Investigating the Synergistic Effects of Combined Modified Alginates on Macrophage Phenotype
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
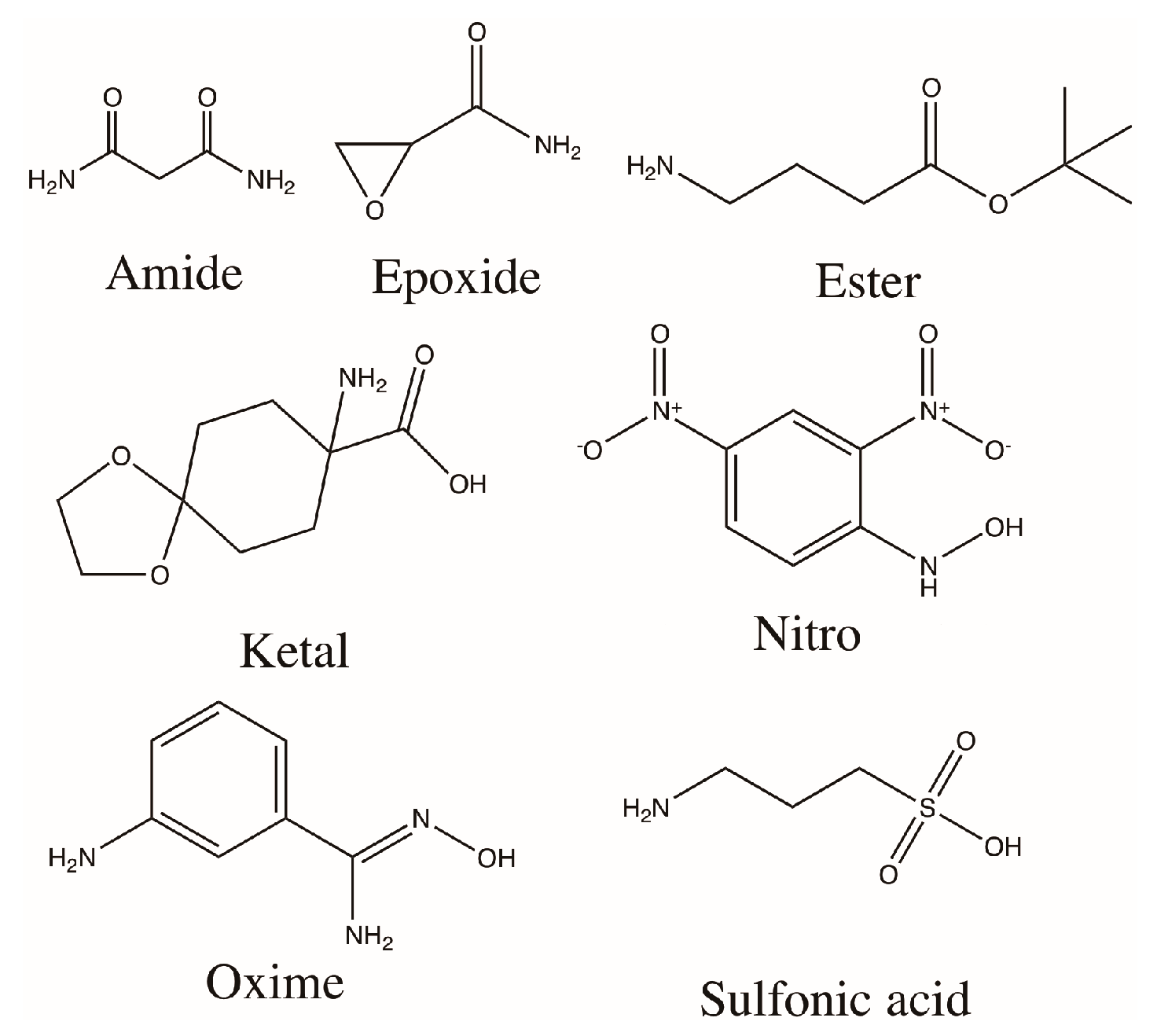
2.2. Alginate Modification
2.3. Elemental Analysis
2.4. Water Contact Angle (WCA)
2.5. Compression Modulus
2.6. Cell Culture
2.7. Cell Viability
2.8. TNF-α ELISA
2.9. Urea Assay
2.10. Griess Reagent Assay
2.11. Immunocytochemistry
2.12. TNF-α Diffusion
2.13. Statistics and Data Analysis
3. Results
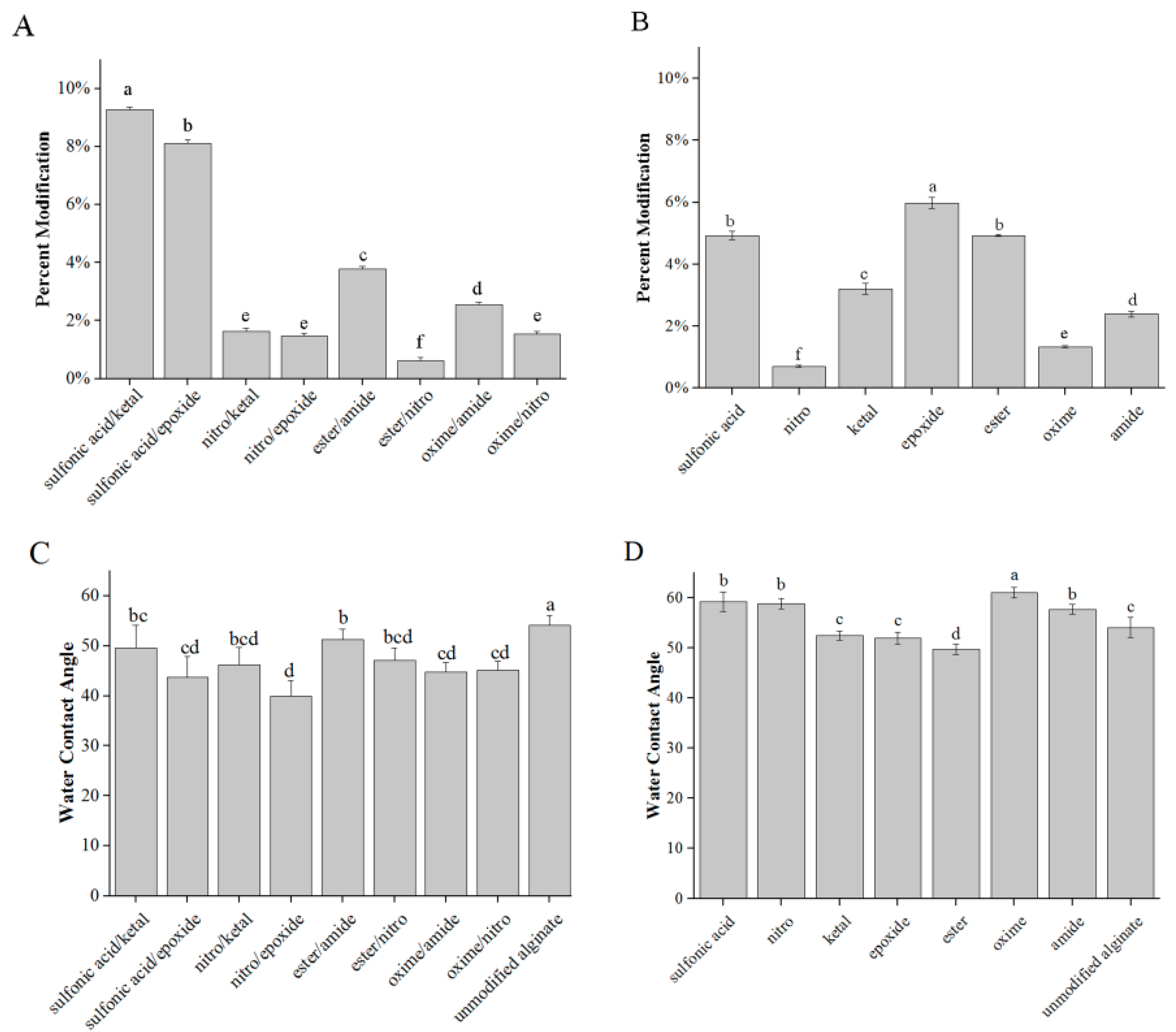
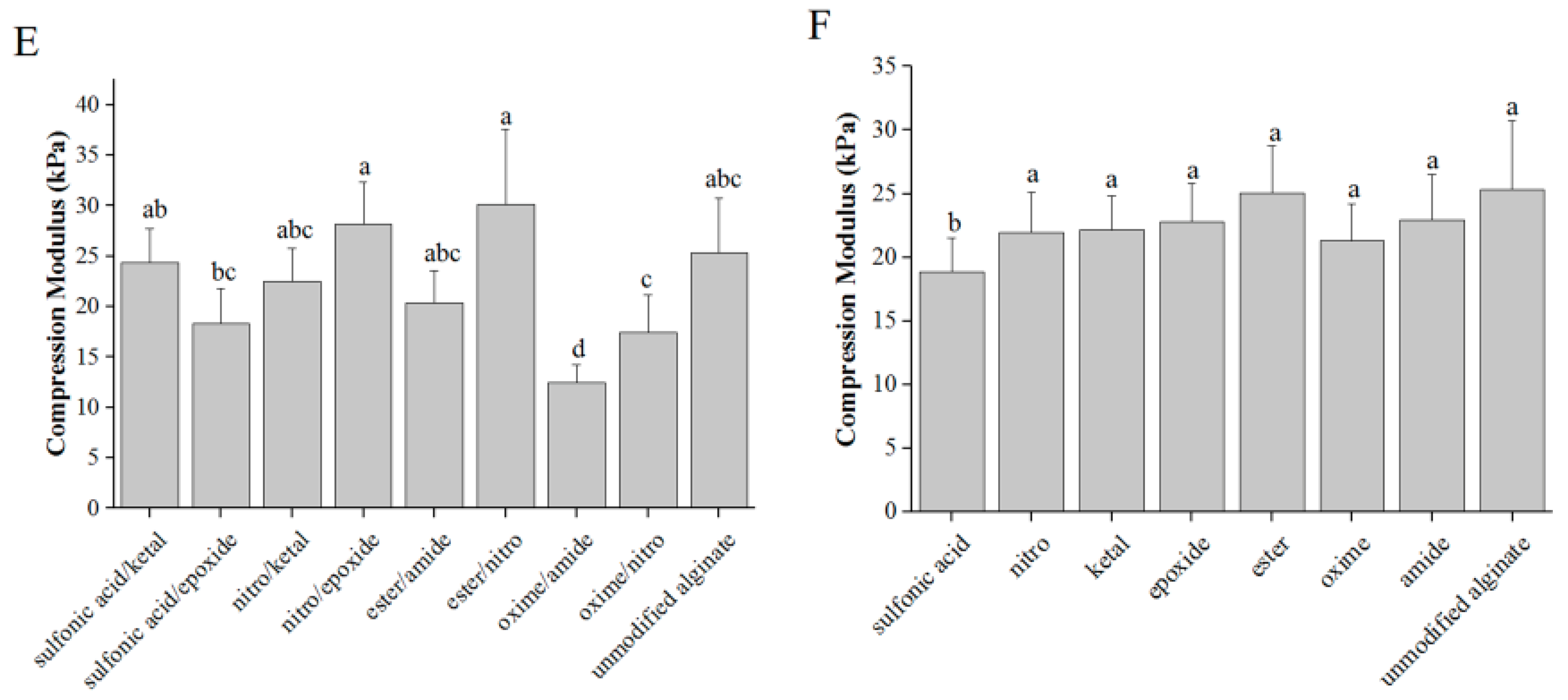
3.1. Modification and Characterization
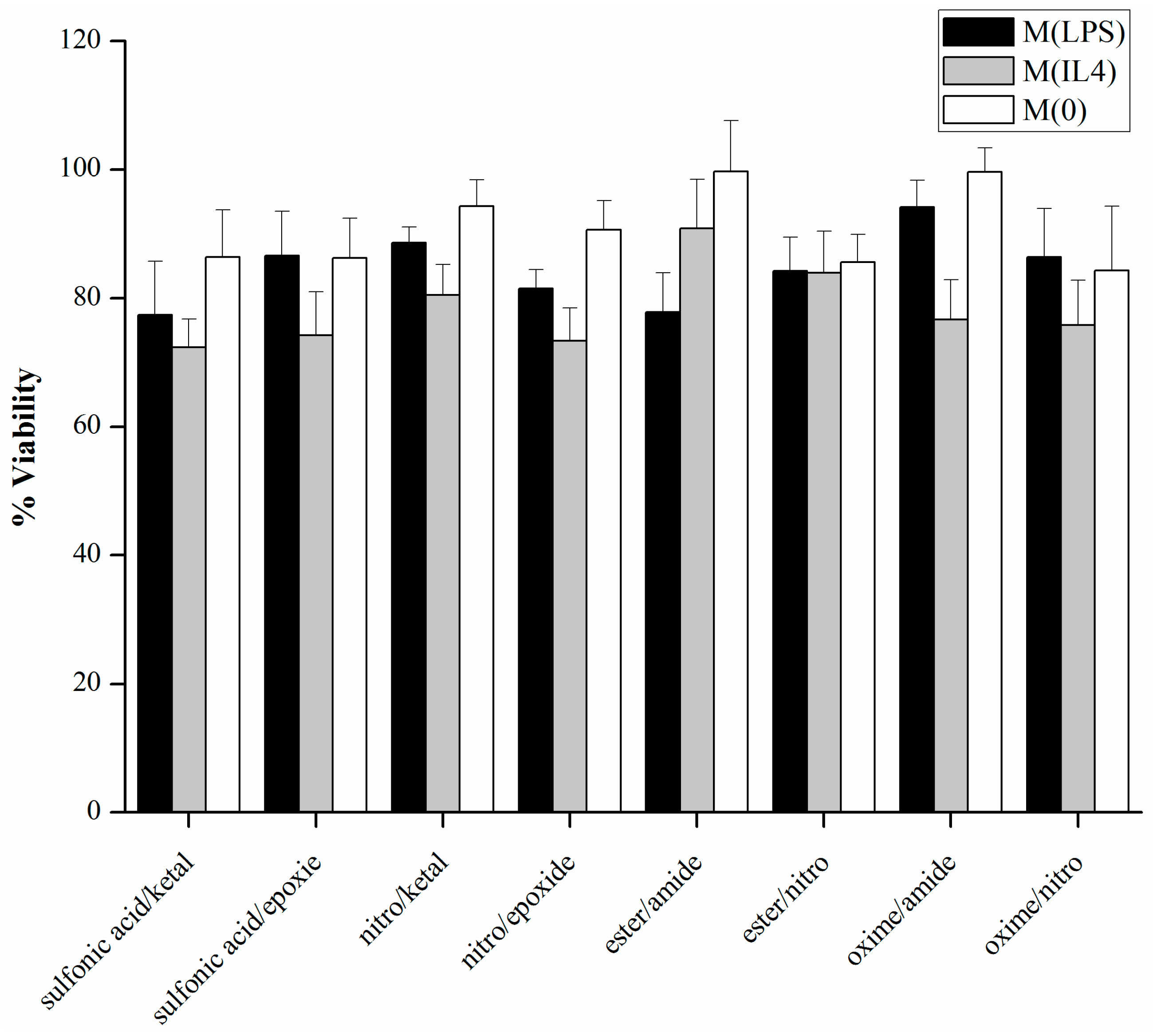
3.2. Cell Viability
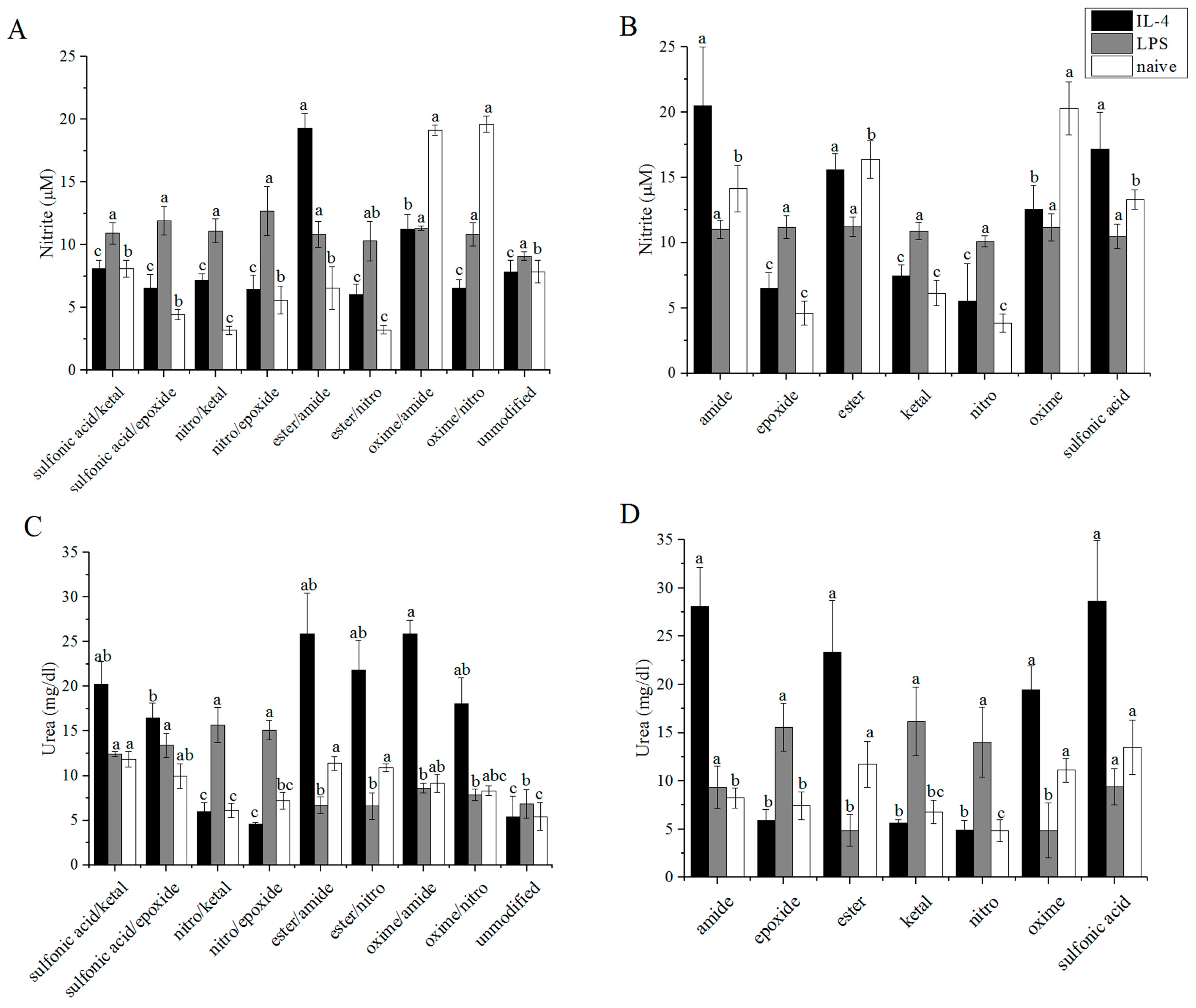
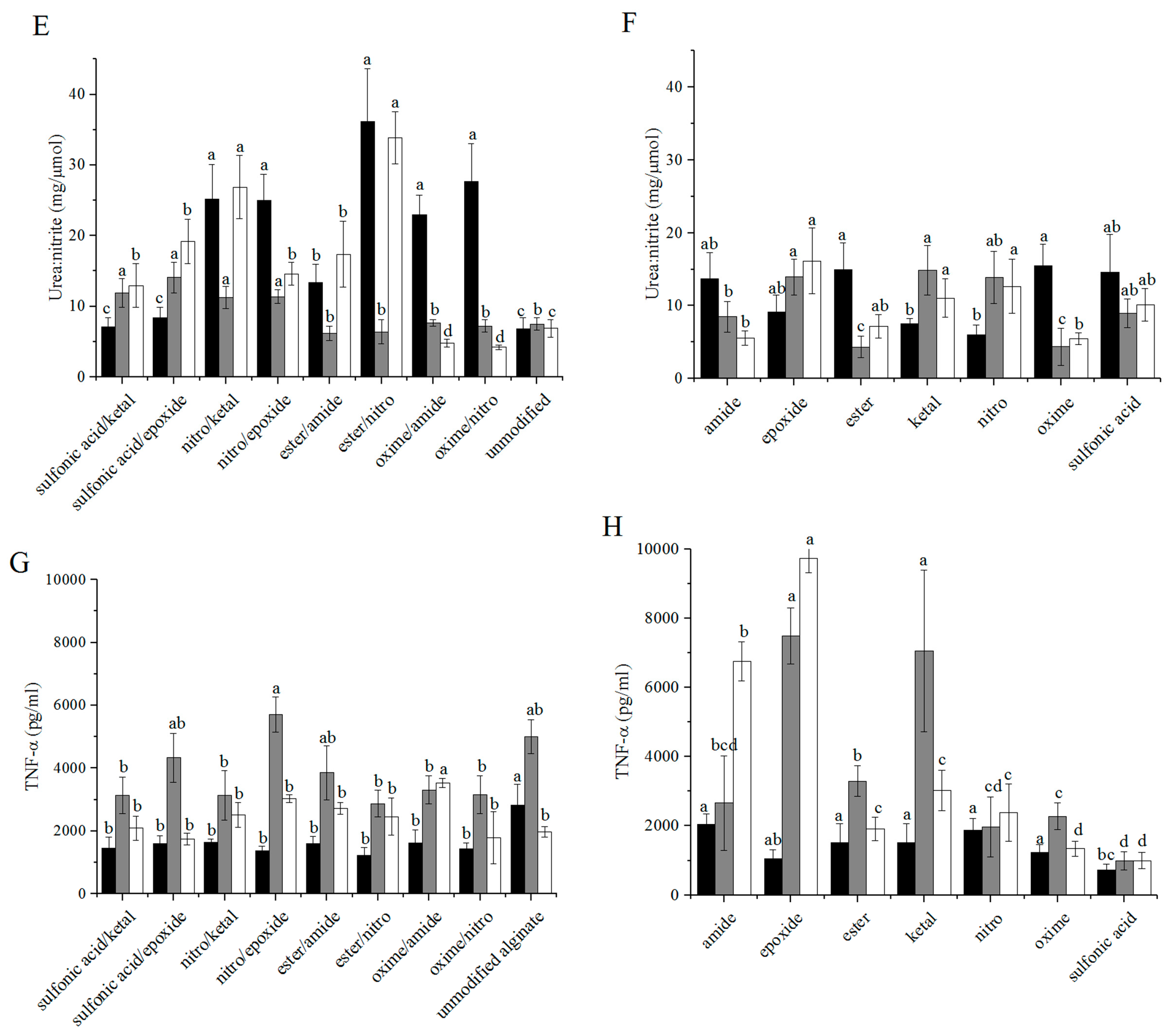
3.3. Arginase/iNOS
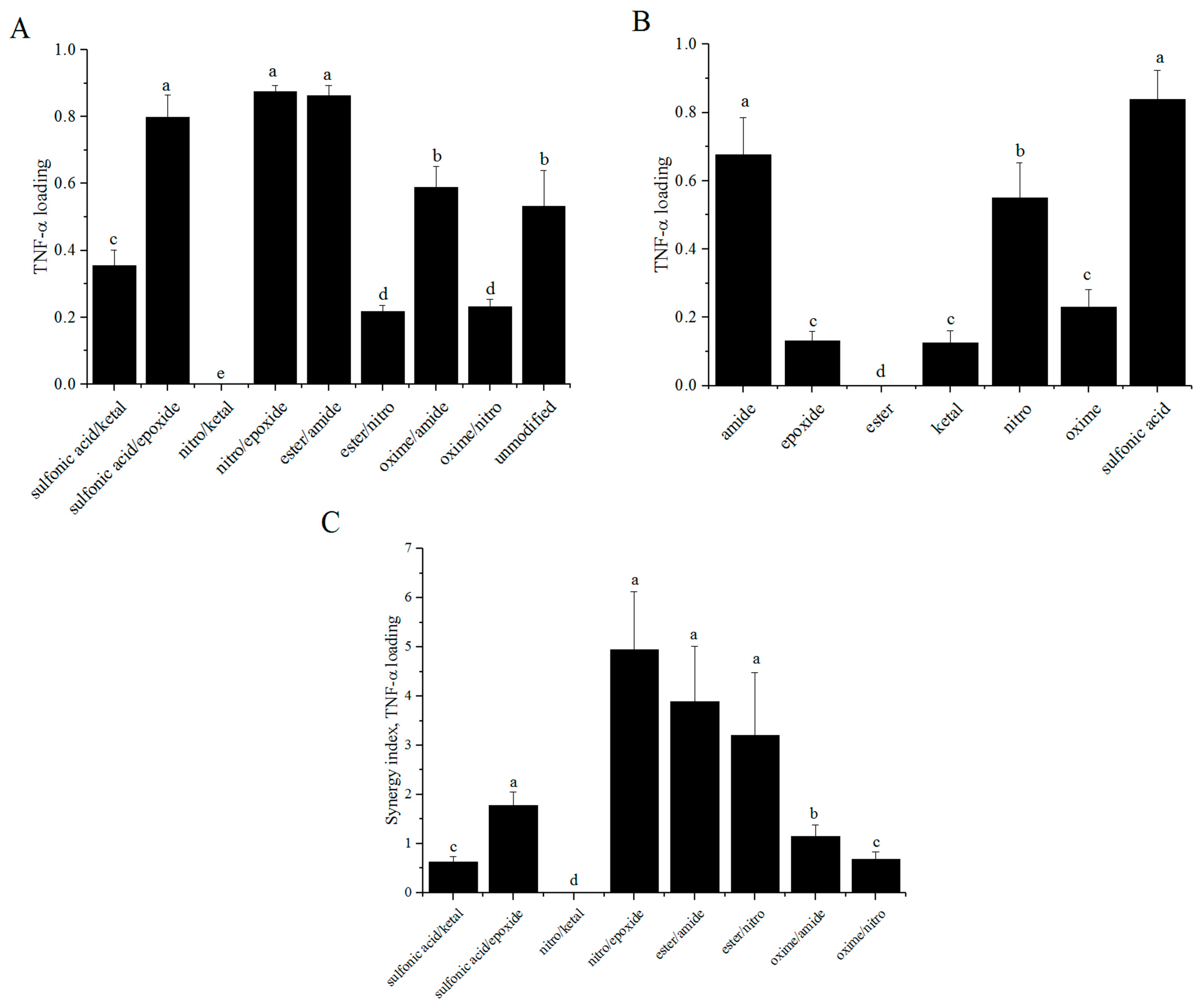
3.4. TNF-α ELISA
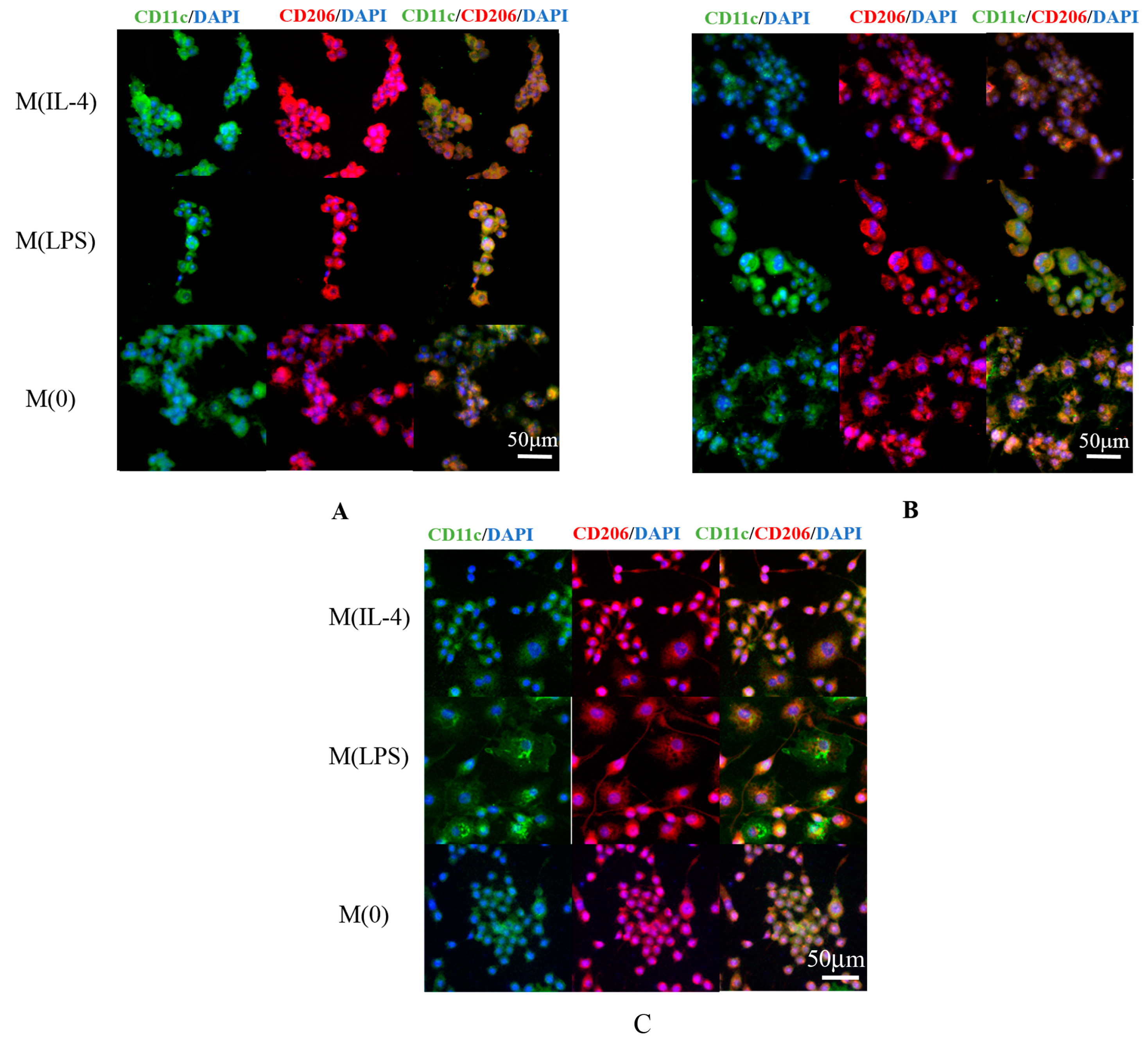
3.5. Fluorescent Imaging
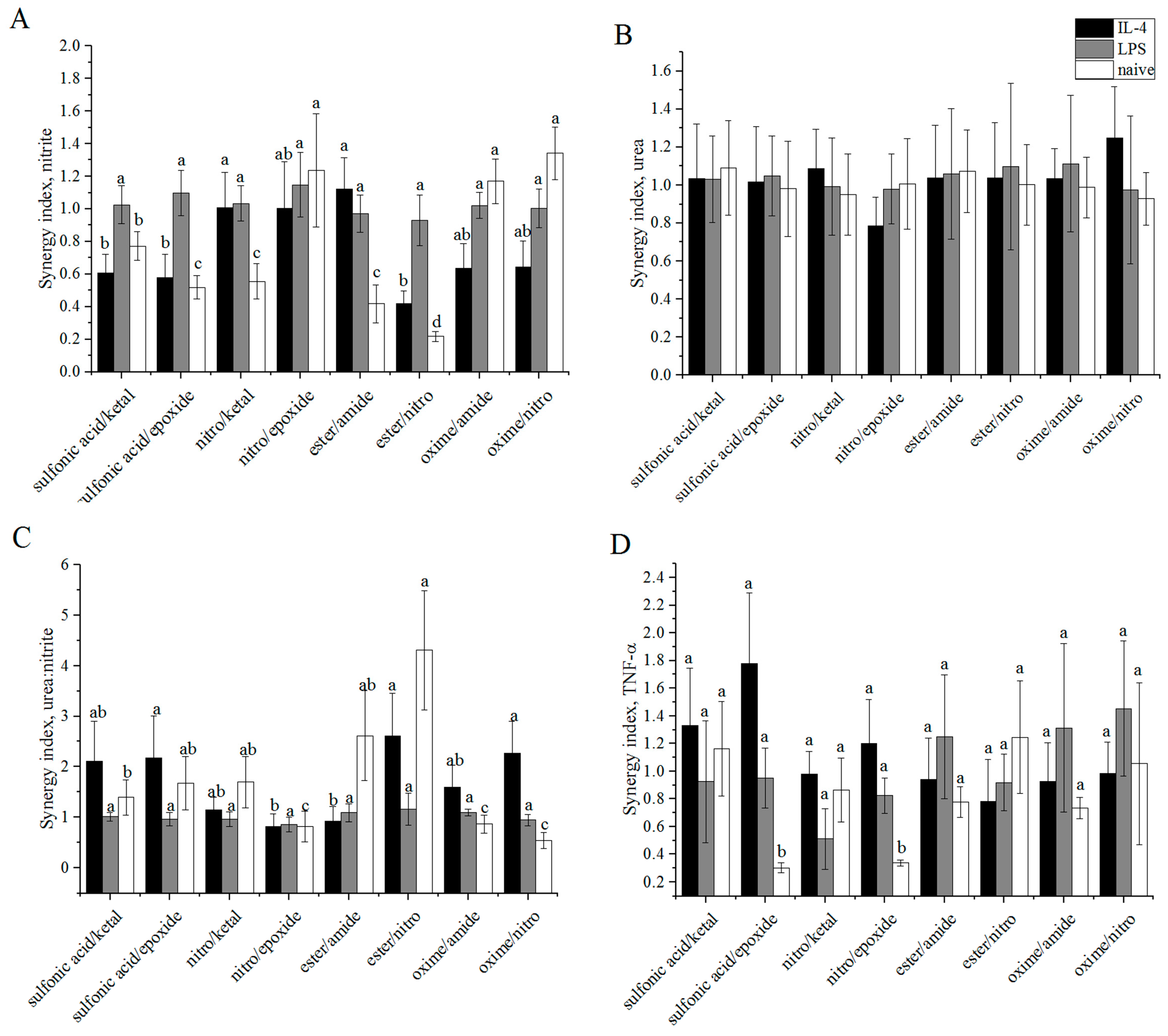
3.6. Synergy
3.7. TNF-α Diffusion
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Mantovani, A.; Sica, A. Macrophages, innate immunity and cancer: Balance, tolerance, and diversity. Curr. Opin. Immunol. 2010, 22, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Tran, T.-H.; Amiji, M. Macrophage repolarization with targeted alginate nanoparticles containing IL-10 plasmid DNA for the treatment of experimental arthritis. Biomaterials 2015, 61, 162–177. [Google Scholar] [CrossRef] [PubMed]
- Quatromoni, J.G.; Eruslanov, E. Tumor-associated macrophages: Function, phenotype, and link to prognosis in human lung cancer. Am. J. Transl. Res. 2012, 4, 376–389. [Google Scholar] [PubMed]
- Wynn, T.A.; Barron, L.; Thompson, R.W.; Madala, S.K.; Wilson, M.S.; Cheever, A.W.; Ramalingam, T. Quantitative assessment of macrophage functions in repair and fibrosis. Curr. Protoc. Immunol. 2011, 2011. [Google Scholar] [CrossRef]
