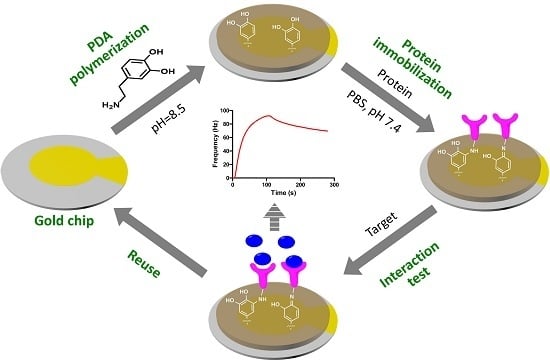
QCM Biosensor Based on Polydopamine Surface for Real-Time Analysis of the Binding Kinetics of Protein-Protein Interactions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. PDA Coating
2.3. Contact Angle Measurement
2.4. X-ray Photoelectron Spectroscopy (XPS) Measurement
2.5. Protein Immobilization
2.6. QCM Analysis of Protein-Protein Interactions
2.7. Reuse of the Sensor Chip
3. Results and Discussion
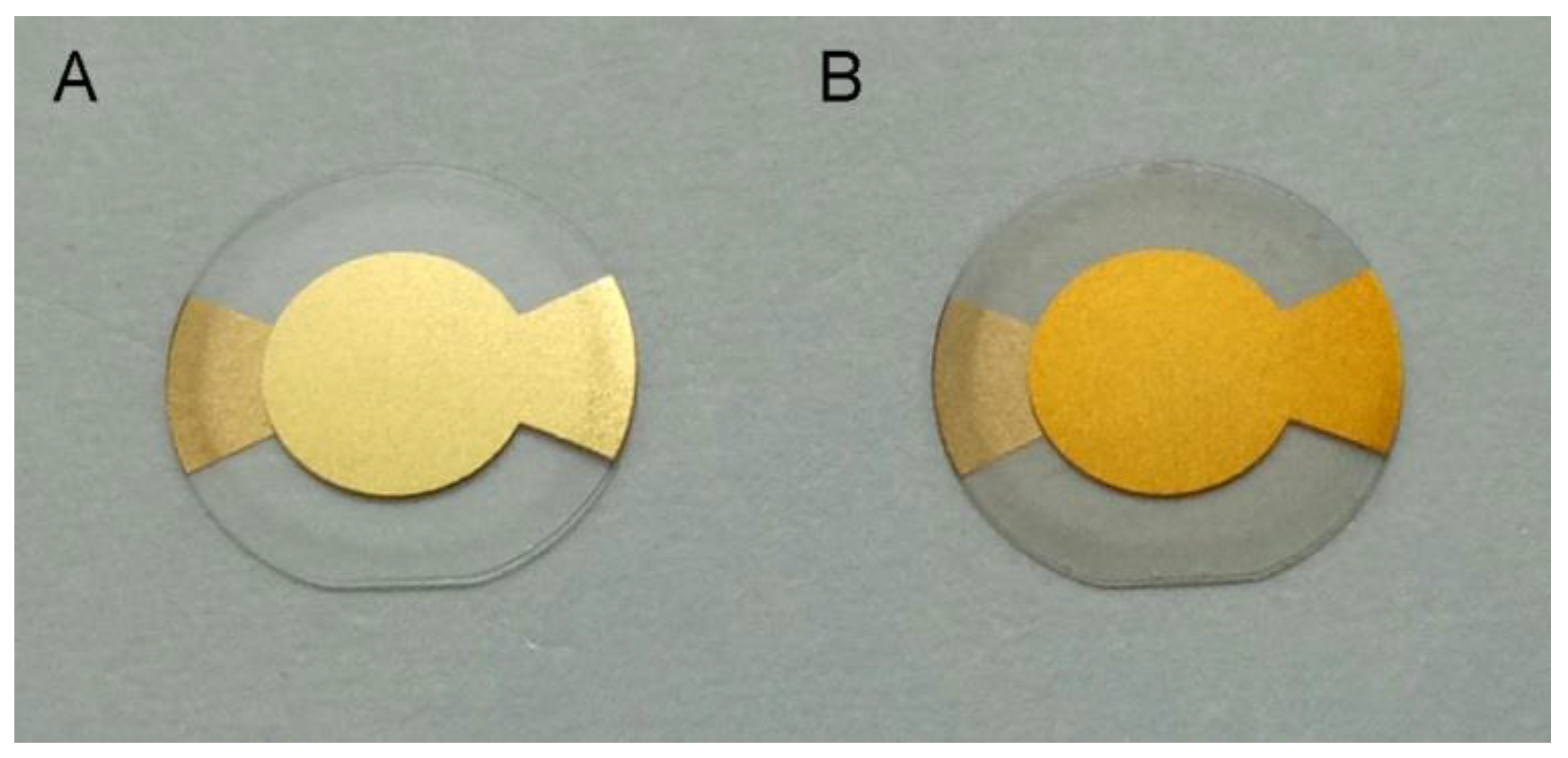
3.1. PDA Coating
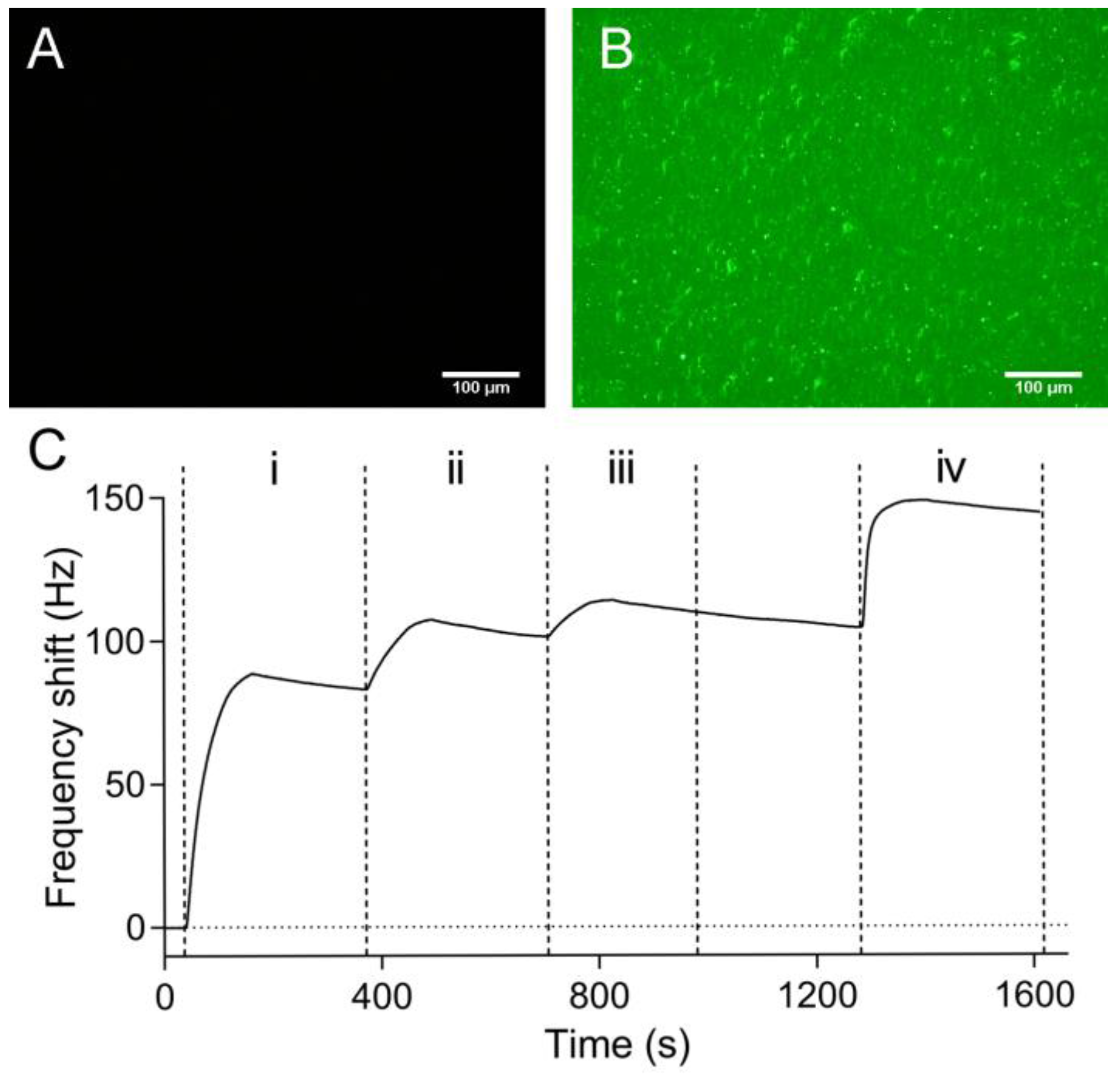
3.2. Protein Immobilization on the PDA-Coated Sensor Chip Surface
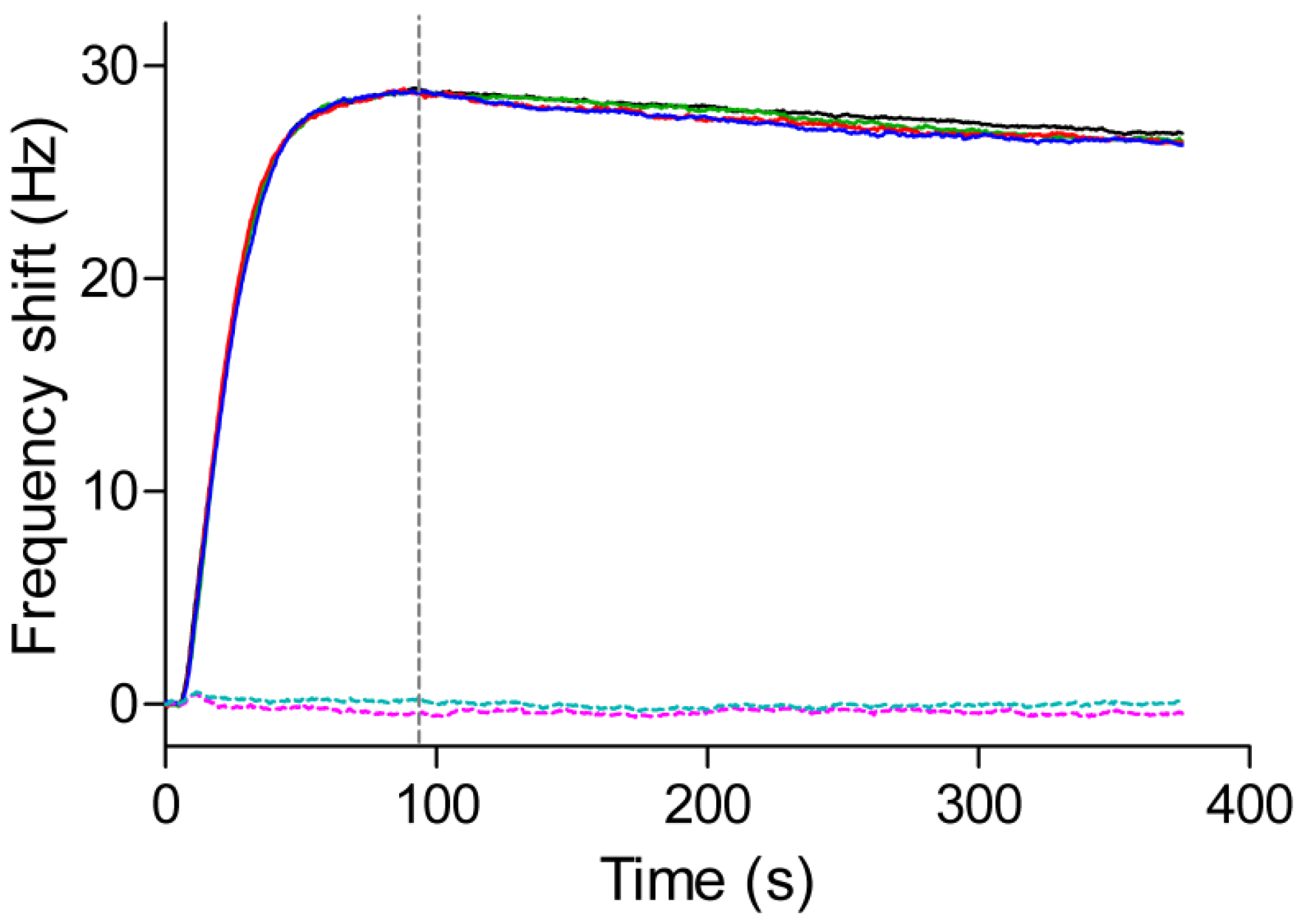
3.3. QCM Measurements of Protein-Protein Interaction
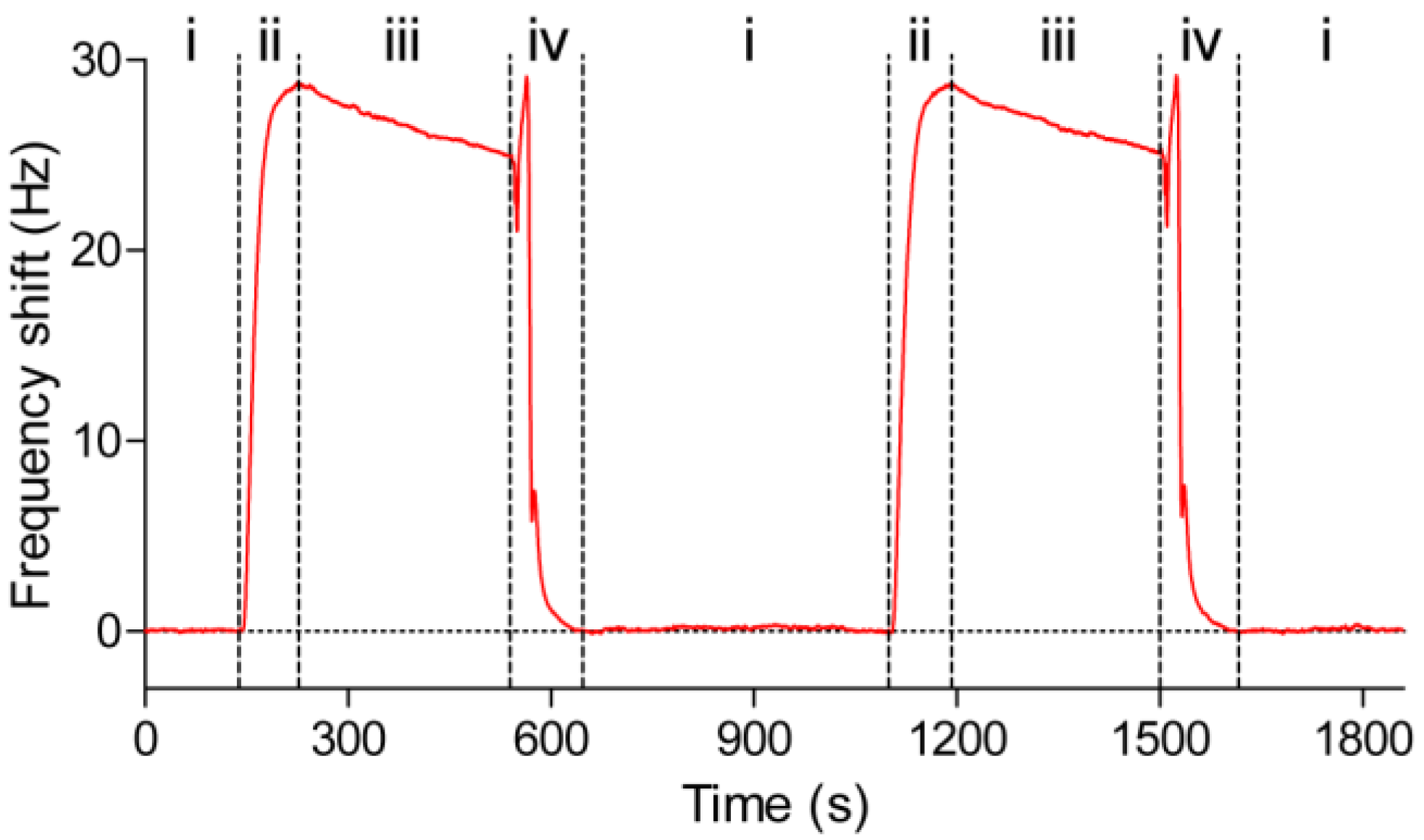
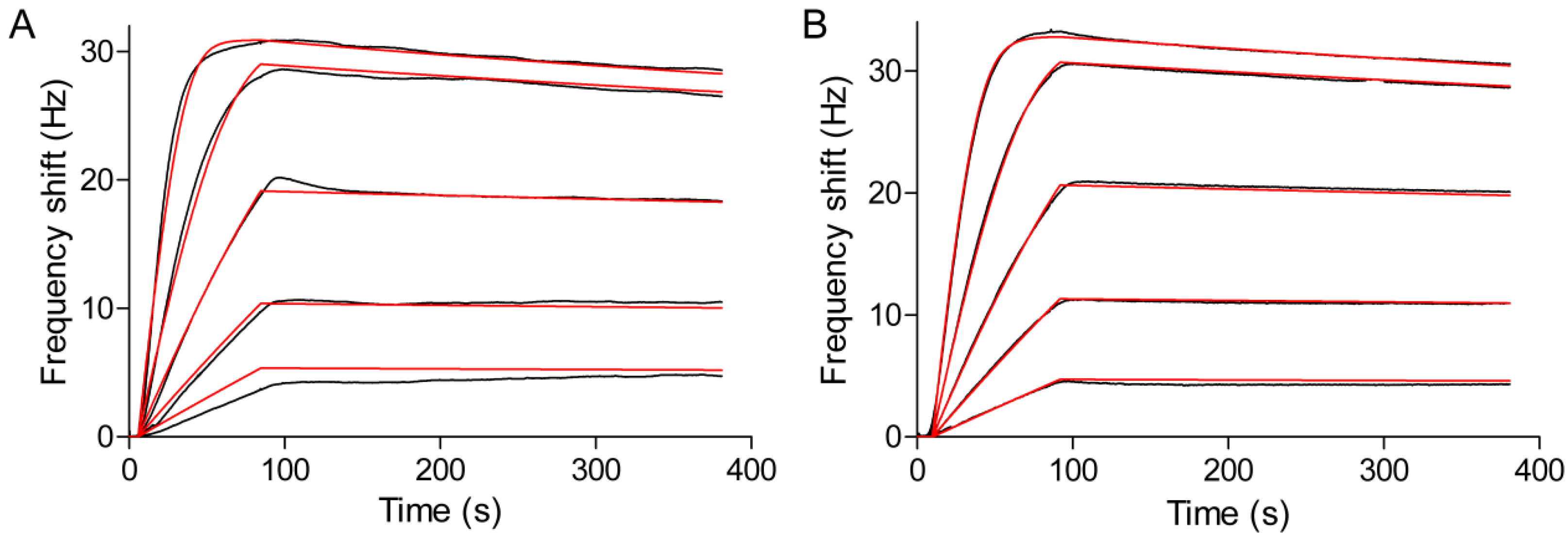
3.4. Kinetic and Affinity Studies
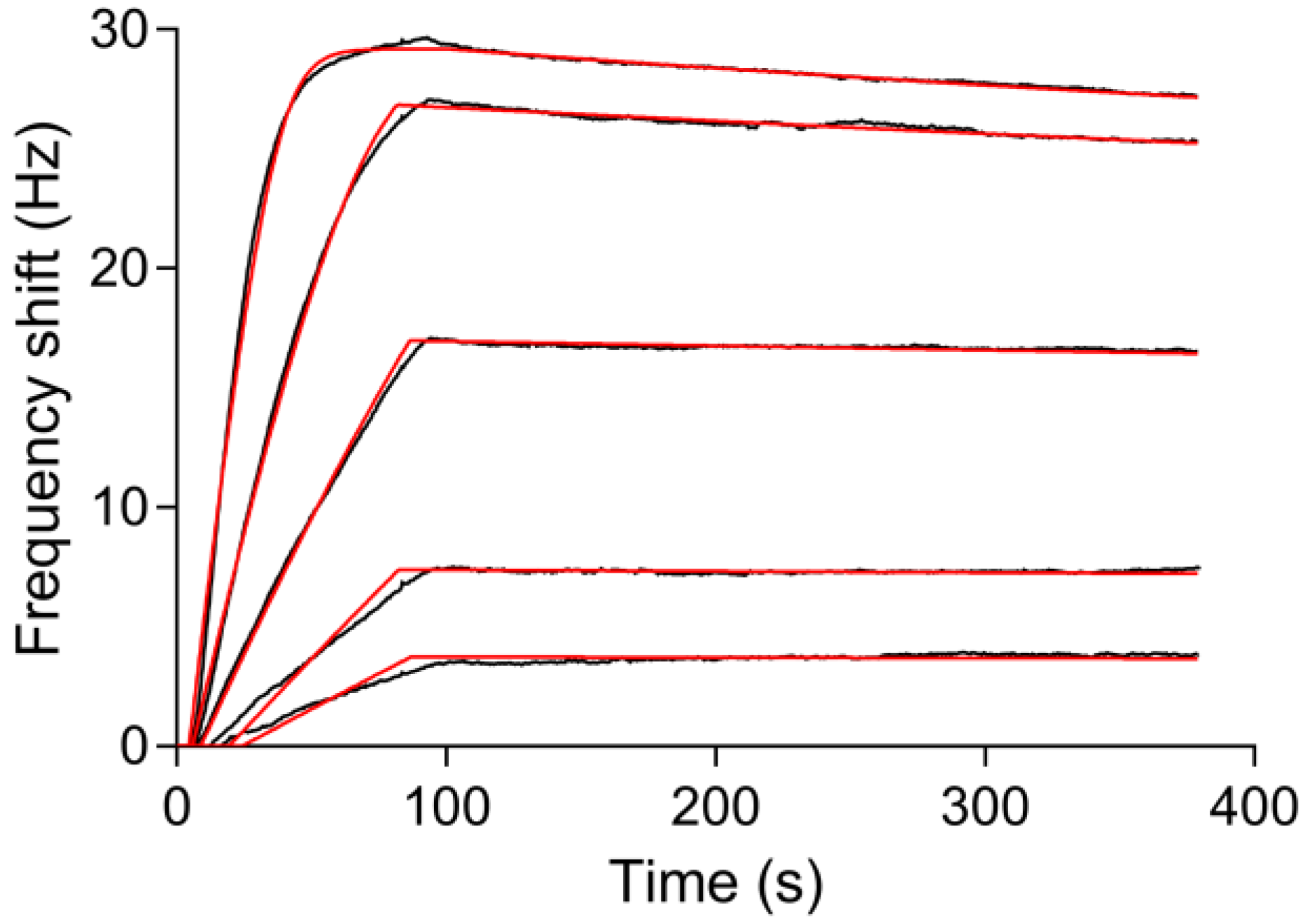
3.5. Reuse of the Sensor Chip for Knietic and Affinity Studies
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Li, X.; Song, S.; Shuai, Q.; Pei, Y.; Aastrup, T.; Pei, Y.; Pei, Z. Real-time and label-free analysis of binding thermodynamics of carbohydrate-protein interactions on unfixed cancer cell surfaces using a QCM biosensor. Sci. Rep. 2015, 5, 14066. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Pei, Y.; Zhang, R.; Shuai, Q.; Wang, F.; Aastrup, T.; Pei, Z. A suspension-cell biosensor for real-time determination of binding kinetics of protein-carbohydrate interactions on cancer cell surfaces. Chem. Commun. 2013, 49, 9908–9910. [Google Scholar] [CrossRef] [PubMed]
- Pei, Z.; Saint-Guirons, J.; Kack, C.; Ingemarsson, B.; Aastrup, T. Real-time analysis of the carbohydrates on cell surfaces using a QCM biosensor: A lectin-based approach. Biosens. Bioelectron. 2012, 35, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Pei, Z.; Larsson, R.; Aastrup, T.; Anderson, H.; Lehn, J.M.; Ramstrom, O. Quartz crystal microbalance bioaffinity sensor for rapid identification of glycosyldisulfide lectin inhibitors from a dynamic combinatorial library. Biosens. Bioelectron. 2006, 22, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Konradi, R.; Textor, M.; Reimhult, E. Using complementary acoustic and optical techniques for quantitative monitoring of biomolecular adsorption at interfaces. Biosensors 2012, 2, 341–376. [Google Scholar] [CrossRef] [PubMed]
