Metal and Ligand Effects on the Construction of Divalent Coordination Polymers Based on bis-Pyridyl-bis-amide and Polycarboxylate Ligands
Abstract
:1. Introduction
2. Experimental Section
2.1. Materials
2.2. Instruments
2.3. Preparations
2.3.1. General Procedure
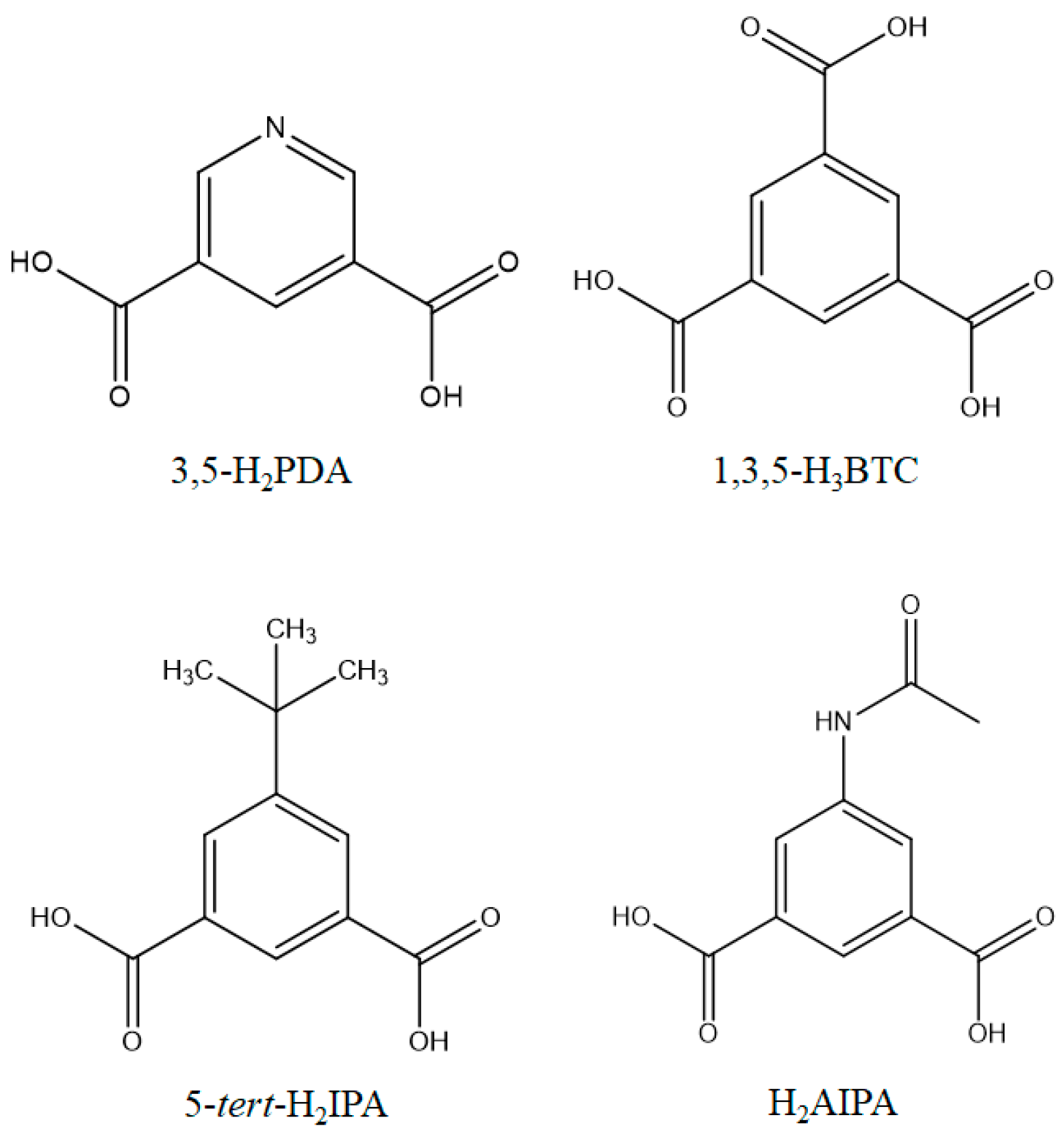
2.3.2. {[Ni(L1)(3,5-PDA)(H2O)3]·2H2O}n, c1
2.3.3. {[Ni2(L1)2(1,3,5-HBTC)2(H2O)4]·H2O}n, c2
2.3.4. {[Ni(L2)(5-tert-IPA)(H2O)2]·2H2O}n, c3
2.3.5. [Ni(L3)1.5(5-tert-IPA)]n, c4
2.3.6. [Co(L1)(1,3,5-HBTC)(H2O)]n, c5
2.3.7. {[Co3(L1)3(1,3,5-BTC)2(H2O)2]·6H2O}n, c6
2.3.8. [Cu(L4)(AIPA)]n, c7
2.3.9. {[Cu(L2)0.5(AIPA)]·MeOH}n, c8
2.3.10. {[Zn(L4)(AIPA)]·2H2O}n, c9
2.3.11. {[Zn(L2)(AIPA)]·2H2O}n, c10
2.4. Photodegradation Experiment
2.5. Single-Crystal X-ray Analysis
3. Results and Discussion
3.1. Structural Descriptions
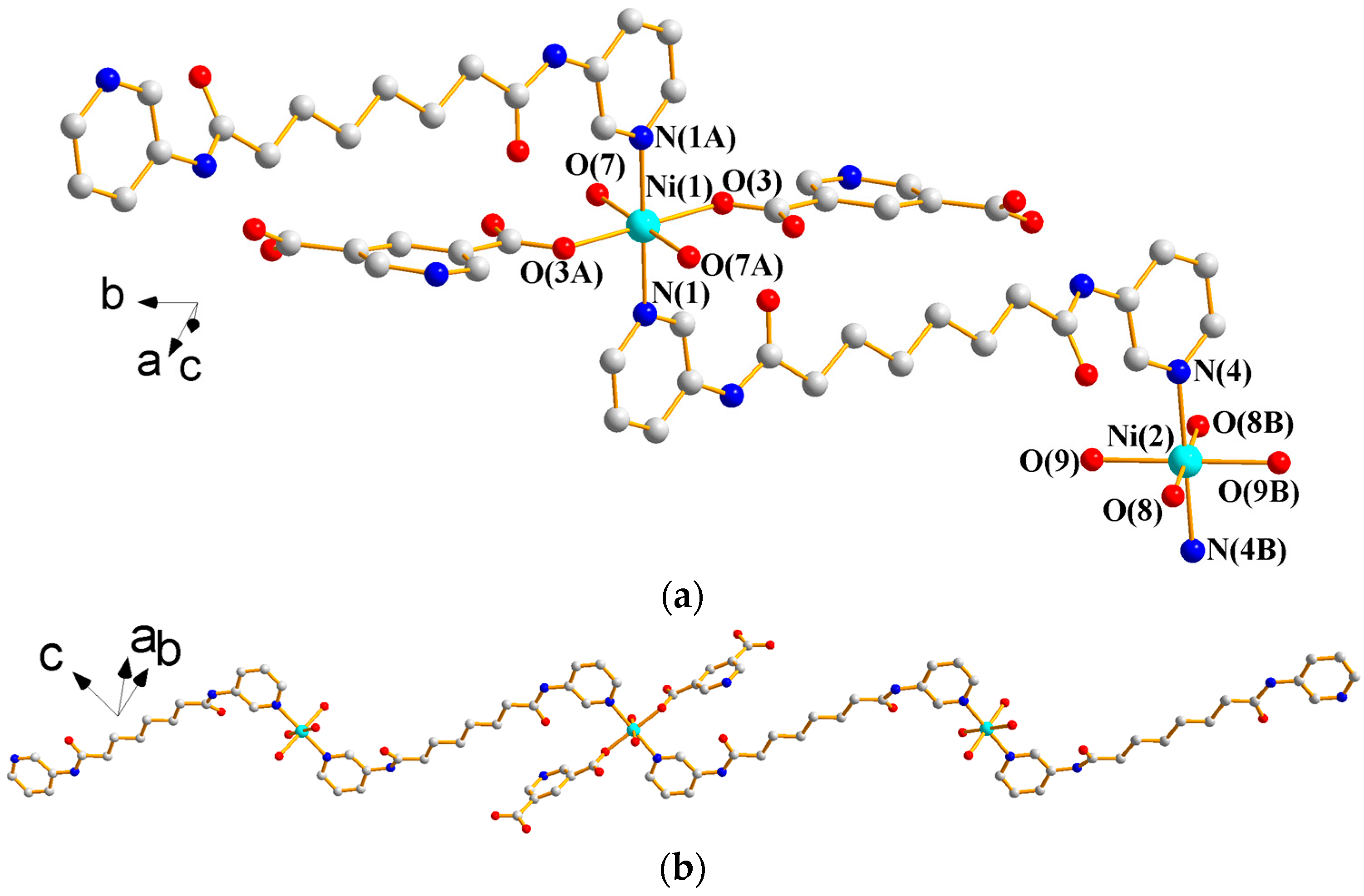
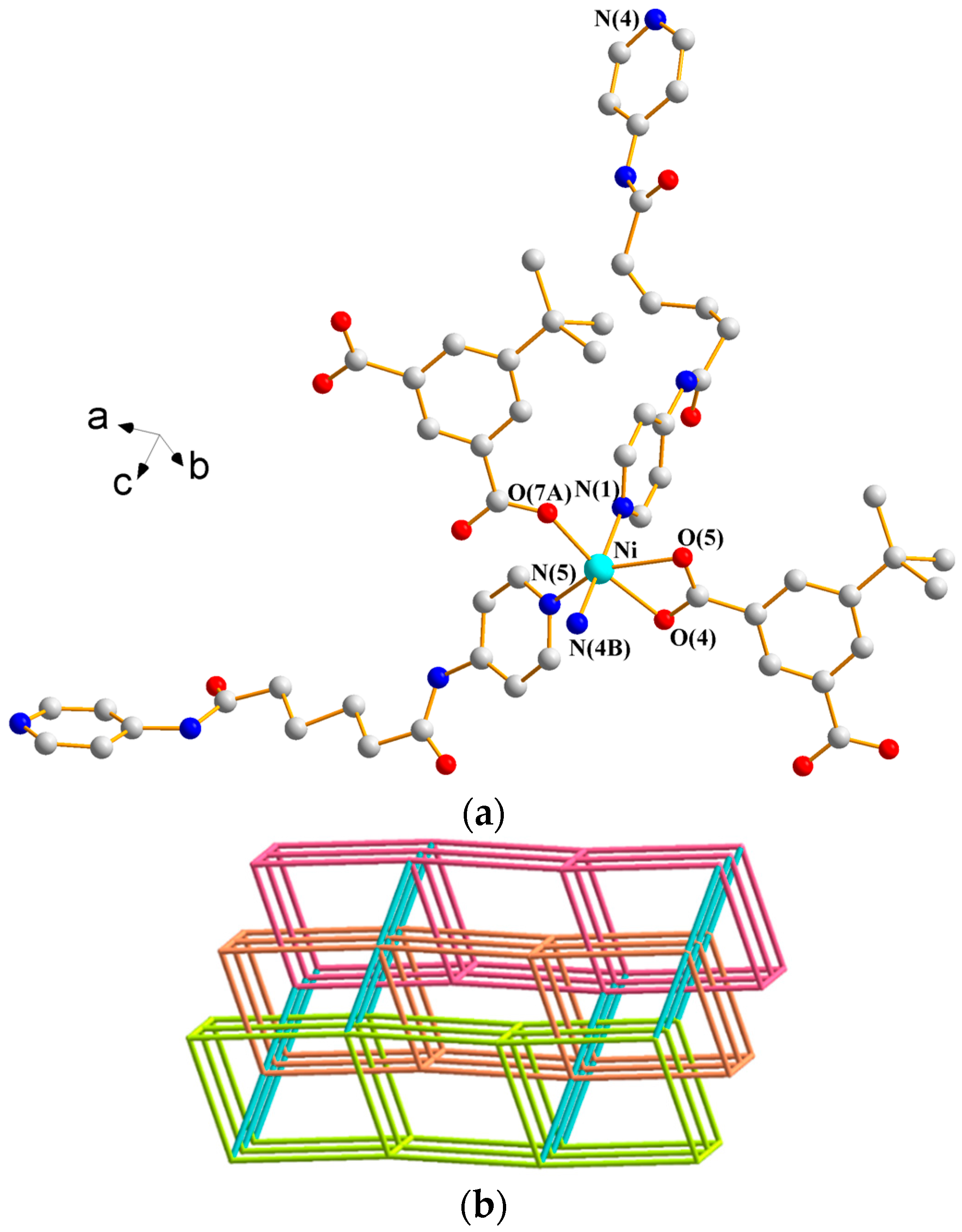
3.1.1. Structure of {[Ni(L1)(3,5-PDA)(H2O)3]·2H2O}n, 1
