholar] [CrossRef]
- Bedrov, D.; Borodin, O.; Smith, G.D. Molecular dynamics simulations of 1,2-dimethoxyethane/water solutions: 1. Conformational and structural properties. J. Phys. Chem. B 1998, 102, 5683–5690. [Google Scholar] [CrossRef]
- Smith, G.D.; Bedrov, D.; Borodin, O. Molecular dynamics simulation study of hydrogen bonding in aqueous poly(ethylene oxide) solutions. Phys. Rev. Lett. 2000, 85, 5583–5586. [Google Scholar] [CrossRef]
- Bedrov, D.; Pekny, M.; Smith, G.D. Quantum-chemistry-based force field for 1,2-dimethoxyethane and poly(ethylene oxide) in aqueous solution. J. Phys. Chem. B 1998, 102, 996–1001. [Google Scholar] [CrossRef]
- Smith, G.D.; Yoon, D.Y.; Jaffe, R.L.; Colby, R.H.; Krishnamoorti, R.; Fetters, L.J. Conformations and structures of poly(oxyethylene) melts from molecular dynamics simulations and small-angle neutron scattering experiments. Macromolecules 1996, 29, 3462–3469. [Google Scholar] [CrossRef]
- Smith, G.D.; Borodin, O.; Bedrov, D. A revised quantum chemistry-based potential for poly(ethylene oxide) and its oligomers in aqueous solution. J. Comput. Chem. 2002, 23, 1480–1488. [Google Scholar] [CrossRef]
- Smith, G.D.; Bedrov, D.; Borodin, O. Conformations and chain dimensions of poly(ethylene oxide) in aqueous solution: A molecular dynamics simulation study. J. Am. Chem. Soc. 2000, 122, 9548–9549. [Google Scholar] [CrossRef]
- Dong, H.; Hyun, J.K.; Durham, C.; Wheeler, R.A. Molecular dynamics simulations and structural comparisons of amorphous poly(ethylene oxide) and poly(ethylenimine) models. Polymer 2001, 42, 7809–7817. [Google Scholar] [CrossRef]
- Fischer, J.; Paschek, D.; Geiger, A.; Sadowski, G. Modeling of aqueous poly(oxyethylene) solutions: 1. Atomistic simulations. J. Phys. Chem. B 2008, 112, 2388–2398. [Google Scholar] [CrossRef]
- Tritopoulou, E.A.; Economou, I.G. Molecular simulation of structure and thermodynamic properties of pure tri- and tetra-ethylene glycols and their aqueous mixtures. Fluid Phase Equilib. 2006, 248, 134–146. [Google Scholar] [CrossRef]
- Winger, M.; de Vries, A.H.; van Gunsteren, W.F. Force-field dependence of the conformational properties of α,ω-dimethoxypolyethylene glycol. Mol. Phys. 2009, 107, 1313–1321. [Google Scholar] [CrossRef]
- Neyertz, S.; Brown, D.; Thomas, J.O. Molecular dynamics simulation of crystalline poly(ethylene oxide). J. Chem. Phys. 1994, 101, 10064–10073. [Google Scholar] [CrossRef]
- Lin, B.; Boinske, P.T.; Halley, J.W. A molecular dynamics model of the amorphous regions of polyethylene oxide. J. Chem. Phys. 1996, 105, 1668–1681. [Google Scholar] [CrossRef]
- Vorobyov, I.; Anisimov, V.M.; Greene, S.; Venable, R.M.; Moser, A.; Pastor, R.W.; MacKerell, A.D. Additive and classical drude polarizable force fields for linear and cyclic ethers. J. Chem. Theory Comput. 2007, 3, 1120–1133. [Google Scholar] [CrossRef]
- Lee, H.; Venable, R.M.; MacKerell, A.D.; Pastor, R.W. Molecular dynamics studies of polyethylene oxide and polyethylene glycol: Hydrodynamic radius and shape anisotropy. Biophys. J. 2008, 95, 1590–1599. [Google Scholar] [CrossRef]
- Stepniewski, M.; Pasenkiewicz-Gierula, M.; Rog, T.; Danne, R.; Orlowski, A.; Karttunen, M.; Urtti, A.; Yliperttula, M.; Vuorimaa, E.; Bunker, A. Study of PEGylated lipid layers as a model for PEGylated liposome surfaces: Molecular dynamics simulation and langmuir monolayer studies. Langmuir 2011, 27, 7788–7798. [Google Scholar] [CrossRef]
- Bedrov, D.; Ayyagari, C.; Smith, G.D. Multiscale modeling of poly(ethylene oxide)–poly(propylene oxide)–poly(ethylene oxide) triblock copolymer micelles in aqueous solution. J. Chem. Theory Comput. 2006, 2, 598–606. [Google Scholar] [CrossRef]
- Fischer, J.; Paschek, D.; Geiger, A.; Sadowski, G. Modeling of aqueous poly(oxyethylene) solutions: 2. Mesoscale simulations. J. Phys. Chem. B 2008, 112, 13561–13571. [Google Scholar] [CrossRef]
- Chen, T.; Hynninen, A.P.; Prud'homme, R.K.; Kevrekidis, I.G.; Panagiotopoulos, A.Z. Coarse-grained simulations of rapid assembly kinetics for polystyrene-b-poly(ethylene oxide) copolymers in aqueous solutions. J. Phys. Chem. B 2008, 112, 16357–16366. [Google Scholar]
- Srinivas, G.; Shelley, J.C.; Nielsen, S.O.; Discher, D.E.; Klein, M.L. Simulation of diblock copolymer self-assembly, using a coarse-grain model. J. Phys. Chem. B 2004, 108, 8153–8160. [Google Scholar] [CrossRef]