- Keller, C.A.; Glasmastar, K.; Zhdanov, V.P.; Kasemo, B. Formation of supported membranes from vesicles. Phys. Rev. Lett. 2000, 84, 5443–5446. [Google Scholar] [CrossRef] [PubMed]
- Bailey, L.E.; Kambhampati, D.; Kanazawa, K.K.; Knoll, W.; Frank, C.W. Using surface plasmon resonance and the quartz crystal microbalance to monitor in situ the interfacial behavior of thin organic films. Langmuir 2002, 18, 479–489. [Google Scholar] [CrossRef]
- Bund, A.; Baba, A.; Berg, S.; Johannsmann, D.; Lubben, J.; Wang, Z.; Knoll, W. Combining surface plasmon resonance and quartz crystal microbalance for the in situ investigation of the electropolymerization and doping/dedoping of poly(pyrrole). J. Phys. Chem. B 2003, 107, 6743–6747. [Google Scholar] [CrossRef]
- Reimhult, E.; Larsson, C.; Kasemo, B.; Hook, F. Simultaneous surface plasmon resonance and quartz crystal microbalance with dissipation monitoring measurements of biomolecular adsorption events involving structural transformations and variations in coupled water. Anal. Chem. 2004, 76, 7211–7220. [Google Scholar] [CrossRef] [PubMed]
- Jachimska, B.; Tokarczyk, K. Combining surface plasmon resonance and quartz crystal microbalance to determine hydration of dendrimer monolayers. J. Phys. Chem. C 2016, 120, 19678–19685. [Google Scholar] [CrossRef]
- Larsson, E.M.; Edvardsson, M.E.M.; Langhammer, C.; Zoric, I.; Kasemo, B. A combined nanoplasmonic and electrodeless quartz crystal microbalance setup. Rev. Sci. Instrum. 2009, 80, 125105. [Google Scholar] [CrossRef] [PubMed]
- Ferhan, A.R.; Jackman, J.A.; Cho, N.J. Integration of quartz crystal microbalance-dissipation and reflection-mode localized surface plasmon resonance sensors for biomacromolecular interaction analysis. Anal. Chem. 2016, 88, 12524–12531. [Google Scholar] [CrossRef] [PubMed]