- Mantovani, A.; Biswas, S.K.; Galdiero, M.R.; Sica, A.; Locati, M. Macrophage plasticity and polarization in tissue repair and remodelling. J. Pathol. 2013, 229, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Peer, D.; Karp, J.M.; Hong, S.; Farokhzad, O.C.; Margalit, R.; Langer, R. Nanocarriers as an emerging platform for cancer therapy. Nat. Nanotechnol. 2007, 2, 751–760. [Google Scholar] [CrossRef] [PubMed]
- Kelly, C.; Jefferies, C.; Cryan, S. Targeted liposomal drug delivery to monocytes and macrophages. J. Drug Deliv. 2011, 2011, 727241. [Google Scholar] [CrossRef] [PubMed]
- Bygd, H.C.; Forsmark, K.D.; Bratlie, K.M. The significance of macrophage phenotype in cancer and biomaterials. Clin. Transl. Med. 2014, 3. [Google Scholar] [CrossRef] [PubMed]
- Bygd, H.C.; Bratlie, K.M. The effect of chemically modified alginates on macrophage phenotype and biomolecule transport. J. Biomed. Mater. Res. Part A 2016, 104, 1707–1719. [Google Scholar] [CrossRef] [PubMed]
- Xia, Z.; Triffitt, J.T. A review on macrophage responses to biomaterials. Biomed. Mater. 2006, 1, R1–R9. [Google Scholar] [CrossRef] [PubMed]
- Ward, W.K. A Review of the Foreign-body Response to Subcutaneously-implanted Devices: The Role of Macrophages and Cytokines in Biofouling and Fibrosis. J. Diabetes Sci. Technol. 2008, 2, 768–777. [Google Scholar] [CrossRef]
- Murray, P.J.; Allen, J.E.; Biswas, S.K.; Fisher, E.A.; Gilroy, D.W.; Goerdt, S.; Gordon, S.; Hamilton, J.A.; Ivashkiv, L.B.; Lawrence, T.; et al. Macrophage Activation and polarization: Nomenclature and experimental guidelines. Immunity 2014, 41, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Martinez, F.O.; Gordon, S. The M1 and M2 paradigm of macrophage activation: Time for reassessment. F1000Prime Rep. 2014, 6, 13. [Google Scholar] [CrossRef] [PubMed]