- Srinivas, G.; Klein, M.L. Coarse-grain molecular dynamics simulations of diblock copolymer surfactants interacting with a lipid bilayer. Mol. Phys. 2004, 102, 883–889. [Google Scholar] [CrossRef]
- Srinivas, G.; Discher, D.E.; Klein, M.L. Self-assembly and properties of diblock copolymers by coarse-grain molecular dynamics. Nat. Mater. 2004, 3, 638–644. [Google Scholar] [CrossRef]
- Marrink, S.J.; Risselada, H.J.; Yefimov, S.; Tieleman, D.P.; de Vries, A.H. The martini force field: Coarse grained model for biomolecular simulations. J. Phys. Chem. B 2007, 111, 7812–7824. [Google Scholar] [CrossRef]
- Marrink, S.J.; de Vries, A.H.; Mark, A.E. Coarse grained model for semiquantitative lipid simulations. J. Phys. Chem. B 2004, 108, 750–760. [Google Scholar] [CrossRef]
- Lee, H.; de Vries, A.H.; Marrink, S.J.; Pastor, R.W. A coarse-grained model for polyethylene oxide and polyethylene glycol: Conformation and hydrodynamics. J. Phys. Chem. B 2009, 113, 13186–13194. [Google Scholar] [CrossRef]
- Lee, H.; Pastor, R.W. Coarse-grained model for PEGylated lipids: Effect of PEGylation on the size and shape of self-assembled structures. J. Phys. Chem. B 2011, 115, 7830–7837. [Google Scholar] [CrossRef]
- Lee, H.; Kim, H.R.; Larson, R.G.; Park, J.C. Effects of the size, shape, and structural transition of thermosensitive polypeptides on the stability of lipid bilayers and liposomes. Macromolecules 2012, 45, 7304–7312. [Google Scholar] [CrossRef]
- Rossi, G.; Fuchs, P.F.J.; Barnoud, J.; Monticelli, L. A coarse-grained martini model of polyethylene glycol and of polyoxyethylene alkyl ether surfactants. J. Phys. Chem. B 2012, 116, 14353–14362. [Google Scholar] [CrossRef]
- Wang, Q.; Keffer, D.J.; Nicholson, D.M. A coarse-grained model for polyethylene glycol polymer. J. Chem. Phys. 2011, 135, 214903:1–214903:10. [Google Scholar]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef]
- Matsuzaki, K. Control of cell selectivity of antimicrobial peptides. Biochim. Biophys. Acta 2009, 1788, 1687–1692. [Google Scholar] [CrossRef]
- Matsuzaki, K. Why and how are peptide-lipid interactions utilized for self-defense? Magainins and tachyplesins as archetypes. Biochim. Biophys. Acta 1999, 1462, 1–10. [Google Scholar] [CrossRef]
- Guiotto, A.; Pozzobon, M.; Canevari, M.; Manganelli, R.; Scarin, M.; Veronese, F.M. PEGylation of the antimicrobial peptide nisin A: Problems and perspectives. Farmaco 2003, 58, 45–50. [Google Scholar] [CrossRef]
- Mátyus, E.; Kandt, C.; Tieleman, D.P. Computer simulation of antimicrobial peptides. Curr. Med. Chem. 2007, 14, 2789–2798. [Google Scholar] [CrossRef]
- Rzepiela, A.J.; Sengupta, D.; Goga, N.; Marrink, S.J. Membrane poration by antimicrobial peptides combining atomistic and coarse-grained descriptions. Faraday Discuss. 2010, 144, 431–443. [Google Scholar] [CrossRef]
- Leontiadou, H.; Mark, A.E.; Marrink, S.J. Antimicrobial peptides in action. J. Am. Chem. Soc. 2006, 128, 12156–12161. [Google Scholar] [CrossRef]
- Wu, X.; Chang, H.; Mello, C.; Nagarajan, R.; Narsimhan, G. Effect of interaction with coesite silica on the conformation of cecropin p1 using explicit solvent molecular dynamics simulation. J. Chem. Phys. 2013. [Google Scholar] [CrossRef]
- Han, E.; Lee, H. Effects of PEGylation on the binding interaction of magainin 2 and tachyplesin I with lipid bilayer surface. Langmuir 2013, 29, 14214–14221. [Google Scholar] [CrossRef]
- Lupas, A.N.; Gruber, M. The Structure of Alpha-Helical Coiled Coils. In Fibrous Proteins: Coiled-Coils, Collagen and Elastomers; Elsevier Academic Press Inc.: San Diego, CA, USA, 2005; Volume 70, pp. 37–78. [Google Scholar]
- Woolfson, D.N. The Design of Coiled-Coil Structures and Assemblies. In Fibrous Proteins: Coiled-Coils, Collagen and Elastomers; Elsevier Academic Press Inc.: San Diego, CA, USA, 2005; Volume 70, pp. 79–112. [Google Scholar]
- Gruber, M.; Lupas, A.N. Historical review: Another 50th anniversary—New periodicities in coiled coils. Trends Biochem. Sci. 2003, 28, 679–685. [Google Scholar] [CrossRef]
- Woolfson, D.N.; Ryadnov, M.G. Peptide-based fibrous biomaterials: Some things old, new and borrowed. Curr. Opin. Chem. Biol. 2006, 10, 559–567. [Google Scholar] [CrossRef]
- Woolfson, D.N.; Mahmoud, Z.N. More than just bare scaffolds: Towards multi-component and decorated fibrous biomaterials. Chem. Soc. Rev. 2010, 39, 3464–3479. [Google Scholar] [CrossRef]
