A Cationic Smart Copolymer for DNA Binding
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
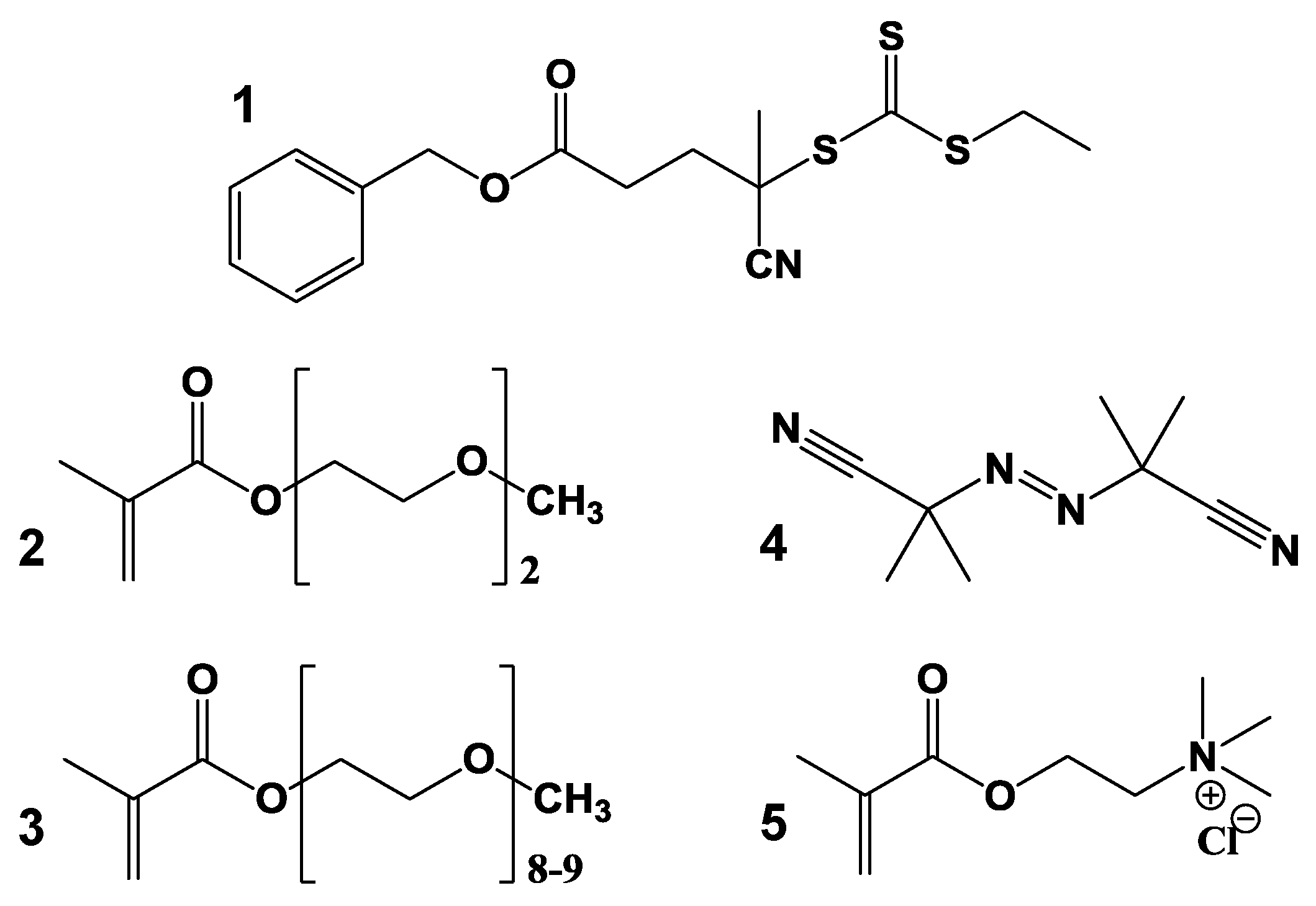
2.2. Synthesis of the Thermoresponsive Block of the Copolymer by RAFT Polymerization
2.3. Synthesis of the Charged Block of the Copolymer by RAFT Polymerization
2.4. Preparation of Block Copolymer-DNA Coacervates
2.5. Size Exclusion Chromatography (SEC)
2.6. Nuclear Magnetic Resonance (NMR)
2.7. Dynamic Light Scattering (DLS)
2.8. Fluorescence Correlation Spectroscopy
2.9. UV–Vis Absorption Spectroscopy
2.10. Fluorescence Spectroscopy
3. Results and Discussion
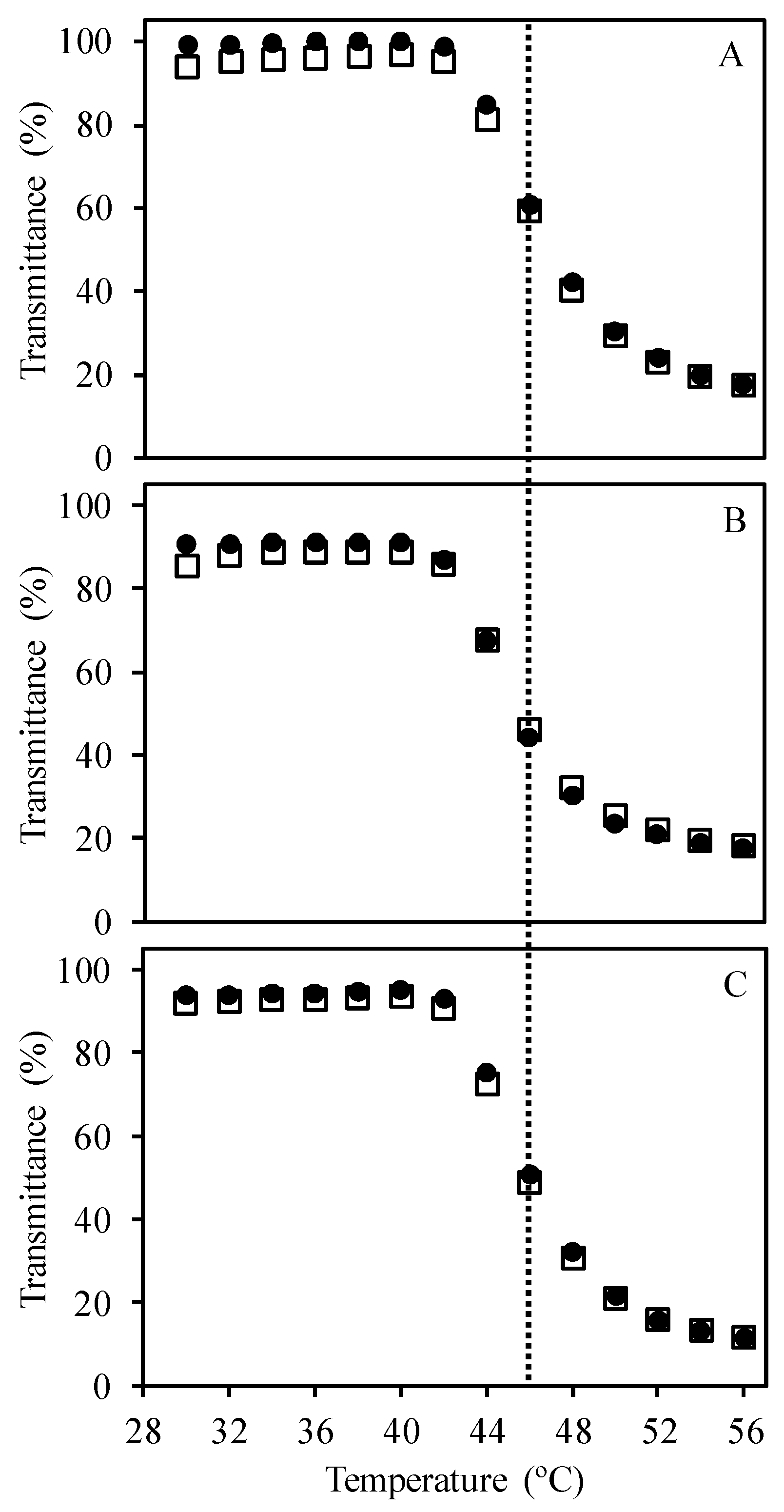
3.1. RAFT Cationic Stimuli-Responsive Copolymer
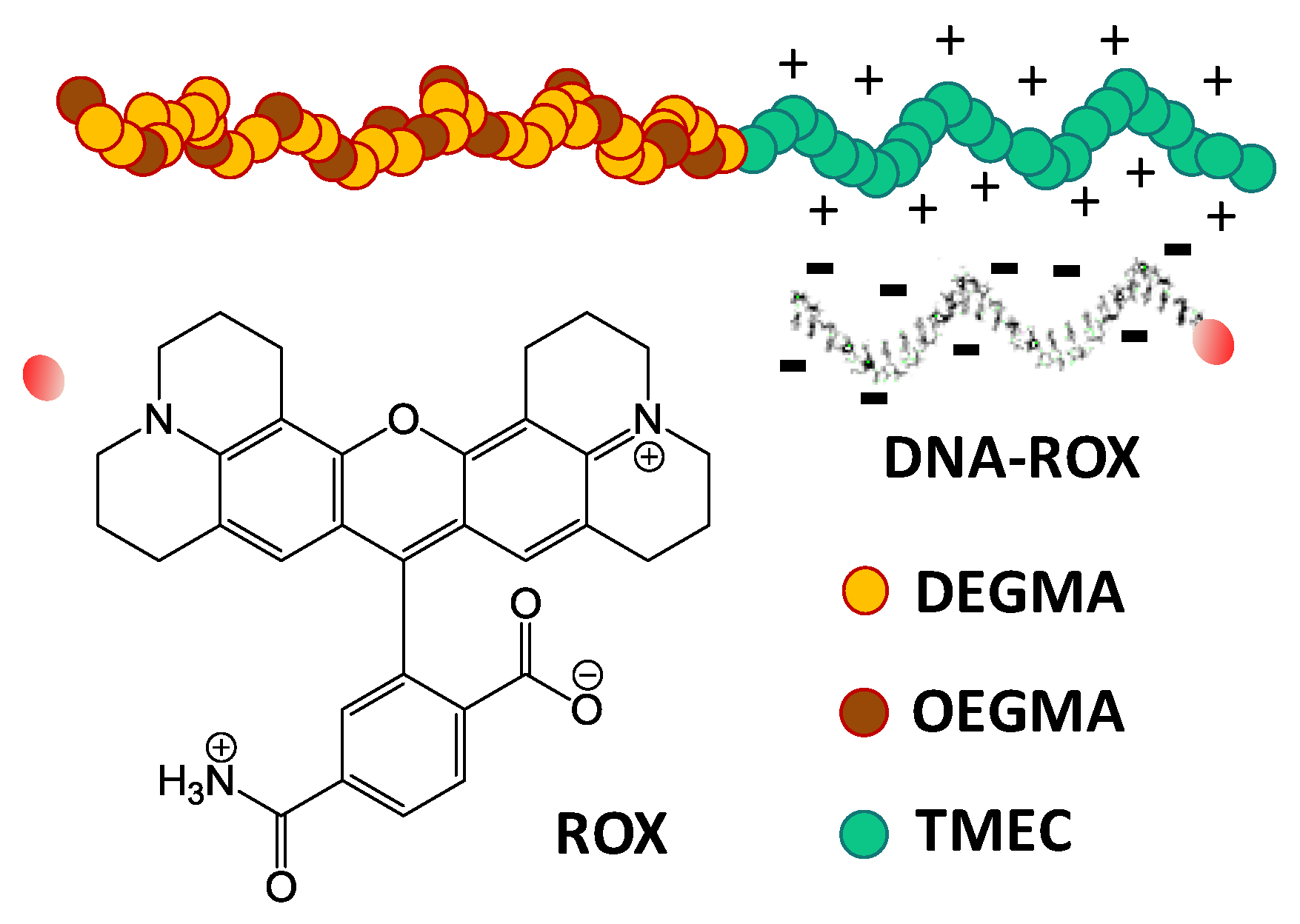
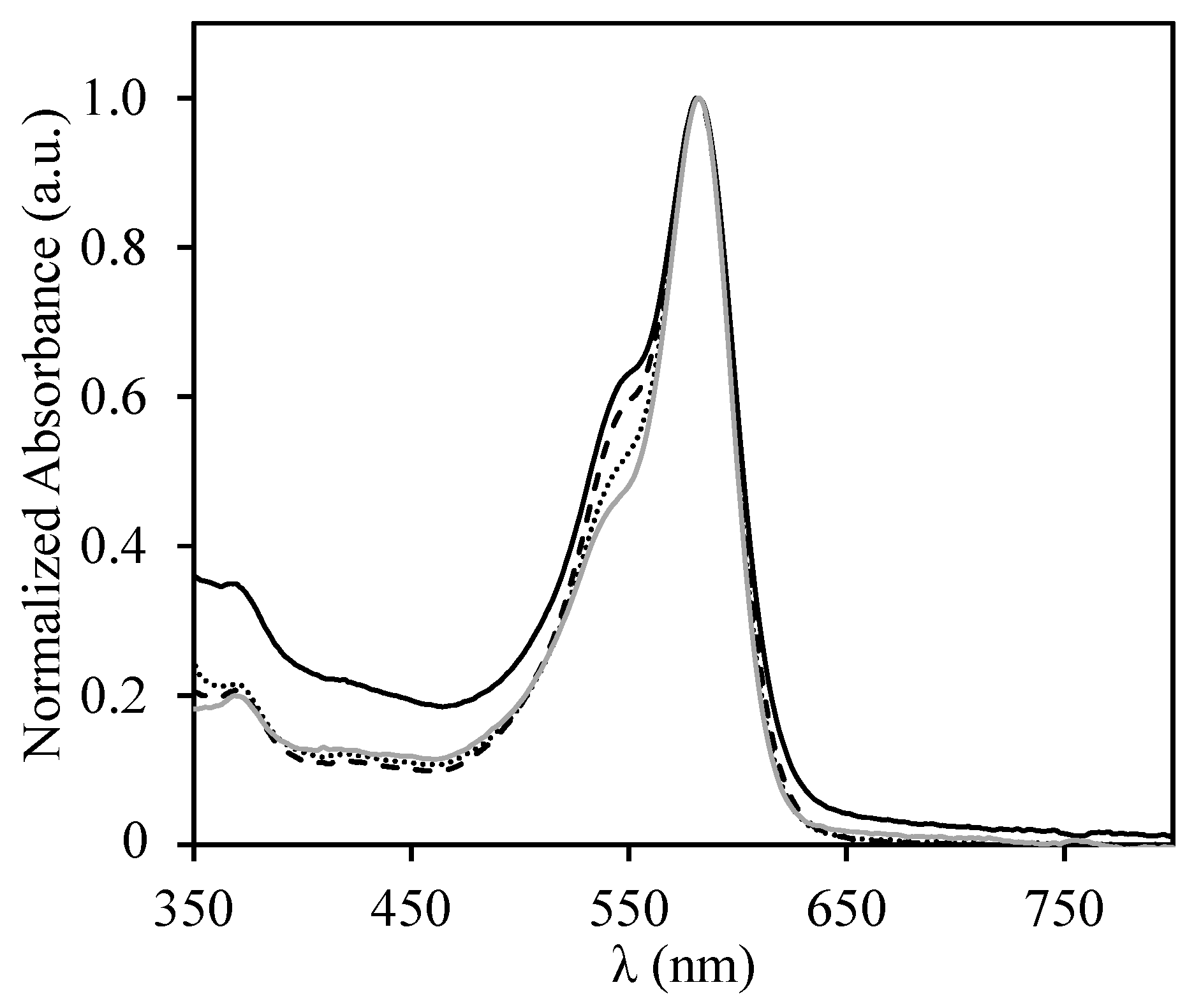
3.2. Interaction of the Block Copolymer Chains with a Fluorescently-Labeled Oligonucleotide
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Pinheiro, J.P.; Moura, L.; Fokkink, R.; Farinha, J.P.S. Preparation and Characterization of Low Dispersity Anionic Multiresponsive Core–Shell Polymer Nanoparticles. Langmuir 2012, 28, 5802–5809. [Google Scholar] [CrossRef] [PubMed]