- Akilbekova, D.; Philiph, R.; Graham, A.; Bratlie, K.M. Macrophage reprogramming: Influence of latex beads with various functional groups on macrophage phenotype and phagocytic uptake in vitro. J. Biomed. Mater. Res. Part A 2015, 103, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Bygd, H.C.; Forsmark, K.D.; Bratlie, K.M. Altering in vivo macrophage responses with modified polymer properties. Biomaterials 2015, 56, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Sica, A.; Mantovani, A. Macrophage plasticity and polarization: In vivo veritas. J. Clin. Investig. 2012, 122, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, A.; Meyerson, H.; Anderson, J.M. Quantitative in vivo cytokine analysis at synthetic biomaterial implant sites. J. Biomed. Mater. Res. A 2009, 89, 152–159. [Google Scholar] [PubMed]
- Suwandi, J.S.; Toes, R.E.M.; Nikolic, T.; Roep, B.O. Inducing tissue specific tolerance in autoimmune disease with tolerogenic dendritic cells. Clin. Exp. Rheumatol. 2015, 33, 97–103. [Google Scholar]
- Mesure, L.; De Visscher, G.; Vranken, I.; Lebacq, A.; Flameng, W. Gene expression study of monocytes/macrophages during early foreign body reaction and identification of potential precursors of myofibroblasts. PLoS ONE 2010, 5, e12949. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S. Alternative activation of macrophages. Nat. Rev. Immunol. 2003, 3, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.; Martinez, F.O. Alternative activation of macrophages: Mechanism and functions. Immunity 2010, 32, 593–604. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Mei, Y.; Hook, A.L.; Taylor, M.; Urquhart, A.J.; Bogatyrev, S.R.; Langer, R.; Anderson, D.G.; Davies, M.C.; Alexander, M.R. Polymer surface functionalities that control human embryoid body cell adhesion revealed by high throughput surface characterization of combinatorial material microarrays. Biomaterials 2010, 31, 8827–8838. [Google Scholar] [CrossRef] [PubMed]