3.1.2. Structure of {[Ni2(L1)2(1,3,5-HBTC)2(H2O)4]·H2O}n, 2
3.1.3. Structure of {[Ni(L2)(5-tert-IPA)(H2O)2]·2H2O}n, 3
3.1.4. Structure of [Ni(L3)1.5(5-tert-IPA)]n, 4
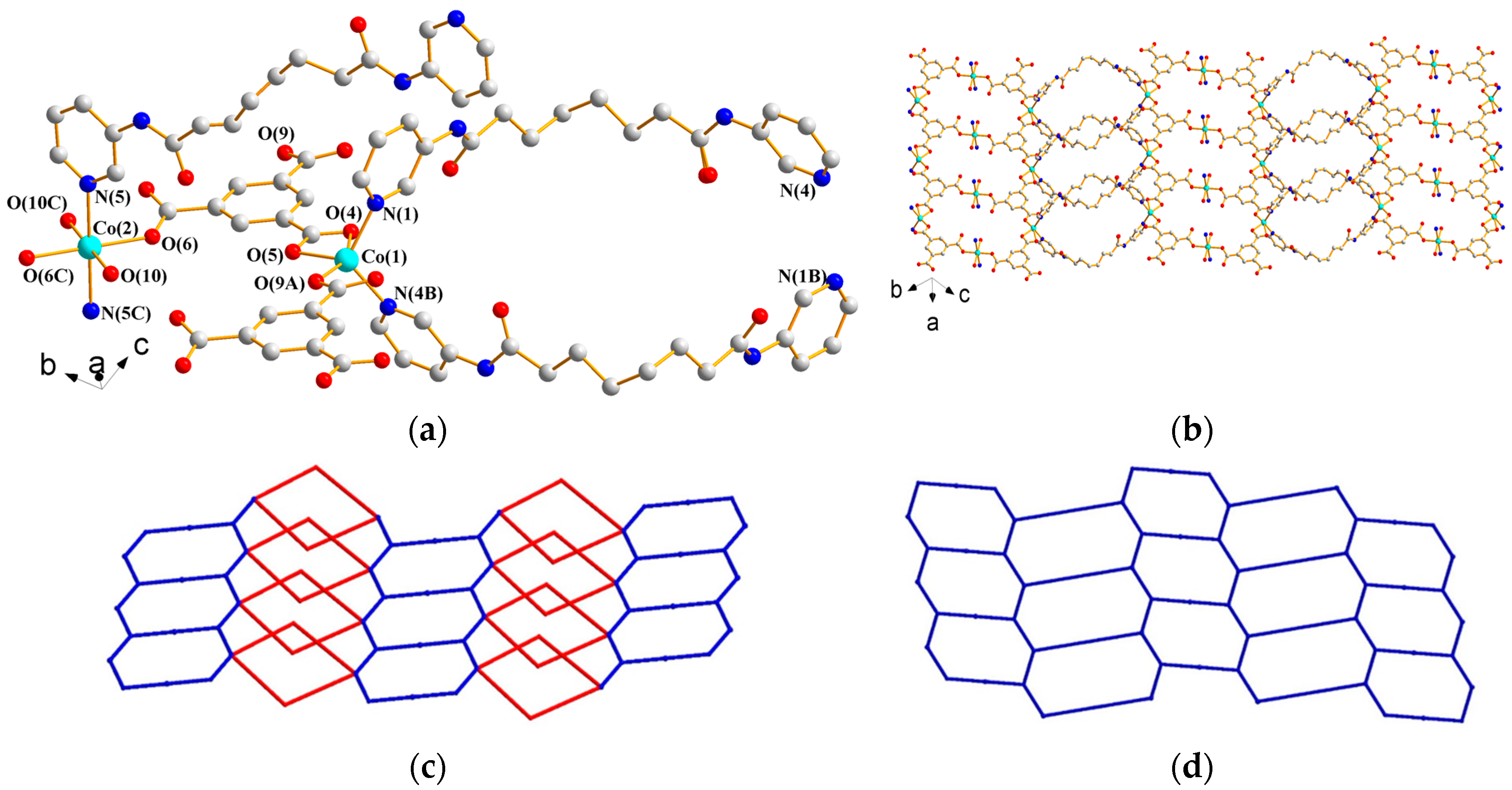
3.1.5. Structure of [Co(L1)(1,3,5-HBTC)(H2O)]n, 5
3.1.6. Structure of {[Co3(L1)3(1,3,5-BTC)2(H2O)2]·6H2O}n, 6
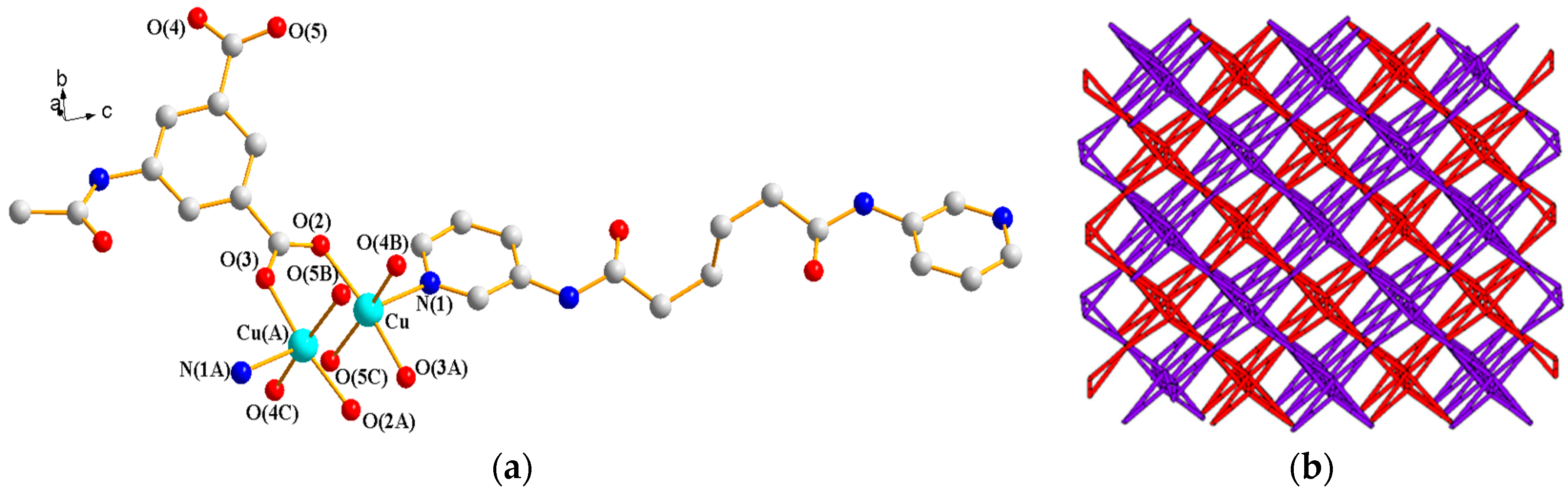
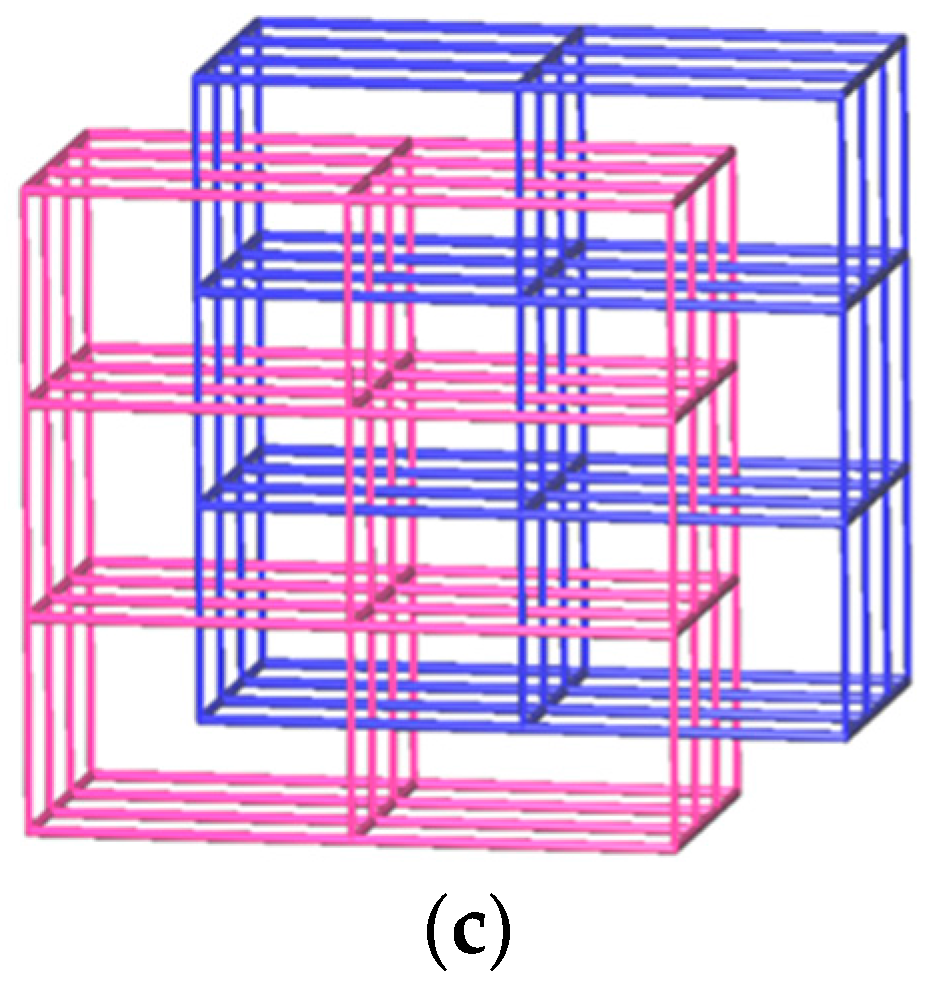
3.1.7. Structure of [Cu(L4)(AIPA)]n, 7
3.1.8. Structure of {[Cu(L2)0.5(AIPA)]·MeOH}n, 8
3.1.9. Structure of {[Zn(L4)(AIPA)]·2H2O}n, 9
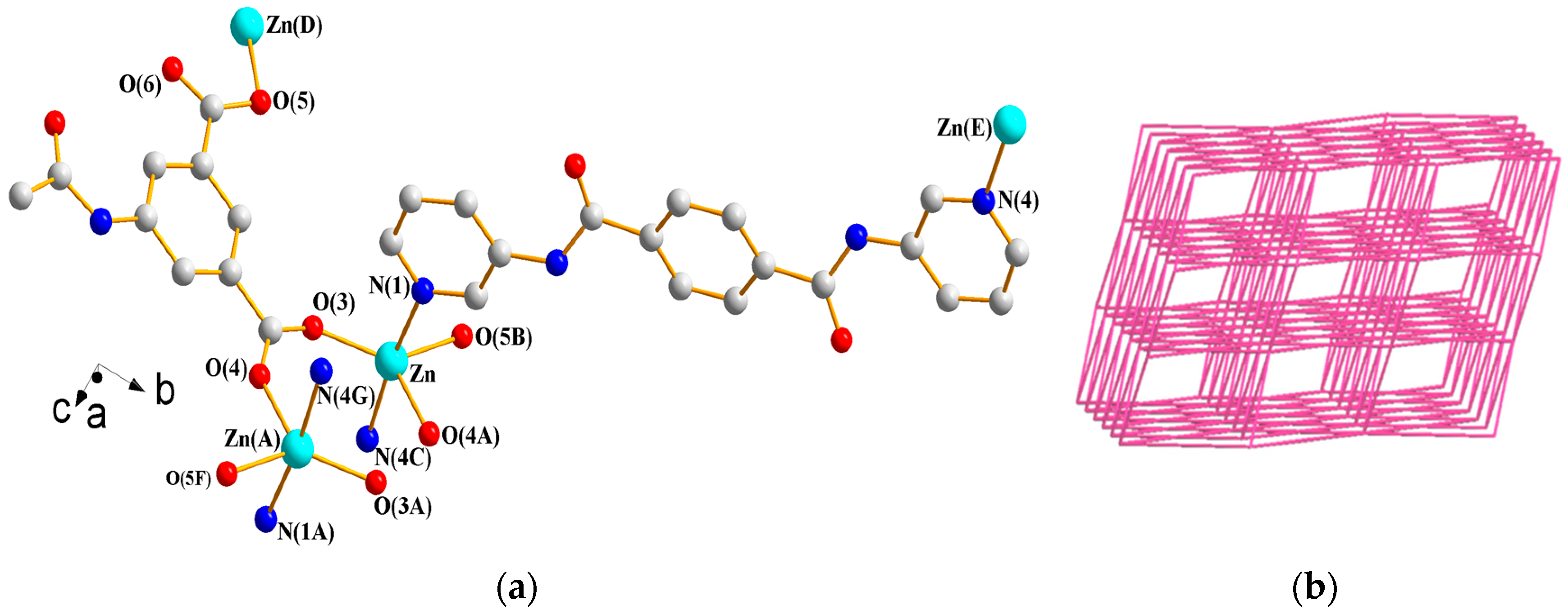
3.1.10. Structure of {[Zn(L2)(AIPA)]·2H2O}n 10
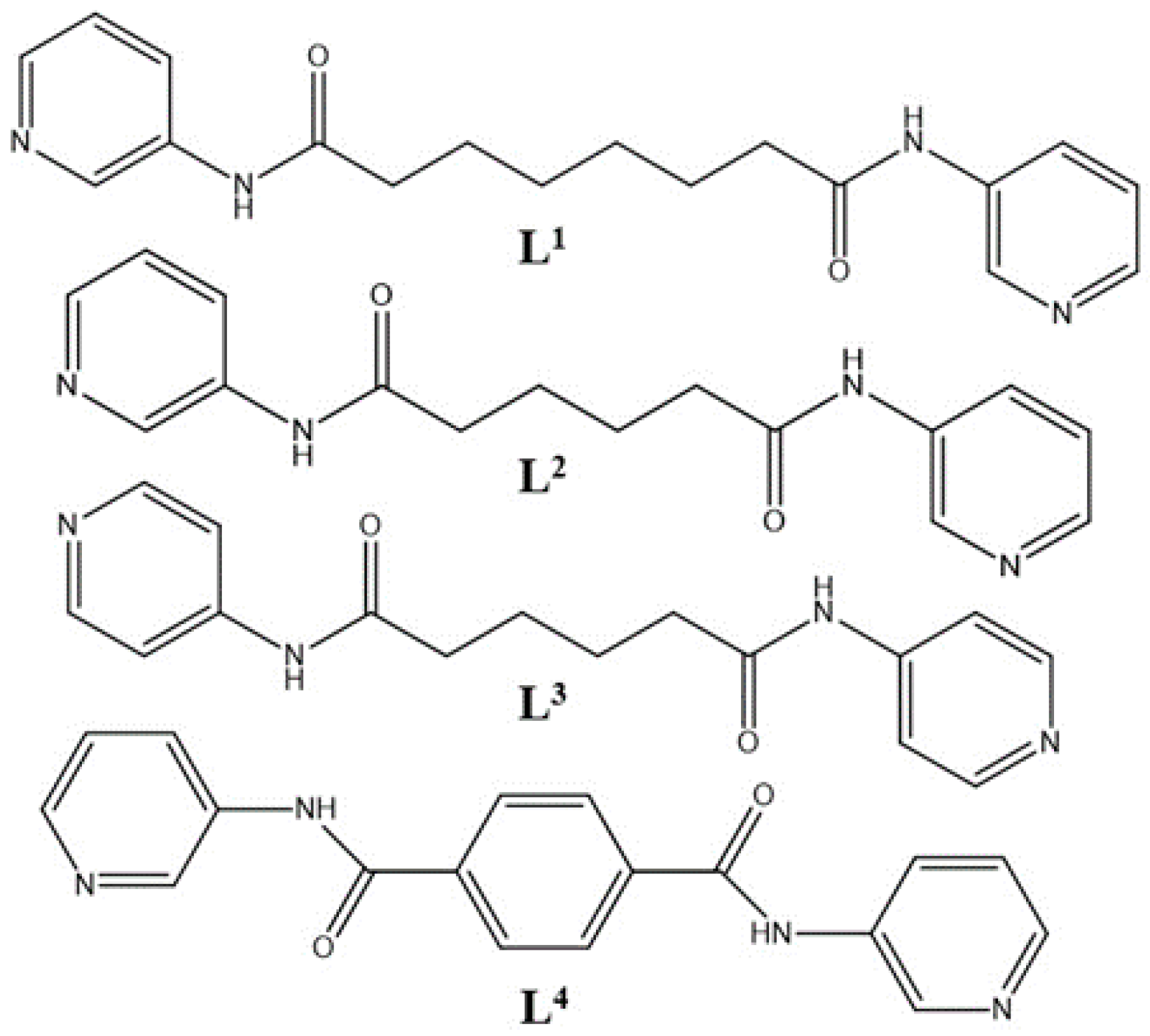
3.2. Ligand Conformations and Bonding Modes
3.3. Structural Comparisons
3.4. Luminescent Properties
3.5. Photocatalytic Properties
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Tiekink, E.R.T.; Vittal, J.J. Frontiers in Crystal Engineering; John Wiley & Sons, Ltd.: Chichester, UK, 2006. [Google Scholar]