- Eslahi, N.; Abdorahim, M.; Simchi, A. Smart Polymeric Hydrogels for Cartilage Tissue Engineering: A Review on the Chemistry and Biological Functions. Biomacromolecules 2016, 17, 3441–3463. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Gao, D.; Cao, Z.; Zhang, C.; Cheng, C.; Liu, J.; Shuai, X. Drug and gene co-delivery systems for cancer treatment. Biomater. Sci. 2015, 3, 1035–1049. [Google Scholar] [CrossRef] [PubMed]
- Moghanjoughi, A.A.; Khoshnevis, D.; Zarrabi, A. A concise review on smart polymers for controlled drug release. Drug Deliv. Transl. Res. 2016, 6, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Galaev, I.Y.; Mattiasson, B. ‘Smart’ polymers and what they could do in biotechnology and medicine. Trends Biotechnol. 1999, 17, 335–340. [Google Scholar] [CrossRef]
- Halperin, A.; Kröger, M.; Winnik, F.M. Poly(N-isopropylacrylamide) Phase Diagrams: Fifty Years of Research. Angew. Chem. Int. Ed. Engl. 2015, 54, 15342–15367. [Google Scholar] [CrossRef] [PubMed]
- Hoare, T.R.; Kohane, D.S. Hydrogels in drug delivery: Progress and challenges. Polymer 2008, 49, 1993–2007. [Google Scholar] [CrossRef]