- McNally, A.K.; Anderson, J.M. Complement C3 participation in monocyte adhesion to different surfaces. Proc. Natl. Acad. Sci. USA 1994, 91, 10119–10123. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Jones, K.S. Effect of alginate on innate immune activation of macrophages. J. Biomed. Mater. Res. A 2009, 90, 411–418. [Google Scholar] [CrossRef] [PubMed]
- MacLauchlan, S.; Skokos, E.A.; Meznarich, N.; Zhu, D.H.; Raoof, S.; Shipley, J.M.; Senior, R.M.; Bornstein, P.; Kyriakides, T.R. Macrophage fusion, giant cell formation, and the foreign body response require matrix metalloproteinase 9. J. Leukoc. Biol. 2009, 85, 617–626. [Google Scholar] [CrossRef] [PubMed]
- Mosser, D.M.; Edwards, J.P. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2008, 8, 958–969. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Y.P.; Zheng, G.; Lee, V.W.S.; Ouyang, L.; Chang, D.H.H.; Mahajan, D.; Coombs, J.; Wang, Y.M.; Alexander, S.I.; et al. Ex vivo programmed macrophages ameliorate experimental chronic inflammatory renal disease. Kidney Int. 2007, 72, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.M. Biological responses to materials. Annu. Rev. Mater. Res. 2001, 31, 81–110. [Google Scholar] [CrossRef]
- Lu, Y.; Huang, J.; Yu, G.; Cardenas, R.; Wei, S.; Wujcik, E.K.; Guo, Z. Coaxial electrospun fibers: Applications in drug delivery and tissue engineering. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2016, 8, 654–677. [Google Scholar] [CrossRef] [PubMed]
- Wujcik, E.K.; Monty, C.N. Nanotechnology for implantable sensors: Carbon nanotubes and graphene in medicine. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2013, 5, 233–249. [Google Scholar] [CrossRef] [PubMed]
- Panilaitis, B.; Altman, G.H.; Chen, J.; Jin, H.J.; Karageorgiou, V.; Kaplan, D.L. Macrophage responses to silk. Biomaterials 2003, 24, 3079–3085. [Google Scholar] [CrossRef]