- Deacon, S.P.E.; Apostolovic, B.; Carbajo, R.J.; Schott, A.K.; Beck, K.; Vicent, M.J.; Pineda-Lucena, A.; Klok, H.A.; Duncan, R. Polymer coiled-coil conjugates: Potential for development as a new class of therapeutic “molecular switch”. Biomacromolecules 2011, 12, 19–27. [Google Scholar] [CrossRef]
- Vandermeulen, G.W.M.; Tziatzios, C.; Duncan, R.; Klok, H.A. PEG-based hybrid block copolymers containing α-helical coiled coil peptide sequences: Control of self-assembly and preliminary biological evaluation. Macromolecules 2005, 38, 761–769. [Google Scholar] [CrossRef]
- Klok, H.A.; Vandermeulen, G.W.M.; Nuhn, H.; Rösler, A.; Hamley, I.W.; Castelletto, V.; Xu, H.; Sheiko, S.S. Peptide mediated formation of hierarchically organized solution and solid state polymer nanostructures. Faraday Discuss. 2005, 128, 29–41. [Google Scholar] [CrossRef]
- Vandermeulen, G.W.M.; Hinderberger, D.; Xu, H.; Sheiko, S.S.; Jeschke, G.; Klok, H.A. Structure and dynamics of self-assembled poly(ethylene glycol) based coiled-coil nano-objects. Chem. Phys. Chem. 2004, 5, 488–494. [Google Scholar] [CrossRef]
- Vandermeulen, G.W.M.; Tziatzios, C.; Klok, H.A. Reversible self-organization of poly(ethylene glycol)-based hybrid block copolymers mediated by a de novo four-stranded α-helical coiled coil motif. Macromolecules 2003, 36, 4107–4114. [Google Scholar] [CrossRef]
- Zheng, T.; Voskuhl, J.; Versluis, F.; Zope, H.R.; Tomatsu, I.; Marsden, H.R.; Kros, A. Controlling the rate of coiled coil driven membrane fusion. Chem. Commun. 2013, 49, 3649–3651. [Google Scholar]
- Martelli, G.; Zope, H.R.; Bròvia Capell, M.; Kros, A. Coiled-coil peptide motifs as thermoresponsive valves for mesoporous silica nanoparticles. Chem. Commun. 2013, 49, 9932–9934. [Google Scholar] [CrossRef]
- Tomatsu, I.; Marsden, H.R.; Rabe, M.; Versluis, F.; Zheng, T.; Zope, H.; Kros, A. Influence of PEGylation on peptide-mediated liposome fusion. J. Mater. Chem. 2011, 21, 18927–18933. [Google Scholar] [CrossRef]
- Robson Marsden, H.; Handgraaf, J.W.; Nudelman, F.; Sommerdijk, N.A.J.M.; Kros, A. Uniting polypeptides with sequence-designed peptides: Synthesis and assembly of poly(γ-benzyl l-glutamate)-b-coiled-coil peptide copolymers. J. Am. Chem. Soc. 2010, 132, 2370–2377. [Google Scholar] [CrossRef]
- Marsden, H.R.; Korobko, A.V.; van Leeuwen, E.N.M.; Pouget, E.M.; Veen, S.J.; Sommerdijk, N.A.J.M.; Kros, A. Noncovalent triblock copolymers based on a coiled-coil peptide motif. J. Am. Chem. Soc. 2008, 130, 9386–9393. [Google Scholar] [CrossRef]
- Shu, J.Y.; Lund, R.; Xu, T. Solution structural characterization of coiled-coil peptide-polymer side-conjugates. Biomacromolecules 2012, 13, 1945–1955. [Google Scholar] [CrossRef]
- Dong, H.; Dube, N.; Shu, J.Y.; Seo, J.W.; Mahakian, L.M.; Ferrara, K.W.; Xu, T. Long-circulating 15 nm micelles based on amphiphilic 3-helix peptide-PEG conjugates. ACS Nano 2012, 6, 5320–5329. [Google Scholar] [CrossRef]
- Shu, J.Y.; Huang, Y.J.; Tan, C.; Presley, A.D.; Chang, J.; Xu, T. Amphiphilic peptide-polymer conjugates based on the coiled-coil helix bundle. Biomacromolecules 2010, 11, 1443–1452. [Google Scholar] [CrossRef]
- Shu, J.Y.; Tan, C.; DeGrado, W.F.; Xu, T. New design of helix bundle peptide-polymer conjugates. Biomacromolecules 2008, 9, 2111–2117. [Google Scholar] [CrossRef]
- Dong, H.; Shu, J.Y.; Dube, N.; Ma, Y.; Tirrell, M.V.; Downing, K.H.; Xu, T. 3-Helix micelles stabilized by polymer springs. J. Am. Chem. Soc. 2012, 134, 11807–11814. [Google Scholar] [CrossRef]
- Lupas, A. Predicting coiled-coil regions in proteins. Curr. Opin. Struct. Biol. 1997, 7, 388–393. [Google Scholar] [CrossRef]
- Lupas, A.; van Dyke, M.; Stock, J. Predicting coiled coils from protein sequences. Science 1991, 252, 1162–1164. [Google Scholar] [CrossRef]
- Berger, B.; Wilson, D.B.; Wolf, E.; Tonchev, T.; Milla, M.; Kim, P.S. Predicting coiled coils by use of pairwise residue correlations. Proc. Natl. Acad. Sci. USA 1995, 92, 8259–8263. [Google Scholar] [CrossRef]
- Woolfson, D.N.; Alber, T. Predicting oligomerization states of coiled coils. Protein Sci. 1995, 4, 1596–1607. [Google Scholar] [CrossRef]
- Wolf, E.; Kim, P.S.; Berger, B. Multicoil: A program for predicting two- and three-stranded coiled coils. Protein Sci. 1997, 6, 1179–1189. [Google Scholar] [CrossRef]