- Knop, K.; Hoogenboom, R.; Fischer, D.; Schubert, U.S. Poly(ethylene glycol) in drug delivery: Pros and cons as well as potential alternatives. Angew. Chem. Int. Ed. Engl. 2010, 49, 6288–6308. [Google Scholar] [CrossRef] [PubMed]
- Lutz, J.-F. Polymerization of Oligo(Ethylene Glycol) (Meth)Acrylates: Toward New Generations of Smart Biocompatible Materials. J. Polym. Sci. A 2008, 46, 3459–3470. [Google Scholar] [CrossRef]
- Lutz, J.-F. Thermo-Switchable Materials Prepared Using the OEGMA-Platform. Adv. Mater. 2011, 23, 2237–2247. [Google Scholar] [CrossRef]
- Lutz, J.-F.; Akdemir, Ö.; Hoth, A. Point by Point Comparison of Two Thermosensitive Polymers Exhibiting a Similar LCST: Is the Age of Poly(NIPAM) over? J. Am. Chem. Soc. 2006, 128, 13046–13047. [Google Scholar] [CrossRef] [PubMed]
- Truong, N.P.; Quinn, J.F.; Anastasaki, A.; Haddleton, D.M.; Whittaker, M.R.; Davis, T.P. Facile access to thermoresponsive filomicelles with tuneable cores. Chem. Commun. 2016, 52, 4497–4500. [Google Scholar] [CrossRef] [PubMed]