- Wang, D.; Bratlie, K.M. Influence of polymer chemistry on cytokine secretion from polarized macrophages. ACS Biomater. Sci. Eng. 2015, 1, 166–174. [Google Scholar] [CrossRef]
- Wang, D.; Phan, N.; Isely, C.; Bruene, L.; Bratlie, K.M. Effect of surface modification and macrophage phenotype on particle internalization. Biomacromolecules 2014, 15, 4102–4110. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, M.; Mahon, K.; Anderson, D. Combinatorial and rational approaches to polymer synthesis for medicine. Adv. Drug Deliv. Rev. 2008, 60, 971–978. [Google Scholar] [CrossRef] [PubMed]
- Brocchini, S. Combinatorial chemistry and biomedical polymer development. Adv. Drug Deliv. Rev. 2001, 53, 123–130. [Google Scholar] [CrossRef]
- Liu, E.; Treiser, M.D.; Patel, H.; Sung, H.J.; Roskov, K.E.; Kohn, J.; Becker, M.L.; Moghe, P.V. High-content profiling of cell responsiveness to graded substrates based on combinatorially variant polymers. Comb. Chem. High Throughput Screen. 2009, 12, 645–655. [Google Scholar] [CrossRef]
- Tallarida, R.J. Drug synergism: Its detection and applications. J. Pharmacol. Exp. Ther. 2001, 298, 865–872. [Google Scholar] [PubMed]
- Mitragotri, S. Synergistic effect of enhancers for transdermal drug delivery. Pharm. Res. 2000, 17, 1354–1359. [Google Scholar] [CrossRef] [PubMed]
- Chou, T.C. Drug combination studies and their synergy quantification using the chou-talalay method. Cancer Res. 2010, 70, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Lehár, J.; Krueger, A.S.; Avery, W.; Heilbut, A.M.; Johansen, L.M.; Price, E.R.; Rickles, R.J.; Short, G.F.; Staunton, J.E.; Jin, X.; et al. Synergistic drug combinations tend to improve therapeutically relevant selectivity. Nat. Biotechnol. 2009, 27, 659–666. [Google Scholar] [CrossRef] [PubMed]
- Levinson, S.S. Kinetic centrifugal analyzer and manual determination of serum urea nitrogen, with use of o-phthaldialdehyde reagent. Clin. Chem. 1978, 24, 2199–2202. [Google Scholar] [PubMed]