- Walshaw, J.; Woolfson, D.N. Socket: A program for identifying and analysing coiled-coil motifs within protein structures. J. Mol. Biol. 2001, 307, 1427–1450. [Google Scholar] [CrossRef]
- Gruber, M.; Söding, J.; Lupas, A.N. Repper—Repeats and their periodicities in fibrous proteins. Nucleic Acids Res. 2005, 33, W239–W243. [Google Scholar] [CrossRef]
- Rozzelle, J.E., Jr.; Tropsha, A.; Erickson, B.W. Rational design of a three-heptad coiled-coil protein and comparison by molecular dynamics simulation with the GCN4 coiled coil: Presence of interior three-center hydrogen bonds. Protein Sci. 1994, 3, 345–355. [Google Scholar]
- Zhong, Q.; Jiang, Q.; Moore, P.B.; Newns, D.M.; Klein, M.L. Molecular dynamics simulation of a synthetic ion channel. Biophys. J. 1998, 74, 3–10. [Google Scholar] [CrossRef]
- Orzechowski, M.; Cieplak, P.; Piela, L. Theoretical calculation of the coiled-coil stability in water in the context of its possible use as a molecular rack. J. Comput. Chem. 2002, 23, 106–110. [Google Scholar] [CrossRef]
- Danciulescu, C.; Nick, B.; Wortmann, F.J. Structural stability of wild type and mutated α-keratin fragments: Molecular dynamics and free energy calculations. Biomacromolecules 2004, 5, 2165–2175. [Google Scholar] [CrossRef]
- Missimer, J.H.; Steinmetz, M.O.; Jahnke, W.; Winkler, F.K.; van Gunsteren, W.F.; Daura, X. Molecular-dynamics simulations of C- and N-terminal peptide derivatives of GCN4-p1 in aqueous solution. Chem. Biodivers. 2005, 2, 1086–1104. [Google Scholar] [CrossRef]
- Pagel, K.; Seeger, K.; Seiwert, B.; Villa, A.; Mark, A.E.; Berger, S.; Koksch, B. Advanced approaches for the characterization of a de novo designed antiparallel coiled coil peptide. Org. Biomol. Chem. 2005, 3, 1189–1194. [Google Scholar] [CrossRef]
- Piñeiro, Á.; Villa, A.; Vagt, T.; Koksch, B.; Mark, A.E. A molecular dynamics study of the formation, stability, and oligomerization state of two designed coiled coils: Possibilities and limitations. Biophys. J. 2005, 89, 3701–3713. [Google Scholar] [CrossRef]
- Kelly, E.; Privé, G.G.; Tieleman, D.P. Molecular models of lipopeptide detergents: Large coiled-coils with hydrocarbon interiors. J. Am. Chem. Soc. 2005, 127, 13446–13447. [Google Scholar] [CrossRef]
- Lee, H.; Larson, R.G. Prediction of the stability of coiled coils using molecular dynamics simulations. Mol. Simul. 2007, 33, 463–473. [Google Scholar] [CrossRef]
- Oshaben, K.M.; Salari, R.; McCaslin, D.R.; Chong, L.T.; Horne, W.S. The native GCN4 leucine-zipper domain does not uniquely specify a dimeric oligomerization state. Biochemistry 2012, 51, 9581–9591. [Google Scholar] [CrossRef]
- Jain, A.; Ashbaugh, H.S. Helix stabilization of poly(ethylene glycol)–peptide conjugates. Biomacromolecules 2011, 12, 2729–2734. [Google Scholar] [CrossRef]
- Hamed, E.; Xu, T.; Keten, S. Poly(ethylene glycol) conjugation stabilizes the secondary structure of α-helices by reducing peptide solvent accessible surface area. Biomacromolecules 2013, 14, 4053–4060. [Google Scholar] [CrossRef]
- Ruiz, L.; Keten, S. Directing the self-assembly of supra-biomolecular nanotubes using entropic forces. Soft Matter 2014, 10, 851–861. [Google Scholar] [CrossRef]
- Majoros, I.J.; Williams, C.R.; Baker, J.R. Current dendrimer applications in cancer diagnosis and therapy. Curr. Top. Med. Chem. 2008, 8, 1165–1179. [Google Scholar] [CrossRef]
- Tian, W.D.; Ma, Y.Q. Theoretical and computational studies of dendrimers as delivery vectors. Chem. Soc. Rev. 2013, 42, 705–727. [Google Scholar] [CrossRef]
- Tu, C.K.; Chen, K.; Tian, W.D.; Ma, Y.Q. Computational investigations of a peptide-modified dendrimer interacting with lipid membranes. Macromol. Rapid Commun. 2013, 34, 1237–1242. [Google Scholar] [CrossRef]
- Lee, H.; Larson, R.G. Multiscale modeling of dendrimers and their interactions with bilayers and polyelectrolytes. Molecules 2009, 14, 423–438. [Google Scholar] [CrossRef]
- Kelly, C.V.; Liroff, M.G.; Triplett, L.D.; Leroueil, P.R.; Mullen, D.G.; Wallace, J.M.; Meshinchi, S.; Baker, J.R.; Orr, B.G.; Holl, M.M.B. Stoichiometry and structure of poly(amidoamine) dendrimer-lipid complexes. ACS Nano 2009, 3, 1886–1896. [Google Scholar] [CrossRef]
- Lee, H.; Larson, R.G. Lipid bilayer curvature and pore formation induced by charged linear polymers and dendrimers: The effect of molecular shape. J. Phys. Chem. B 2008, 112, 12279–12285. [Google Scholar] [CrossRef]