- Truong, N.P.; Quinn, J.F.; Anastasaki, A.; Rolland, M.; Vu, M.N.; Haddleton, D.M.; Whittaker, M.R.; Davis, T.P. Surfactant-free RAFT emulsion polymerization using a novel biocompatible thermoresponsive polymer. Polym. Chem. 2017, 8, 1353–1363. [Google Scholar] [CrossRef]
- Zhang, Y.; Aigner, A.; Agarwal, S. Degradable and Biocompatible Poly(N,N-dimethylaminoethyl Methacrylate-co-caprolactone)s as DNA Transfection Agents. Macromol. Biosci. 2013, 13, 1267–1275. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Jiang, S.; Li, Z.; Loh, X.J. Conjugation of poly(ethylene glycol) to poly(lactide)-based polyelectrolytes: An effective method to modulate cytotoxicity in gene delivery. Mater. Sci. Eng. C 2017, 73, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Adolph, E.J.; Nelson, C.E.; Werfel, T.A.; Guo, R.; Davidson, J.M.; Guelcher, S.A.; Duval, C.L. Enhanced performance of plasmid DNA polyplexes stabilized by a combination of core hydrophobicity and surface PEGylation. J. Mater. Chem. B 2014, 2, 8154–8164. [Google Scholar] [CrossRef] [PubMed]