- Kulseng, B.; Thu, B.; Espevik, T.; Skjåk-Braek, G. Alginate poly-lysine microcapsules as immune barrier: Permeability of cytokines and immunoglobins over the capsule membrane. Cell Transplant. 1997, 6, 387–394. [Google Scholar] [CrossRef]
- Karande, P.; Jain, A.; Mitragotri, S. Insights into synergistic interactions in binary mixtures of chemical permeation enhancers for transdermal drug delivery. J. Control. Release 2006, 115, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.Y.; Mooney, D.J. Alginate: Properties and biomedical applications. Prog. Polym. Sci. 2012, 37, 106–126. [Google Scholar] [CrossRef] [PubMed]
- Gombotz, W.R.; Wee, S.F. Protein release from alginate matrices. Adv. Drug Deliv. Rev. 2012, 64, 194–205. [Google Scholar] [CrossRef]
- Elliott, R.B.; Escobar, L.; Tan, P.L.; Muzina, M.; Zwain, S.; Buchanan, C. Live encapsulated porcine islets from a type 1 diabetic patient 9.5 yr after xenotransplantation. Xenotransplantation 2007, 14, 157–161. [Google Scholar] [CrossRef] [PubMed]
- De Vos, P.; van Hoogmoed, C.G.; van Zanten, J.; Netter, S.; Strubbe, J.H.; Busscher, H.J. Long-term biocompatibility, chemistry, and function of microencapsulated pancreatic islets. Biomaterials 2003, 24, 305–312. [Google Scholar] [CrossRef]
- Tam, S.K.; Bilodeau, S.; Dusseault, J.; Langlois, G.; Hallé, J.P.; Yahia, L.H. Biocompatibility and physicochemical characteristics of alginate-polycation microcapsules. Acta Biomater. 2011, 7, 1683–1692. [Google Scholar] [CrossRef] [PubMed]
- De Vos, P.; De Haan, B.; Van Schilfgaarde, R. Effect of the alginate composition on the biocompatibility of alginate-polylysine microcapsules. Biomaterials 1997, 18, 273–278. [Google Scholar] [CrossRef]