- Lee, H.; Larson, R.G. Coarse-grained molecular dynamics studies of the concentration and size dependence of fifth- and seventh-generation pamam dendrimers on pore formation in dmpc bilayer. J. Phys. Chem. B 2008, 112, 7778–7784. [Google Scholar] [CrossRef]
- Lee, H.; Larson, R.G. Molecular dynamics simulations of pamam dendrimer-induced pore formation in DPPC bilayers with a coarse-grained model. J. Phys. Chem. B 2006, 110, 18204–18211. [Google Scholar] [CrossRef]
- Lee, H.; Baker, J.R.; Larson, R.G. Molecular dynamics studies of the size, shape, and internal structure of 0% and 90% acetylated fifth-generation polyamidoamine dendrimers in water and methanol. J. Phys. Chem. B 2006, 110, 4014–4019. [Google Scholar] [CrossRef]
- Lee, H.; Choi, J.S.; Larson, R.G. Molecular dynamics studies of the size and internal structure of the pamam dendrimer grafted with arginine and histidine. Macromolecules 2011, 44, 8681–8686. [Google Scholar] [CrossRef]
- Chen, J.M.; Hessler, J.A.; Putchakayala, K.; Panama, B.K.; Khan, D.P.; Hong, S.; Mullen, D.G.; DiMaggio, S.C.; Som, A.; Tew, G.N.; et al. Cationic nanoparticles induce nanoscale disruption in living cell plasma membranes. J. Phys. Chem. B 2009, 113, 11179–11185. [Google Scholar] [CrossRef]
- Leroueil, P.R.; Berry, S.A.; Duthie, K.; Han, G.; Rotello, V.M.; McNerny, D.Q.; Baker, J.R.; Orr, B.G.; Holl, M.M.B. Wide varieties of cationic nanoparticles induce defects in supported lipid bilayers. Nano Lett. 2008, 8, 420–424. [Google Scholar] [CrossRef]
- Kelly, C.V.; Leroueil, P.R.; Nett, E.K.; Wereszczynski, J.M.; Baker, J.R.; Orr, B.G.; Holl, M.M.B.; Andricioaei, I. Poly(amidoamine) dendrimers on lipid bilayers I: Free energy and conformation of binding. J. Phys. Chem. B 2008, 112, 9337–9345. [Google Scholar] [CrossRef]
- Kandasamy, S.K.; Lee, H.; Larson, R.G. Computer Simulations of Dendrimers. In Dendrimer-Based Nanomedicine; Majoros, I.J., Baker, J.R.J., Eds.; Pan Stanford Publishing: Singapore, 2008; pp. 331–354. [Google Scholar]
- Leroueil, P.R.; Hong, S.Y.; Mecke, A.; Baker, J.R.; Orr, B.G.; Holl, M.M.B. Nanoparticle interaction with biological membranes: Does nanotechnology present a janus face? Acc. Chem. Res. 2007, 40, 335–342. [Google Scholar] [CrossRef]
- Hong, S.P.; Leroueil, P.R.; Janus, E.K.; Peters, J.L.; Kober, M.M.; Islam, M.T.; Orr, B.G.; Baker, J.R.; Holl, M.M.B. Interaction of polycationic polymers with supported lipid bilayers and cells: Nanoscale hole formation and enhanced membrane permeability. Bioconjugate Chem. 2006, 17, 728–734. [Google Scholar] [CrossRef]
- Shukla, R.; Thomas, T.P.; Peters, J.; Kotlyar, A.; Myc, A.; Baker, J.R., Jr. Tumor angiogenic vasculature targeting with pamam dendrimer-rgd conjugates. Chem. Commun. 2005, 5739–5741. [Google Scholar]
- Patri, A.K.; Kukowska-Latallo, J.F.; Baker, J.R. Targeted drug delivery with dendrimers: Comparison of the release kinetics of covalently conjugated drug and non-covalent drug inclusion complex. Adv. Drug Deliv. Rev. 2005, 57, 2203–2214. [Google Scholar] [CrossRef]
- Mecke, A.; Majoros, I.J.; Patri, A.K.; Baker, J.R.; Holl, M.M.B.; Orr, B.G. Lipid bilayer disruption by polycationic polymers: The roles of size and chemical functional group. Langmuir 2005, 21, 10348–10354. [Google Scholar] [CrossRef]
- Choi, Y.; Thomas, T.; Kotlyar, A.; Islam, M.T.; Baker, J.R. Synthesis and functional evaluation of DNA-assembled polyamidoamine dendrimer clusters for cancer cell-specific targeting. Chem. Biol. 2005, 12, 35–43. [Google Scholar] [CrossRef]
- Mecke, A.; Uppuluri, S.; Sassanella, T.M.; Lee, D.K.; Ramamoorthy, A.; Baker, J.R.; Orr, B.G.; Holl, M.M.B. Direct observation of lipid bilayer disruption by poly(amidoamine) dendrimers. Chem. Phys. Lipids 2004, 132, 3–14. [Google Scholar] [CrossRef]
- Mecke, A.; Lee, I.; Baker, J.R.; Holl, M.M.B.; Orr, B.G. Deformability of poly(amidoamine) dendrimers. Eur. Phys. J. E 2004, 14, 7–16. [Google Scholar] [CrossRef]
- Hong, S.P.; Bielinska, A.U.; Mecke, A.; Keszler, B.; Beals, J.L.; Shi, X.Y.; Balogh, L.; Orr, B.G.; Baker, J.R.; Holl, M.M.B. Interaction of poly(amidoamine) dendrimers with supported lipid bilayers and cells: Hole formation and the relation to transport. Bioconj. Chem. 2004, 15, 774–782. [Google Scholar] [CrossRef]
- Choi, Y.S.; Mecke, A.; Orr, B.G.; Holl, M.M.B.; Baker, J.R. DNA-directed synthesis of generation 7 and 5 pamam dendrimer nanoclusters. Nano Lett. 2004, 4, 391–397. [Google Scholar] [CrossRef]