- Truong, N.P.; Gu, W.; Prasadam, I.; Jia, Z.; Crawford, R.; Xiao, Y.; Monteiro, M.J. An influenza virus-inspired polymer system for the timed release of siRNA. Nat. Commun. 2013, 4, 1902. [Google Scholar] [CrossRef] [PubMed]
- Truong, N.P.; Jia, Z.; Burgess, M.; Payne, L.; McMillan, N.A.J.; Monteiro, M.J. Self-Catalyzed Degradable Cationic Polymer for Release of DNA. Biomacromolecules 2011, 12, 3540–3548. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, A.M.; Calejo, T.; Morais, C.M.; Cardoso, A.L.; Cruz, R.; Zhu, K.; Pedroso de Lima, M.C.; Jurado, A.S.; Nyström, B. Application of Thermoresponsive PNIPAAM-b-PAMPTMA Diblock Copolymers in siRNA Delivery. Mol. Pharm. 2014, 11, 819–827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.; Tian, H.; Chen, X. Recent progress in cationic polymeric gene carriers for cancer therapy. Sci. China Chem. 2017, 60, 319–328. [Google Scholar] [CrossRef]
- Giron-Gonzalez, M.D.; Salto-Gonzalez, R.; Lopez-Jaramillo, F.J.; Salinas-Castillo, A.; Jodar-Reyes, A.B.; Ortega-Muñoz, M.; Hernandez-Mateo, F.; Santoyo-Gonzalez, F. Polyelectrolyte Complexes of Low Molecular Weight PEI and Citric Acid as Efficient and Nontoxic Vectors for in Vitro and in Vivo Gene Delivery. Bioconjug. Chem. 2016, 27, 549–561. [Google Scholar] [CrossRef] [PubMed]