- Mallett, A.G.; Korbutt, G.S. Alginate modification improves long-term survival and function of transplanted encapsulated islets. Tissue Eng. Part A 2009, 15, 1301–1309. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-P.; Chu, I.-M.; Shiao, M.-Y.; Hsu, B.R.-S.; Fu, S.-H. Microencapsulation of islets in PEG-amine modified alginate-poly(l-Lysine )-alginate microcapsules for constructing bioartificial pancreas. J. Ferment. Bioeng. 1998, 86, 185–190. [Google Scholar] [CrossRef]
- Jork, A.; Thürmer, F.; Cramer, H.; Zimmermann, G.; Gessner, P.; Hämel, K.; Hofmann, G.; Kuttler, B.; Hahn, H.J.; Josimovic-Alasevic, O.; et al. Biocompatible alginate from freshly collected Laminaria pallida for implantation. Appl. Microbiol. Biotechnol. 2000, 53, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Petruzzo, P.; Cappai, A.; Ruiu, G.; Dessy, E.; Rescigno, A.; Brotzu, G. Development of biocompatible barium alginate microcapsules. Transplant. Proc. 1997, 1345, 2129–2130. [Google Scholar] [CrossRef]
- Schneider, S.; Feilen, P.J.; Kraus, O.; Haase, T.; Sagban, T.A.; Lehr, H.A.; Beyer, J.; Pommersheim, R.; Weber, M.M. Biocompatibility of alginates for grafting: Impact of alginate molecular weight. Artif. Cells Blood Substit. Biotechnol. 2003, 31, 383–394. [Google Scholar] [CrossRef]
- Zimmermann, U.; Thürmer, F.; Jork, A.; Weber, M.; Mimietz, S.; Hillgärtner, M.; Brunnenmeier, F.; Zimmermann, H.; Westphal, I.; Fuhr, G.; et al. A novel class of amitogenic alginate microcapsules for long-term immunoisolated transplantation. Ann. N. Y. Acad. Sci. 2006, 944, 199–215. [Google Scholar] [CrossRef]
- Steele, J.A.M.; Hallé, J.-P.; Poncelet, D.; Neufeld, R.J. Therapeutic cell encapsulation techniques and applications in diabetes. Adv. Drug Deliv. Rev. 2014, 67–68, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Paredes-Juarez, G.; de Haan, B.; Faas, M.; de Vos, P. A technology platform to test the efficacy of purification of alginate. Materials 2014, 7, 2087–2103. [Google Scholar] [CrossRef]