- Ferhan, A.R.; Jackman, J.A.; Cho, N.J. Investigating how vesicle size influences vesicle adsorption on titanium oxide: A competition between steric packing and shape deformation. PCCP 2017, 19, 2131–2139. [Google Scholar] [CrossRef] [PubMed]
- Hao, D.; Hu, C.; Grant, J.; Glidle, A.; Cumming, D.R.S. Hybrid localized surface plasmon resonance and quartz crystal microbalance sensor for label free biosensing. Biosens. Bioelectron. 2017, 100, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Hook, F.; Kasemo, B.; Nylander, T.; Fant, C.; Sott, K.; Elwing, H. Variations in coupled water, viscoelastic properties, and film thickness of a Mefp-1 protein film during adsorption and cross-linking: A quartz crystal microbalance with dissipation monitoring, ellipsometry, and surface plasmon resonance study. Anal. Chem. 2001, 73, 5796–5804. [Google Scholar] [CrossRef] [PubMed]
- Zan, G.H.; Cho, N.J. Rupture of zwitterionic lipid vesicles by an amphipathic, alpha-helical peptide: Indirect effects of sensor surface and implications for experimental analysis. Colloid Surf. B 2014, 121, 340–346. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Rodahl, M.; Edvardsson, M.; Svedhem, S.; Ohlsson, G.; Hook, F.; Kasemo, B. A combined reflectometry and quartz crystal microbalance with dissipation setup for surface interaction studies. Rev. Sci. Instrum. 2008, 79, 075107. [Google Scholar] [CrossRef] [PubMed]
- Cho, N.J.; Wang, G.L.; Edvardsson, M.; Glenn, J.S.; Hook, F.; Frank, C.W. Alpha-helical peptide-induced vesicle rupture revealing new insight into the vesicle fusion process as monitored in situ by quartz crystal microbalance-dissipation and reflectometry. Anal. Chem. 2009, 81, 4752–4761. [Google Scholar] [CrossRef] [PubMed]
- Edvardsson, M.; Svedhem, S.; Wang, G.; Richter, R.; Rodahl, M.; Kasemo, B. QCM-D and reflectometry instrument: Applications to supported lipid structures and their biomolecular interactions. Anal. Chem. 2009, 81, 349–361. [Google Scholar] [CrossRef] [PubMed]