- Yaghi, O.M.; Li, H.; Davis, C.; Richardson, D.; Groy, T.L. Synthetic strategies, structure patterns, and emerging properties in the chemistry of modular porous solids. Acc. Chem. Res. 1998, 31, 474–484. [Google Scholar] [CrossRef]
- Batten, S.R.; Neville, S.M.; Turner, D.R. Coordination Polymers: Design, Analysis and Application; Royal Society of Chemistry: Cambridge, UK, 2009. [Google Scholar]
- Carlucci, L.; Ciani, G.; Proserpio, D.M.; Mitina, T.G.; Blatov, V.A. Entangled two-dimensional coordination networks: A general survey. Chem. Rev. 2014, 114, 7557–7580. [Google Scholar] [CrossRef] [PubMed]
- Thapa, K.B.; Chen, J.-D. Crystal engineering of coordination polymers containing flexible bis-pyridyl-bis-amide ligands. CrystEngComm 2015, 17, 4611–4626. [Google Scholar] [CrossRef]
- Zhang, J.-W.; Kan, X.-M.; Li, X.-L.; Luan, J.; Wang, X.-L. Transition metal carboxylate coordination polymers with amide-bridged polypyridine co-ligands: Assemblies and properties. CrystEngComm 2015, 17, 3887–3907. [Google Scholar] [CrossRef]
- Lo, Y.-C.; Hsu, W.; He, H.-Y.; Hyde, S.-T.; Proserpio, D.M.; Chen, J.-D. Structural directing roles of isomeric phenylenediacetate ligands in the formation of coordination networks based on flexible N,N′-di(3-pyridyl)suberoamide. CrystEngComm 2015, 17, 90–97. [Google Scholar] [CrossRef]
- He, H.-Y.; Hsu, C.-H.; Chang, H.-Y.; Yang, H.-K.; Chhetri, P.M.; Proserpio, D.M.; Chen, J.-D. Self-catenated coordination polymers involving bis-pyridyl-bis-amide. Cryst. Growth Des. 2017, 17, 1991–1998. [Google Scholar] [CrossRef]
- Huang, W.-H.; Hsu, C.-J.; Tsai, S.-K.; He, H.-Y.; Ding, J.-J.; Hsu, T.-W.; Yang, C.-C.; Chen, J.-D. Homo- and heterometallic coordination networks based on linear trinuclear Co(II) units: Synthesis, structures and magnetic properties. RSC Adv. 2015, 5, 23374–23382. [Google Scholar] [CrossRef]