- Calafiore, R.; Basta, G. Clinical application of microencapsulated islets: Actual prospectives on progress and challenges. Adv. Drug Deliv. Rev. 2014, 67–68, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Shin, J.; Lee, J.S.; Byun, E.; Ryu, J.H.; Um, S.H.; Kim, D.I.; Lee, H.; Cho, S.W. Bioinspired, calcium-free alginate hydrogels with tunable physical and mechanical properties and improved biocompatibility. Biomacromolecules 2013, 14, 2004–2013. [Google Scholar] [CrossRef] [PubMed]
- Orive, G.; Hernandez, R.M.; Gascon, A.R. Survival of different cell lines in alginate-agarose microcapsules. Eur. J. Pharm. Sci. 2003, 18, 23–30. [Google Scholar] [CrossRef]
- Yang, J.-S.; Xie, Y.-J.; He, W. Research progress on chemical modification of alginate: A review. Carbohydr. Polym. 2011, 84, 33–39. [Google Scholar] [CrossRef]
- Pawar, S.N.; Edgar, K.J. Alginate derivatization: A review of chemistry, properties and applications. Biomaterials 2012, 33, 3279–3305. [Google Scholar] [CrossRef] [PubMed]
- Vegas, A.J.; Veiseh, O.; Doloff, J.C.; Ma, M.; Tam, H.H.; Bratlie, K.; Li, J.; Bader, A.R.; Langan, E.; Olejnik, K.; et al. Combinatorial hydrogel library enables identification of materials that mitigate the foreign body response in primates. Nat. Biotechnol. 2016, 34, 345–352. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bygd, H.C.; Bratlie, K.M. Investigating the Synergistic Effects of Combined Modified Alginates on Macrophage Phenotype. Polymers 2016, 8, 422. https://doi.org/10.3390/polym8120422
Bygd HC, Bratlie KM. Investigating the Synergistic Effects of Combined Modified Alginates on Macrophage Phenotype. Polymers. 2016; 8(12):422. https://doi.org/10.3390/polym8120422
Chicago/Turabian StyleBygd, Hannah C., and Kaitlin M. Bratlie. 2016. "Investigating the Synergistic Effects of Combined Modified Alginates on Macrophage Phenotype" Polymers 8, no. 12: 422. https://doi.org/10.3390/polym8120422