- Cooper, M.A.; Singleton, V.T. A survey of the 2001 to 2005 quartz crystal microbalance biosensor literature: Applications of acoustic physics to the analysis of biomolecular interactions. J. Mol. Recognit. 2007, 20, 154–184. [Google Scholar] [CrossRef] [PubMed]
- Becker, B.; Cooper, M.A. A survey of the 2006–2009 quartz crystal microbalance biosensor literature. J. Mol. Recognit. 2011, 24, 754–787. [Google Scholar] [CrossRef] [PubMed]
- Speight, R.E.; Cooper, M.A. A survey of the 2010 quartz crystal microbalance literature. J. Mol. Recognit. 2012, 25, 451–473. [Google Scholar] [CrossRef] [PubMed]
- Hosseiny, S.S.; van Rijn, P. Surface initiated polymerizations via e-ATRP in pure water. Polymers 2013, 5, 1229–1240. [Google Scholar] [CrossRef]
- Isaksson, S.; Watkins, E.B.; Browning, K.L.; Lind, T.K.; Cardenas, M.; Hedfalk, K.; Hook, F.; Andersson, M. Protein-containing lipid bilayers intercalated with size-matched mesoporous silica thin films. Nano Lett. 2017, 17, 476–485. [Google Scholar] [CrossRef] [PubMed]
- Lundgren, A.; Agnarsson, B.; Zirbs, R.; Zhdanov, V.P.; Reimhult, E.; Hook, F. Nonspecific colloidal-type interaction explains size-dependent specific binding of membrane-targeted nanoparticles. ACS Nano 2016, 10, 9974–9982. [Google Scholar] [CrossRef] [PubMed]
- Jönsson, M.; Anderson, H.; Lindberg, U.; Aastrup, T. Quartz crystal microbalance biosensor designii. Simulation of sample transport. Sens. Actuators B Chem. 2007, 123, 21–26. [Google Scholar] [CrossRef]
- Anderson, H.; Jönsson, M.; Vestling, L.; Lindberg, U.; Aastrup, T. Quartz crystal microbalance sensor designi. Experimental study of sensor response and performance. Sens. Actuators B Chem. 2007, 123, 27–34. [Google Scholar] [CrossRef]
- Li, X.; Song, S.; Pei, Y.; Dong, H.; Aastrup, T.; Pei, Z. Oriented and reversible immobilization of his-tagged proteins on two- and three-dimensional surfaces for study of protein-protein interactions by a QCM biosensor. Sens. Actuators B Chem. 2016, 224, 814–822. [Google Scholar] [CrossRef]
- Pei, Z.; Anderson, H.; Aastrup, T.; Ramstrom, O. Study of real-time lectin-carbohydrate interactions on the surface of a quartz crystal microbalance. Biosens. Bioelectron. 2005, 21, 60–66. [Google Scholar] [CrossRef] [PubMed]