- Venkataraman, S.; Ong, W.L.; Ong, Z.Y.; Loo, S.C.J.; Ee, P.L.R.; Yang, Y.Y. The role of PEG architecture and molecular weight in the gene transfection performance of PEGylated poly(dimethylaminoethyl methacrylate) based cationic polymers. Biomaterials 2011, 32, 2369–2378. [Google Scholar] [CrossRef] [PubMed]
- Luzon, M.; Boyer, C.; Peinado, C.; Corrales, T.; Whittaker, M.; Tao, L.; Davis, T.P. Water-Soluble, Thermoresponsive, Hyperbranched Copolymers Based on PEG-Methacrylates: Synthesis, Characterization, and LCST Behavior. J. Polym. Sci. A 2010, 48, 2783–2792. [Google Scholar] [CrossRef]
- Lutz, J.-F.; Hoth, A. Preparation of Ideal PEG Analogues with a Tunable Thermosensitivity by Controlled Radical Copolymerization of 2-(2-Methoxyethoxy)ethyl Methacrylate and Oligo(ethylene glycol) Methacrylate. Macromolecules 2006, 39, 893–896. [Google Scholar] [CrossRef]
- Farinha, J.P.S.; Martinho, J.M.G.; Xu, H.; Winnik, M.A.; Quirk, R.P. Influence of Intrachain Hydrogen Bonding on the Cyclization of a Polystyrene Chain. J. Polym. Sci. Part B Polym. Phys. 1994, 321, 1635–1642. [Google Scholar] [CrossRef]