- Bruker AXS Inc. Bruker AXS, APEX2, V2008.6, SAD ABS V2008/1, SAINT+ V7.60A, SHELXTL V6.14; Bruker AXS Inc.: Madison, WI, USA, 2008. [Google Scholar]
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. 2008, A64, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Blatov, V.A.; Shevchenko, A.P.; Proserpio, D.M. Applied topological analysis of crystal structures with the program package ToposPro. Cryst. Growth Des. 2014, 14, 3576–3586. [Google Scholar] [CrossRef]
- Zhan, S.-Z.; Li, M.; Zhou, X.-P.; Ni, J.; Huang, X.-C.; Li, D. From simple to complex: Topological evolution and luminescence variation in a copper(I) pyridylpyrazolate system tuned via second ligating spacers. Inorg. Chem. 2011, 50, 8879–8892. [Google Scholar] [CrossRef] [PubMed]
- Addison, A.W.; Rao, T.N.; Reedijik, J.; Rijn, J.V.; Verschoor, G.C. Synthesis, structure, and spectroscopic properties of copper(II) compounds containing nitrogen-sulphur donor ligands; the crystal and molecular structure of aqua[1,7-bis(N-methylbenzimidazol-2′-yl)-2,6-dithiaheptane]copper(II) perchlorate. J. Chem. Soc. Dalton Trans. 1984, 1349–1356. [Google Scholar] [CrossRef]
- Spek, A.L. Structure validation in chemical crystallography. Acta Cryst. 2009, D65, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Yan, Z.-H.; Blatov, V.A.; Wang, L.; Sun, D.-F. Syntheses, topological structures, and photoluminescences of six new Zn(II) coordination polymers based on mixed tripodal imidazole ligand and varied polycarboxylates. Cryst. Growth Des. 2013, 13, 1277–1289. [Google Scholar] [CrossRef]
- Luo, F.; Yuan, Z.-Z.; Feng, X.-F.; Batten, S.R.; Li, J.-Q.; Luo, M.-B.; Liu, S.-J.; Xu, W.-Y.; Sun, G.-M.; Song, Y.-M.; et al. Multifunctional 3-fold interpenetrated porous metal-organic frameworks composed of unprecedented self-catenated networks. Cryst. Growth Des. 2012, 12, 3392–3396. [Google Scholar] [CrossRef]