- D’Ulivo, L.; Saint-Guirons, J.; Ingemarsson, B.; Riekkola, M.L. Quartz crystal microbalance, a valuable tool for elucidation of interactions between apoB-100 peptides and extracellular matrix components. Anal. Bioanal. Chem. 2010, 396, 1373–1380. [Google Scholar] [CrossRef] [PubMed]
- Grellier, B.; Le Pogam, F.; Vitorino, M.; Starck, J.P.; Geist, M.; Duong, V.; Haegel, H.; Menguy, T.; Bonnefoy, J.Y.; Marchand, J.B.; et al. 3D modeling and characterization of the human CD115 monoclonal antibody H27K15 epitope and design of a chimeric CD115 target. MAbs 2014, 6, 533–546. [Google Scholar] [CrossRef] [PubMed]
- Qiu, H.; Yang, B.; Pei, Z.C.; Zhang, Z.; Ding, K. WSS25 inhibits growth of xenografted hepatocellular cancer cells in nude mice by disrupting angiogenesis via blocking bone morphogenetic protein (BMP)/smad/id1 signaling. J. Biol. Chem. 2010, 285, 32638–32646. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wu, C.; Zhang, S. Ultrasensitive detection of DNA and ramos cell using in situ selective crystallization based quartz crystal microbalance. Anal. Chem. 2017, 89, 4309–4313. [Google Scholar] [CrossRef] [PubMed]
- Szabo, J.E.; Nemeth, V.; Papp-Kadar, V.; Nyiri, K.; Leveles, I.; Bendes, A.A.; Zagyva, I.; Rona, G.; Palinkas, H.L.; Besztercei, B.; et al. Highly potent dutpase inhibition by a bacterial repressor protein reveals a novel mechanism for gene expression control. Nucleic Acids Res. 2014, 42, 11912–11920. [Google Scholar] [CrossRef] [PubMed]
- Viitala, T.; Vikholm, I.; Peltonen, J. Protein immobilization to a partially cross-linked organic monolayer. Langmuir 2000, 16, 4953–4961. [Google Scholar] [CrossRef]
- Myrskog, A.; Anderson, H.; Aastrup, T.; Ingemarsson, B.; Liedberg, B. Esterification of self-assembled carboxylic-acid-terminated thiol monolayers in acid environment: A time-dependent study. Langmuir 2010, 26, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Rho, J.; Messersmith, P.B. Facile conjugation of biomolecules onto surfaces via mussel adhesive protein inspired coatings. Adv. Mater. 2009, 21, 431–434. [Google Scholar] [CrossRef] [PubMed]