- Müller, C.B.; Loman, A.; Pacheco, V.; Koberling, F.; Willbold, D.; Richtering, W.; Enderlein, J. Precise measurement of diffusion by multi-color dual-focus fluorescence correlation spectroscopy. Europhys. Lett. 2008, 83, 46001. [Google Scholar] [CrossRef]
- Vieira, T.J.; Farinha, J.P.S.; Martinho, J.M.G. Control of Oligonucleotide Distribution on the Shell of Thermo-responsive Polymer Nanoparticles. J. Phys. Chem. C 2008, 112, 16331–16339. [Google Scholar] [CrossRef]
- Martinho, J.M.G.; Moura, L.; Farinha, J.P.S. Fluorescence of Oligonucleotides Adsorbed onto the Thermo-responsive Poly(isopropyl acrylamide) Shell of Polymer Nanoparticles. Application to Bioassays. Pure Appl. Chem. 2009, 81, 1615–1634. [Google Scholar] [CrossRef]
- Moura, L.; Martinho, J.M.G.; Farinha, J.P.S. DNA Hybridization in Thermoresponsive Polymer Nanoparticles. ChemPhysChem 2010, 11, 1749–1756. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.C.; Rasnik, I.; Cheng, W.; Lohman, T.M.; Ha, T. Probing Single-Stranded DNA Conformational Flexibility Using Fluorescence Spectroscopy. Biophys. J. 2004, 86, 2530–2537. [Google Scholar] [CrossRef]