- Majoros, I.J.; Keszler, B.; Woehler, S.; Bull, T.; Baker, J.R. Acetylation of poly(amidoamine) dendrimers. Macromolecules 2003, 36, 5526–5529. [Google Scholar] [CrossRef]
- Patri, A.K.; Majoros, I.J.; Baker, J.R. Dendritic polymer macromolecular carriers for drug delivery. Curr. Opin. Chem. Biol. 2002, 6, 466–471. [Google Scholar] [CrossRef]
- Choi, S.K.; Myc, A.; Silpe, J.E.; Sumit, M.; Wong, P.T.; McCarthy, K.; Desai, A.M.; Thomas, T.P.; Kotlyar, A.; Holl, M.M.B.; et al. Dendrimer-based multivalent vancomycin nanoplatform for targeting the drug-resistant bacterial surface. ACS Nano 2013, 7, 214–228. [Google Scholar] [CrossRef]
- Zong, H.; Thomas, T.P.; Lee, K.H.; Desai, A.M.; Li, M.H.; Kotlyar, A.; Zhang, Y.; Leroueil, P.R.; Gam, J.J.; Holl, M.M.B.; et al. Bifunctional pamam dendrimer conjugates of folic acid and methotrexate with defined ratio. Biomacromolecules 2012, 13, 982–991. [Google Scholar] [CrossRef]
- Thomas, T.P.; Huang, B.; Choi, S.K.; Silpe, J.E.; Kotlyar, A.; Desai, A.M.; Zong, H.; Gam, J.; Joice, M.; Baker, J.R. Polyvalent dendrimer-methotrexate as a folate receptor-targeted cancer therapeutic. Mol. Pharm. 2012, 9, 2669–2676. [Google Scholar] [CrossRef]
- Mullen, D.G.; Desai, A.; van Dongen, M.A.; Barash, M.; Baker, J.R.; Banaszak Holl, M.M. Best practices for purification and characterization of pamam dendrimer. Macromolecules 2012, 45, 5316–5320. [Google Scholar] [CrossRef]
- Lyulin, S.V.; Vattulainen, I.; Gurtovenko, A.A. Complexes comprised of charged dendrimers, linear polyelectrolytes, and counterions: Insight through coarse-grained molecular dynamics simulations. Macromolecules 2008, 41, 4961–4968. [Google Scholar] [CrossRef]
- Welch, P.; Muthukumar, M. Dendrimer-polyelectrolyte complexation: A model guest-host system. Macromolecules 2000, 33, 6159–6167. [Google Scholar] [CrossRef]
- Hedden, R.C.; Bauer, B.J. Structure and dimensions of PAMAM/PEG dendrimer-star polymers. Macromolecules 2003, 36, 1829–1835. [Google Scholar] [CrossRef]
- Tanis, I.; Karatasos, K. Molecular dynamics simulations of polyamidoamine dendrimers and their complexes with linear poly(ethylene oxide) at different ph conditions: Static properties and hydrogen bonding. Phys. Chem. Chem. Phys. 2009, 11, 10017–10028. [Google Scholar] [CrossRef]
- Lee, H.; Larson, R.G. Molecular dynamics study of the structure and interparticle interactions of polyethylene glycol-conjugated pamam dendrimers. J. Phys. Chem. B 2009, 113, 13202–13207. [Google Scholar] [CrossRef]
- Lee, H.; Larson, R.G. Membrane pore formation induced by acetylated and polyethylene glycol-conjugated polyamidoamine dendrimers. J. Phys. Chem. C 2011, 115, 5316–5322. [Google Scholar] [CrossRef]
- Lee, H.; Larson, R.G. Effects of PEGylation on the size and internal structure of dendrimers: Self-penetration of long PEG chains into the dendrimer core. Macromolecules 2011, 44, 2291–2298. [Google Scholar] [CrossRef]
- Albertazzi, L.; Mickler, F.M.; Pavan, G.M.; Salomone, F.; Bardi, G.; Panniello, M.; Amir, E.; Kang, T.; Killops, K.L.; Bräuchle, C.; et al. Enhanced bioactivity of internally functionalized cationic dendrimers with PEG cores. Biomacromolecules 2012, 13, 4089–4097. [Google Scholar]
- Karatasos, K. Self-association and complexation of the anti-cancer drug doxorubicin with PEGylated hyperbranched polyesters in an aqueous environment. J. Phys. Chem. B 2013, 117, 2564–2575. [Google Scholar] [CrossRef]
- Pavan, G.M.; Barducci, A.; Albertazzi, L.; Parrinello, M. Combining metadynamics simulation and experiments to characterize dendrimers in solution. Soft Matter 2013, 9, 2593–2597. [Google Scholar] [CrossRef]
- Bianco, A.; Kostarelos, K.; Prato, M. Applications of carbon nanotubes in drug delivery. Curr. Opin. Chem. Biol. 2005, 9, 674–679. [Google Scholar] [CrossRef]
- Lacerda, L.; Bianco, A.; Prato, M.; Kostarelos, K. Carbon nanotubes as nanomedicines: From toxicology to pharmacology. Adv. Drug Deliv. Rev. 2006, 58, 1460–1470. [Google Scholar] [CrossRef]
- Liu, Z.; Robinson, J.T.; Tabakman, S.M.; Yang, K.; Dai, H. Carbon materials for drug delivery & cancer therapy. Mater. Today 2011, 14, 316–323. [Google Scholar] [CrossRef]
- Ke, P.C.; Lamm, M.H. A biophysical perspective of understanding nanoparticles at large. Phys. Chem. Chem. Phys. 2011, 13, 7273–7283. [Google Scholar]