- Pei, Z.; Anderson, H.; Myrskog, A.; Duner, G.; Ingemarsson, B.; Aastrup, T. Optimizing immobilization on two-dimensional carboxyl surface: pH dependence of antibody orientation and antigen binding capacity. Anal. Biochem. 2010, 398, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Johnsson, B.; Löfås, S.; Lindquist, G. Immobilization of proteins to a carboxymethyldextran-modified gold surface for biospecific interaction analysis in surface plasmon resonance sensors. Anal. Biochem. 1991, 198, 268–277. [Google Scholar] [CrossRef]
- Della Ventura, B.; Schiavo, L.; Altucci, C.; Esposito, R.; Velotta, R. Light assisted antibody immobilization for bio-sensing. Biomed. Opt. Express 2011, 2, 3223–3231. [Google Scholar] [CrossRef] [PubMed]
- Funari, R.; Della Ventura, B.; Altucci, C.; Offenhausser, A.; Mayer, D.; Velotta, R. Single molecule characterization of UV-activated antibodies on gold by atomic force microscopy. Langmuir 2016, 32, 8084–8091. [Google Scholar] [CrossRef] [PubMed]
- Löfås, S.; Johnsson, B. A novel hydrogel matrix on gold surfaces in surface plasmon resonance sensors for fast and efficient covalent immobilization of ligands. J. Chem. Soc. Chem. Commun. 1990, 0, 1526–1528. [Google Scholar] [CrossRef]
- Duner, G.; Anderson, H.; Pei, Z.; Ingemarsson, B.; Aastrup, T.; Ramstrom, O. Signal enhancement in ligand-receptor interactions using dynamic polymers at quartz crystal microbalance sensors. Analyst 2016, 141, 3993–3996. [Google Scholar] [CrossRef] [PubMed]
- Sivaraman, B.; Fears, K.P.; Latour, R.A. Investigation of the effects of surface chemistry and solution concentration on the conformation of adsorbed proteins using an improved circular dichroism method. Langmuir 2009, 25, 3050–3056. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, K.; Sakiyama, T.; Imamura, K. On the adsorption of proteins on solid surfaces, a common but very complicated phenomenon. J. Biosci. Bioeng. 2001, 91, 233–244. [Google Scholar] [CrossRef]