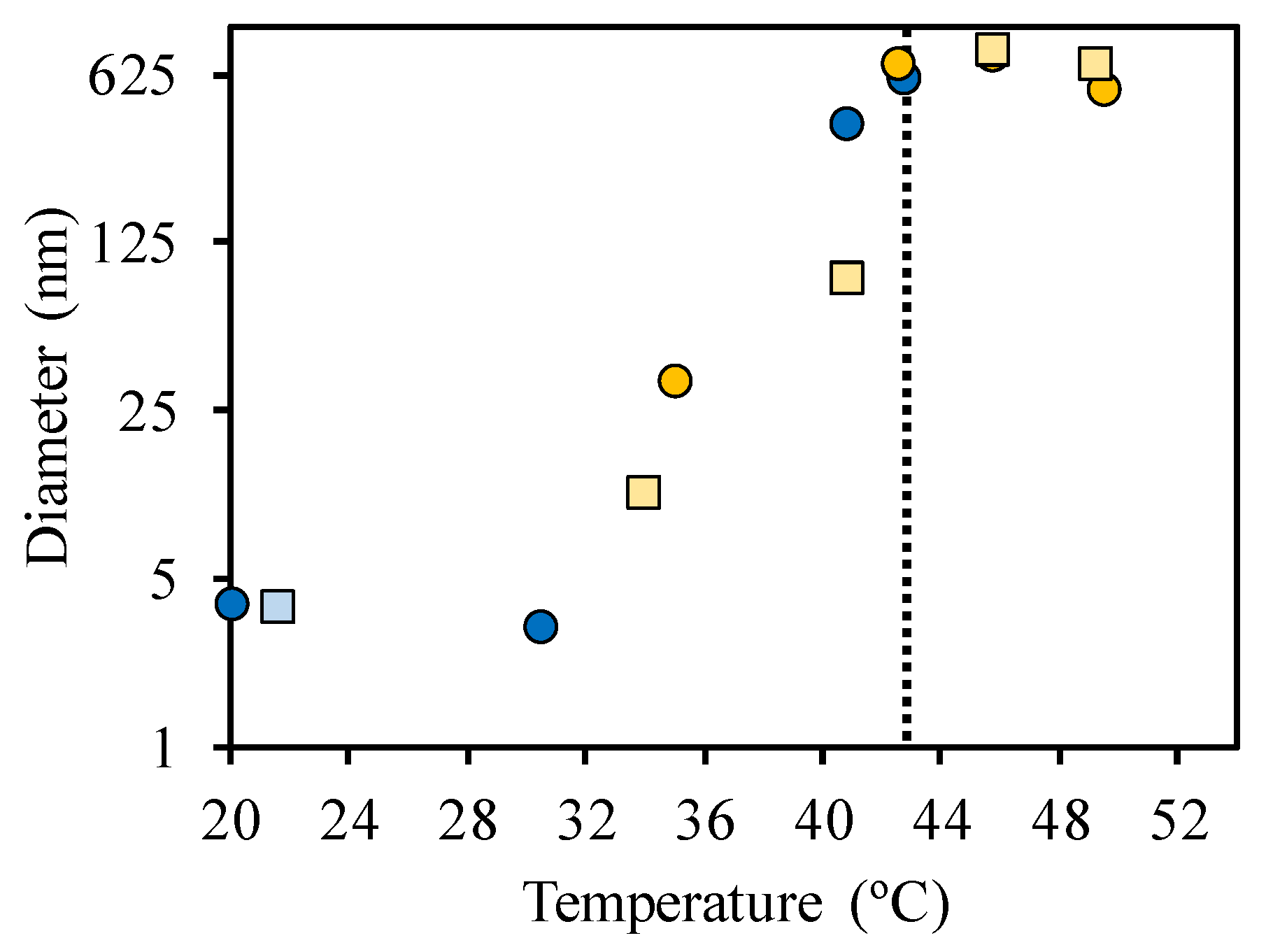
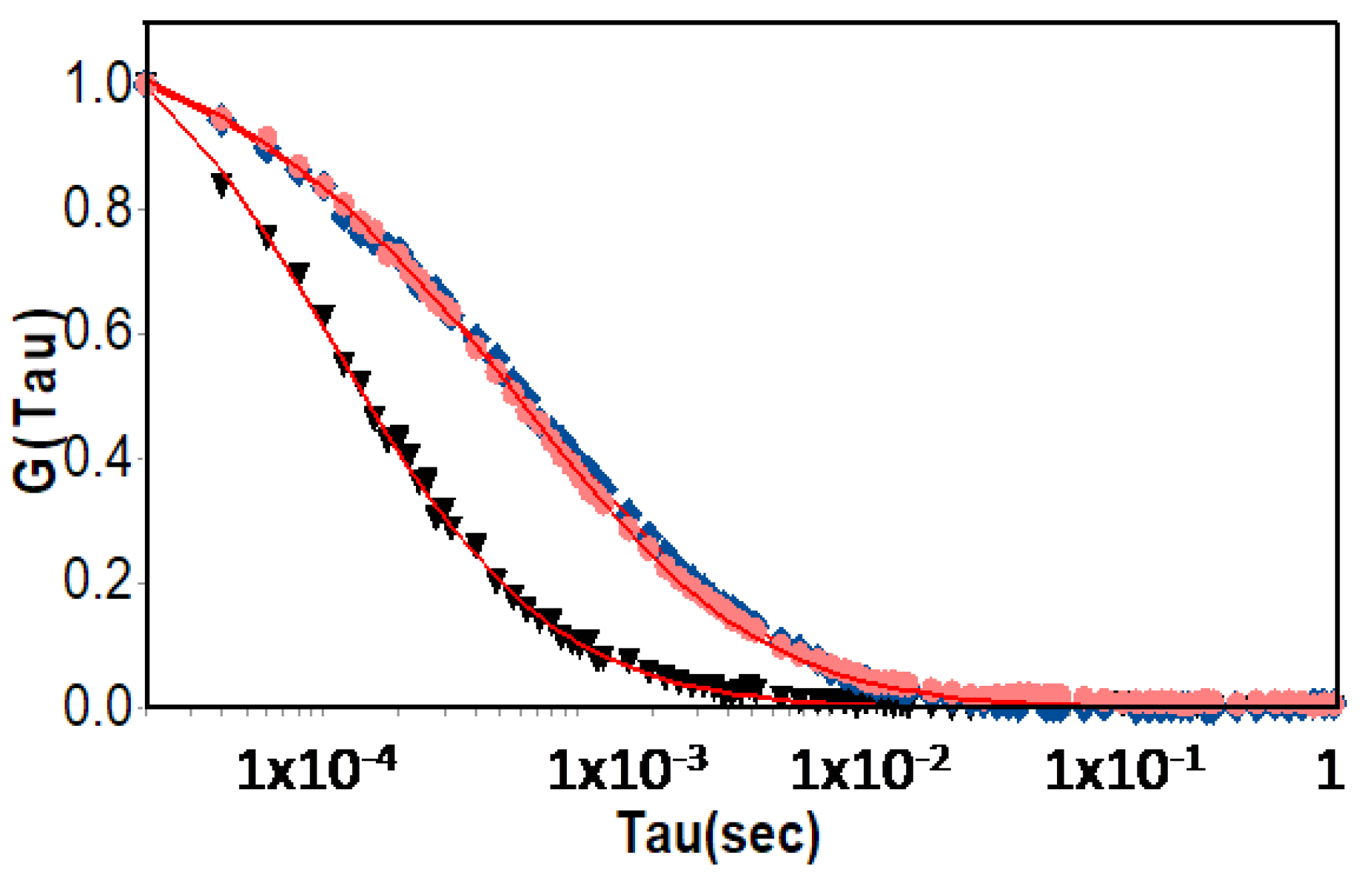
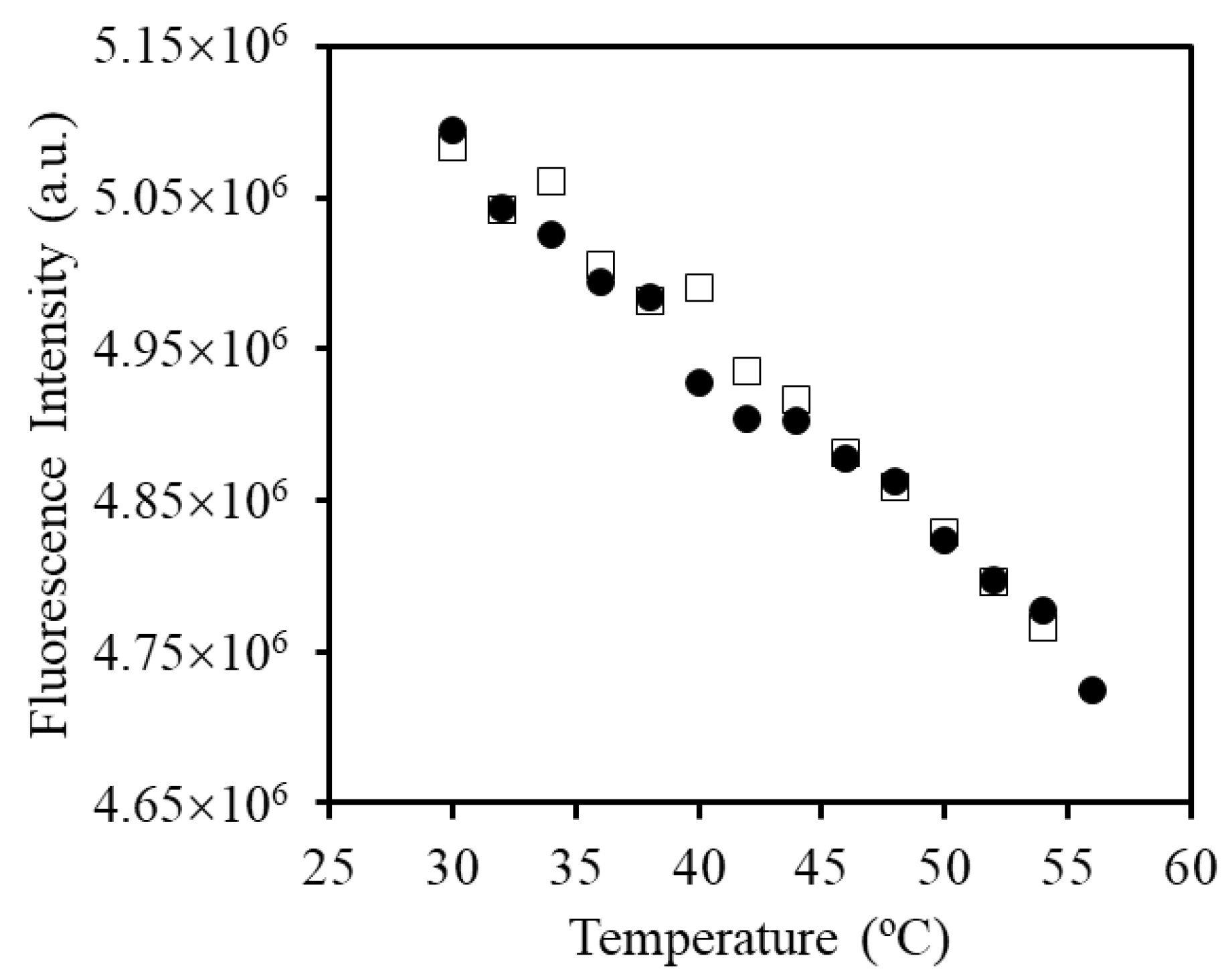
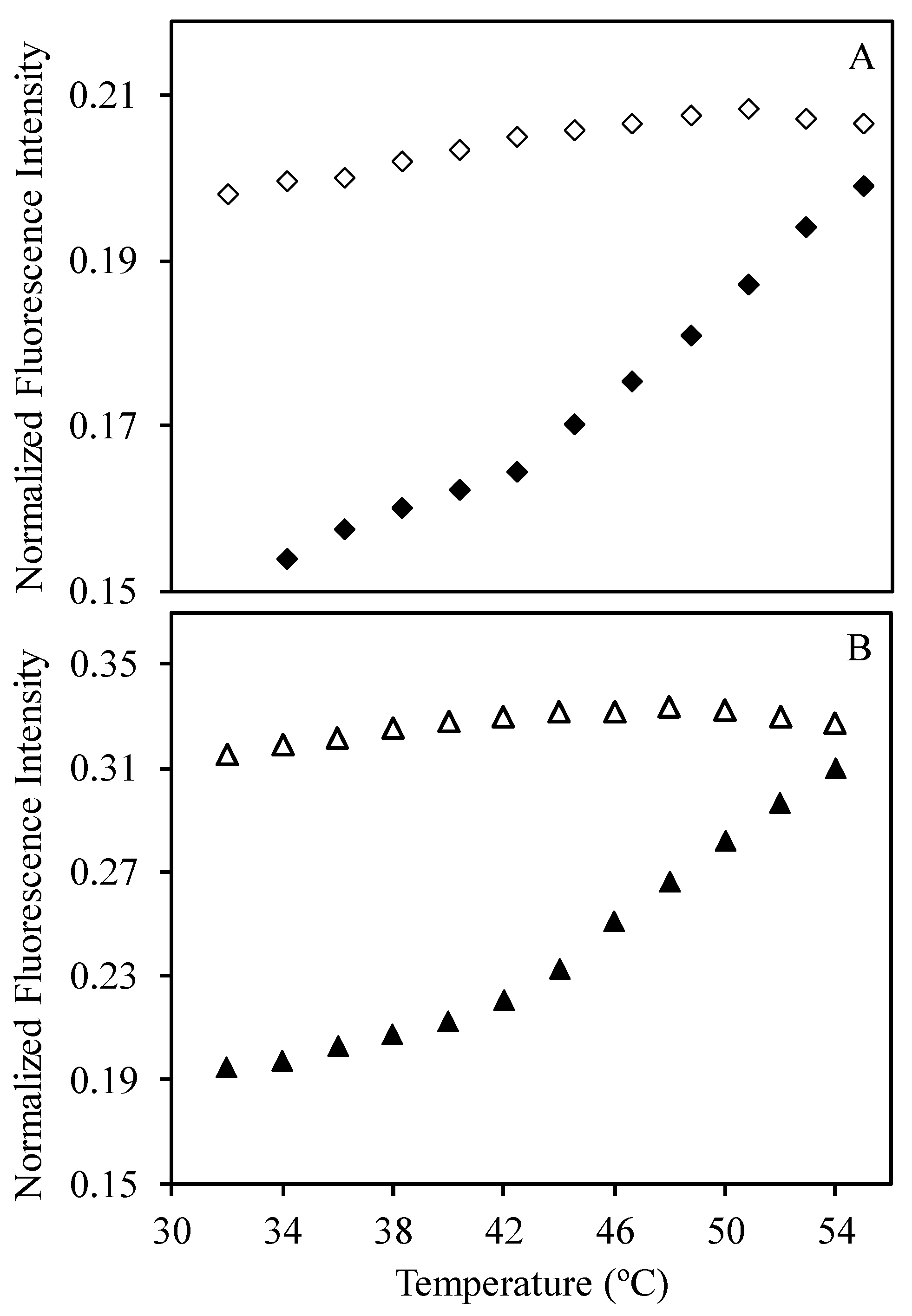
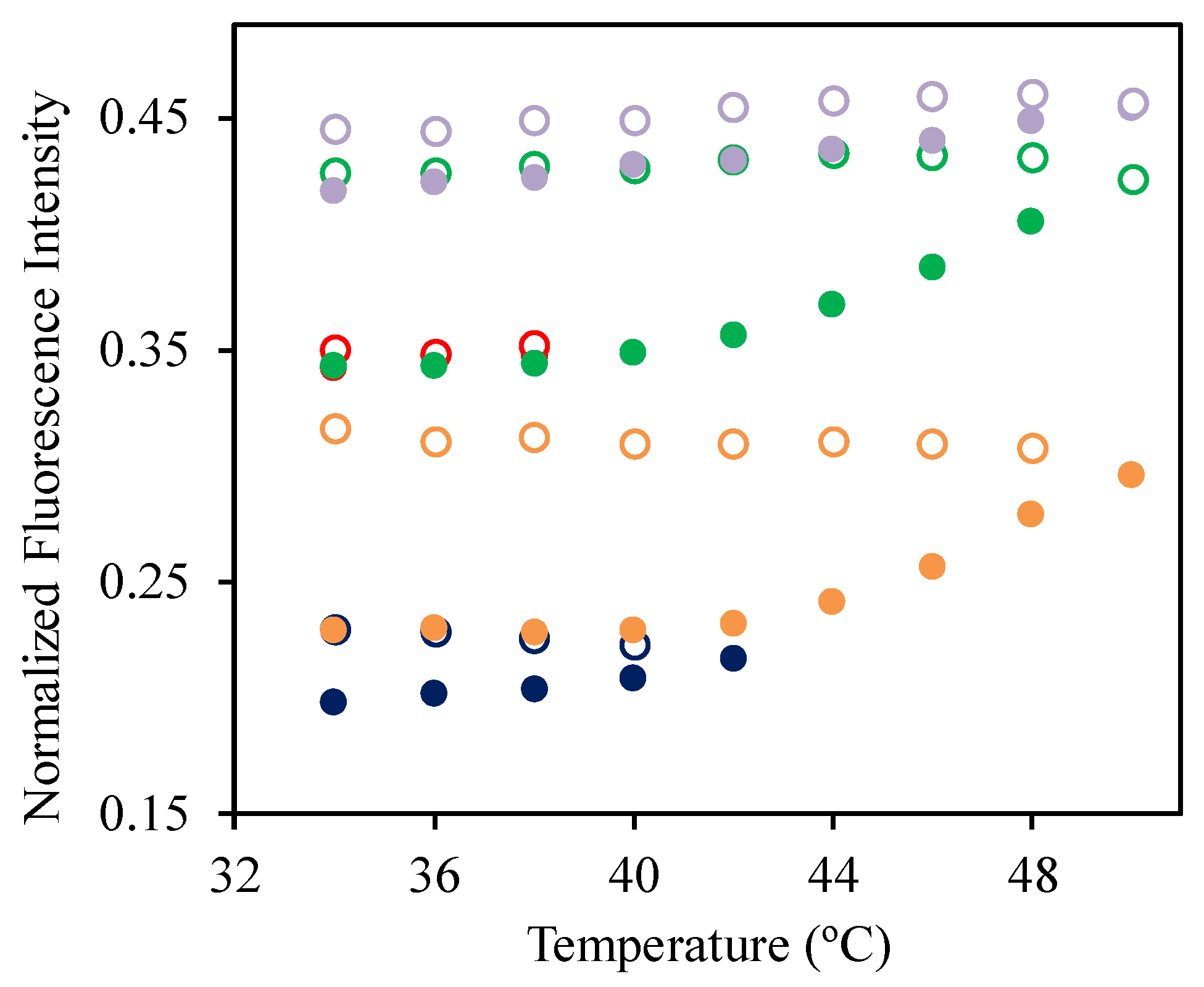
| Sample | Hydrodynamic Diameter (nm) | |
|---|---|---|
| DLS | FCS | |
| SRP | 3.8 ± 0.6 | - |
| DNA-ROX | - | 4.4 ± 0.4 |
| 1:1 SRP:DNA-ROX mixture | 35 ± 6 | 44 ± 5 |
| 2:1 SRP:DNA-ROX mixture | 45 ± 8 | 44 ± 5 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ribeiro, T.; Santiago, A.M.; Gaspar Martinho, J.M.; Farinha, J.P. A Cationic Smart Copolymer for DNA Binding. Polymers 2017, 9, 576. https://doi.org/10.3390/polym9110576
Ribeiro T, Santiago AM, Gaspar Martinho JM, Farinha JP. A Cationic Smart Copolymer for DNA Binding. Polymers. 2017; 9(11):576. https://doi.org/10.3390/polym9110576
Chicago/Turabian StyleRibeiro, Tânia, Ana Margarida Santiago, Jose Manuel Gaspar Martinho, and Jose Paulo Farinha. 2017. "A Cationic Smart Copolymer for DNA Binding" Polymers 9, no. 11: 576. https://doi.org/10.3390/polym9110576