- Yurekli, K.; Mitchell, C.A.; Krishnamoorti, R. Small-angle neutron scattering from surfactant-assisted aqueous dispersions of carbon nanotubes. J. Am. Chem. Soc. 2004, 126, 9902–9903. [Google Scholar] [CrossRef]
- Matarredona, O.; Rhoads, H.; Li, Z.; Harwell, J.H.; Balzano, L.; Resasco, D.E. Dispersion of single-walled carbon nanotubes in aqueous solutions of the anionic surfactant naddbs. J. Phys. Chem. B 2003, 107, 13357–13367. [Google Scholar] [CrossRef]
- Qiao, R.; Ke, P.C. Lipid-carbon nanotube self-assembly in aqueous solution. J. Am. Chem. Soc. 2006, 128, 13656–13657. [Google Scholar] [CrossRef]
- O’Connell, M.J.; Bachilo, S.H.; Huffman, C.B.; Moore, V.C.; Strano, M.S.; Haroz, E.H.; Rialon, K.L.; Boul, P.J.; Noon, W.H.; Kittrell, C.; et al. Band gap fluorescence from individual single-walled carbon nanotubes. Science 2002, 297, 593–596. [Google Scholar] [CrossRef]
- Kam, N.W.S.; Liu, Z.; Dai, H. Functionalization of carbon nanotubes via cleavable disulfide bonds for efficient intracellular delivery of sirna and potent gene silencing. J. Am. Chem. Soc. 2005, 127, 12492–12493. [Google Scholar] [CrossRef]
- Wu, Y.; Hudson, J.S.; Lu, Q.; Moore, J.M.; Mount, A.S.; Rao, A.M.; Alexov, E.; Ke, P.C. Coating single-walled carbon nanotubes with phospholipids. J. Phys. Chem. B 2006, 110, 2475–2478. [Google Scholar]
- Lin, S.; Keskar, G.; Wu, Y.; Wang, X.; Mount, A.S.; Klaine, S.J.; Moore, J.M.; Rao, A.M.; Ke, P.C. Detection of phospholipid-carbon nanotube translocation using fluorescence energy transfer. Appl. Phys. Lett. 2006. [Google Scholar] [CrossRef]
- Liu, X.; Tao, H.; Yang, K.; Zhang, S.; Lee, S.T.; Liu, Z. Optimization of surface chemistry on single-walled carbon nanotubes for in vivo photothermal ablation of tumors. Biomaterials 2011, 32, 144–151. [Google Scholar] [CrossRef]
- Liu, Z.; Robinson, J.T.; Sun, X.; Dai, H. PEGylated nanographene oxide for delivery of water-insoluble cancer drugs. J. Am. Chem. Soc. 2008, 130, 10876–10877. [Google Scholar] [CrossRef]
- Wallace, E.J.; Sansom, M.S.P. Carbon nanotube self-assembly with lipids and detergent: A molecular dynamics study. Nanotechnology 2009, 20, 045101–045101. [Google Scholar] [CrossRef]
- Wallace, E.J.; Sansom, M.S.P. Carbon nanotube/detergent interactions via coarse-grained molecular dynamics. Nano Lett. 2007, 7, 1923–1928. [Google Scholar] [CrossRef]
- Tummala, N.R.; Morrow, B.H.; Resasco, D.E.; Striolo, A. Stabilization of aqueous carbon nanotube dispersions using surfactants: Insights from molecular dynamics simulations. ACS Nano 2010, 4, 7193–7204. [Google Scholar] [CrossRef]
- Xu, Z.; Yang, X.; Yang, Z. A molecular simulation probing of structure and interaction for supramolecular sodium dodecyl sulfate/single-wall carbon nanotube assemblies. Nano Lett. 2010, 10, 985–991. [Google Scholar] [CrossRef]
- Calvaresi, M.; Dallavalle, M.; Zerbetto, F. Wrapping nanotubes with micelles, hemimicelles, and cylindrical micelles. Small 2009, 5, 2191–2198. [Google Scholar] [CrossRef]
- Lopez, C.F.; Nielsen, S.O.; Moore, P.B.; Klein, M.L. Understanding nature's design for a nanosyringe. Proc. Natl. Acad. Sci. USA 2004, 101, 4431–4434. [Google Scholar] [CrossRef]
- Nielsen, S.O.; Ensing, B.; Ortiz, V.; Moore, P.B.; Klein, M.L. Lipid bilayer perturbations around a transmembrane nanotube: A coarse grain molecular dynamics study. Biophys. J. 2005, 88, 3822–3828. [Google Scholar] [CrossRef]
- Lopez, C.F.; Nielsen, S.O.; Ensing, B.; Moore, P.B.; Klein, M.L. Structure and dynamics of model pore insertion into a membrane. Biophys. J. 2005, 88, 3083–3094. [Google Scholar]
- Hofinger, S.; Melle-Franco, M.; Gallo, T.; Cantelli, A.; Calvaresi, M.; Gomes, J.A.N.F.; Zerbetto, F. A computational analysis of the insertion of carbon nanotubes into cellular membranes. Biomaterials 2011, 32, 7079–7085. [Google Scholar] [CrossRef]
- Makarucha, A.J.; Todorova, N.; Yarovsky, I. Nanomaterials in biological environment: A review of computer modelling studies. Eur. Biophys. J. 2011, 40, 103–115. [Google Scholar] [CrossRef]
- Monticelli, L.; Salonen, E.; Ke, P.C.; Vattulainen, I. Effects of carbon nanoparticles on lipid membranes: A molecular simulation perspective. Soft Matter 2009, 5, 4433–4445. [Google Scholar] [CrossRef]
- Shi, X.; Kong, Y.; Gao, H. Coarse grained molecular dynamics and theoretical studies of carbon nanotubes entering cell membrane. Acta Mech. Sin. 2008, 24, 161–169. [Google Scholar] [CrossRef]