- Pei, Y.; Yu, H.; Pei, Z.; Theurer, M.; Ammer, C.; André, S.; Gabius, H.-J.; Yan, M.; Ramström, O. Photoderivatized polymer thin films at quartz crystal microbalance surfaces: Sensors for carbohydrate-protein interactions. Anal. Chem. 2007, 79, 6897–6902. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Dellatore, S.M.; Miller, W.M.; Messersmith, P.B. Mussel-inspired surface chemistry for multifunctional coatings. Science 2007, 318, 426–430. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ai, K.; Lu, L. Polydopamine and its derivative materials: Synthesis and promising applications in energy, environmental, and biomedical fields. Chem. Rev. 2014, 114, 5057–5115. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.; Fan, F.; Zhang, Y.; Pei, Z.; Wang, W.; Pei, Y. A facile approach for fabrication of core-shell magnetic molecularly imprinted nanospheres towards hypericin. Polymers 2017, 9, 135. [Google Scholar] [CrossRef]
- Shi, C.; Dai, J.; Li, C.; Shen, X.; Peng, L.; Zhang, P.; Wu, D.; Sun, D.; Zhao, J. A modified ceramic-coating separator with high-temperature stability for lithium-ion battery. Polymers 2017, 9, 159. [Google Scholar] [CrossRef]
- Wang, K.; Dong, Y.; Zhang, W.; Zhang, S.; Li, J. Preparation of stable superhydrophobic coatings on wood substrate surfaces via mussel-inspired polydopamine and electroless deposition methods. Polymers 2017, 9, 218. [Google Scholar] [CrossRef]
- Del Frari, D.; Bour, J.; Ball, V.; Toniazzo, V.; Ruch, D. Degradation of polydopamine coatings by sodium hypochlorite: A process depending on the substrate and the film synthesis method. Polym. Degrad. Stab. 2012, 97, 1844–1849. [Google Scholar] [CrossRef]
- Sauerbrey, G. Verwendung von schwingquarzen zur wägung dünner schichten und zur mikrowägung. Zeitschrift Physik 1959, 155, 206–222. [Google Scholar] [CrossRef]
- Wei, Q.; Zhang, F.L.; Li, J.; Li, B.J.; Zhao, C.S. Oxidant-induced dopamine polymerization for multifunctional coatings. Polym. Chem. 2010, 1, 1430–1433. [Google Scholar] [CrossRef]
- Hu, M.; Mi, B. Enabling graphene oxide nanosheets as water separation membranes. Environ. Sci. Technol. 2013, 47, 3715–3723. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, D.; Dong, Z.; Zhang, F.; Jin, J. Interface chemistry engineering for stable cycling of reduced Go/SnO2 nanocomposites for lithium ion battery. Nano Lett. 2013, 13, 1711–1716. [Google Scholar] [CrossRef] [PubMed]
- Drake, A.W.; Klakamp, S.L. A strategic and systematic approach for the determination of biosensor regeneration conditions. J. Immunol. Methods 2011, 371, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Loo, L.; Wu, W.; Shih, W.Y.; Shih, W.-H.; Borghaei, H.; Pourrezaei, K.; Adams, G.P. A rapid method to regenerate piezoelectric microcantilever sensors (PEMS). Sensors 2011, 11, 5520–5528. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Lajevardi-Khosh, A.; Choi, S.; Chae, J. Regenerative surface plasmon resonance (SPR) biosensor: Real-time measurement of fibrinogen in undiluted human serum using the competitive adsorption of proteins. Biosens. Bioelectron. 2011, 28, 304–307. [Google Scholar] [CrossRef] [PubMed]
- Vashist, S.K. A method for regenerating gold surface for prolonged reuse of gold-coated surface plasmon resonance chip. Anal. Biochem. 2012, 423, 23–25. [Google Scholar] [CrossRef] [PubMed]



© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, C.; Li, X.; Song, S.; Pei, Y.; Guo, L.; Pei, Z. QCM Biosensor Based on Polydopamine Surface for Real-Time Analysis of the Binding Kinetics of Protein-Protein Interactions. Polymers 2017, 9, 482. https://doi.org/10.3390/polym9100482
Wu C, Li X, Song S, Pei Y, Guo L, Pei Z. QCM Biosensor Based on Polydopamine Surface for Real-Time Analysis of the Binding Kinetics of Protein-Protein Interactions. Polymers. 2017; 9(10):482. https://doi.org/10.3390/polym9100482
Chicago/Turabian StyleWu, Chunli, Xueming Li, Siyu Song, Yuxin Pei, Lili Guo, and Zhichao Pei. 2017. "QCM Biosensor Based on Polydopamine Surface for Real-Time Analysis of the Binding Kinetics of Protein-Protein Interactions" Polymers 9, no. 10: 482. https://doi.org/10.3390/polym9100482