- Kraszewski, S.; Bianco, A.; Tarek, M.; Ramseyer, C. Insertion of short amino-functionalized single-walled carbon nanotubes into phospholipid bilayer occurs by passive diffusion. PLoS ONE 2012. [Google Scholar] [CrossRef]
- Pogodin, S.; Baulin, V.A. Can a carbon nanotube pierce through a phospholipid bilayer? ACS Nano 2010, 4, 5293–5300. [Google Scholar] [CrossRef]
- Skandani, A.A.; Zeineldin, R.; Al-Haik, M. Effect of chirality and length on the penetrability of single-walled carbon nanotubes into lipid bilayer cell membranes. Langmuir 2012, 28, 7872–7879. [Google Scholar] [CrossRef]
- Yang, K.; Ma, Y.Q. Computer simulation of the translocation of nanoparticles with different shapes across a lipid bilayer. Nat. Nanotechnol. 2010, 5, 579–583. [Google Scholar] [CrossRef]
- Wallace, E.J.; Sansom, M.S.P. Blocking of carbon nanotube based nanoinjectors by lipids: A simulation study. Nano Lett. 2008, 8, 2751–2756. [Google Scholar] [CrossRef]
- Shi, X.; von Dem Bussche, A.; Hurt, R.H.; Kane, A.B.; Gao, H. Cell entry of one-dimensional nanomaterials occurs by tip recognition and rotation. Nat. Nanotechnol. 2011, 6, 714–719. [Google Scholar] [CrossRef]
- Baoukina, S.; Monticelli, L.; Tieleman, D.P. Interaction of pristine and functionalized carbon nanotubes with lipid membranes. J. Phys. Chem. B 2013, 117, 12113–12123. [Google Scholar] [CrossRef]
- Lee, H.; Kim, H. Self-assembly of lipids and single-walled carbon nanotubes: Effects of lipid structure and PEGylation. J. Phys. Chem. C 2012, 116, 9327–9333. [Google Scholar] [CrossRef]
- Lee, H. Interparticle dispersion, membrane curvature, and penetration induced by single-walled carbon nanotubes wrapped with lipids and PEGylated lipids. J. Phys. Chem. B 2013, 117, 1337–1344. [Google Scholar] [CrossRef]
- Lee, H. Membrane penetration and curvature induced by single-walled carbon nanotubes: The effect of diameter, length, and concentration. Phys. Chem. Chem. Phy. 2013, 15, 16334–16340. [Google Scholar] [CrossRef]
- Lee, H. Molecular dynamics studies of PEGylated single-walled carbon nanotubes: The effect of PEG size and grafting density. J. Phys. Chem. C 2013, 117, 26334–26341. [Google Scholar] [CrossRef]
- Sacchetti, C.; Motamedchaboki, K.; Magrini, A.; Palmieri, G.; Mattei, M.; Bernardini, S.; Rosato, N.; Bottini, N.; Bottini, M. Surface polyethylene glycol conformation influences the protein corona of polyethylene glycol-modified single-walled carbon nanotubes: Potential implications on biological performance. ACS Nano 2013, 7, 1974–1989. [Google Scholar] [CrossRef]
- De Gennes, P.G. Polymers at an interface—A simplified view. Adv. Colloid Interface Sci. 1987, 27, 189–209. [Google Scholar] [CrossRef]
- Di Crescenzo, A.; Aschi, M.; Fontana, A. Toward a better understanding of steric stabilization when using block copolymers as stabilizers of single-walled carbon nanotubes (SWCNTS) aqueous dispersions. Macromolecules 2012, 45, 8043–8050. [Google Scholar] [CrossRef]
- Aslan, S.; Määttä, J.; Haznedaroglu, B.Z.; Goodman, J.P.M.; Pfefferle, L.D.; Elimelech, M.; Pauthe, E.; Sammalkorpi, M.; van Tassel, P.R. Carbon nanotube bundling: Influence on layer-by-layer assembly and antimicrobial activity. Soft Matter 2013, 9, 2136–2144. [Google Scholar] [CrossRef]
- Skandani, A.A.; Al-Haik, M. Reciprocal effects of the chirality and the surface functionalization on the drug delivery permissibility of carbon nanotubes. Soft Matter 2013, 9, 11645–11649. [Google Scholar] [CrossRef]
- Zeineldin, R.; Al-Haik, M.; Hudson, L.G. Role of polyethylene glycol integrity in specific receptor targeting of carbon nanotubes to cancer cells. Nano Lett. 2009, 9, 751–757. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Lee, H. Molecular Modeling of PEGylated Peptides, Dendrimers, and Single-Walled Carbon Nanotubes for Biomedical Applications. Polymers 2014, 6, 776-798. https://doi.org/10.3390/polym6030776
Lee H. Molecular Modeling of PEGylated Peptides, Dendrimers, and Single-Walled Carbon Nanotubes for Biomedical Applications. Polymers. 2014; 6(3):776-798. https://doi.org/10.3390/polym6030776
Chicago/Turabian StyleLee, Hwankyu. 2014. "Molecular Modeling of PEGylated Peptides, Dendrimers, and Single-Walled Carbon Nanotubes for Biomedical Applications" Polymers 6, no. 3: 776-798. https://doi.org/10.3390/polym6030776