- Wang, F.; Liu, Z.-S.; Yang, H.; Tan, Y.-X.; Zhang, J. Hybrid zeolitic imidazolate frameworks with catalytically active TO4 building blocks. Angew. Chem. Int. Ed. 2011, 50, 450–453. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-L.; Sui, F.-F.; Lin, H.-Y.; Zhang, J.-W.; Liu, G.-C. Multifunctional cobalt(II) coordination polymers tuned by flexible bis(pyridylamide) ligands with different spacers and polycarboxylates. Cryst. Growth Des. 2014, 14, 3438–3452. [Google Scholar] [CrossRef]







| Complex | 1 | 2 | 3 | 4 | 5 |
| Formula | C25H35NiN5O11 | C54H62Ni2N8O21 | C28H38NiN4O10 | C36H39NiN6O7 | C27H28CoN4O9 |
| Formula weight | 640.29 | 1276.53 | 649.33 | 726.44 | 611.46 |
| Crystal system | Triclinic | Triclinic | Triclinic | Monoclinic | Triclinic |
| Space group | Pī | Pī | Pī | C2/c | Pī |
| a, Å | 10.2217(2) | 9.2151(1) | 11.7614(2) | 12.9109(14) | 9.6229(1) |
| b, Å | 12.1140(2) | 15.2396(2) | 12.0360(2) | 16.2298(17) | 10.2784(1) |
| c, Å | 13.1648(3) | 20.1781(2) | 12.2961(2) | 33.736(4) | 14.7068(2) |
| α, ° | 67.612(1) | 105.765(1) | 78.548(1) | 90 | 87.707(1) |
| β, ° | 76.879(1) | 96.500(1) | 67.752(1) | 95.395(7) | 89.509(1) |
| γ, ° | 70.475(1) | 90.244(1) | 86.067(1) | 90 | 73.917(1) |
| V, Å3 | 1411.27(5) | 2707.76(6) | 1578.94(5) | 7037.8(13) | 1396.56(3) |
| Z | 2 | 2 | 2 | 8 | 2 |
| dcalc, mg/m3 | 1.507 | 1.566 | 1.366 | 1.371 | 1.454 |
| F(000) | 672 | 1332 | 684 | 3048 | 634 |
| µ(Mo Kα), mm−1 | 0.756 | 0.786 | 0.674 | 0.609 | 0.674 |
| Range (2θ) for data collection | 3.36 to 56.68 | 3.92 to 56.72 | 3.46 to 56.59 | 4.14 to 56.92 | 4.12 to 56.56 |
| Independent reflections | 7003 | 13388 | 7815 | 8791 | 6910 |
| [R(int) = 0.0355] | [R(int) = 0.0253] | [R(int) = 0.0245] | [R(int) = 0.0337] | [R(int) = 0.0185] | |
| Data/restraints/parameters | 7003/0/412 | 13388/1369/819 | 7815/0/395 | 8791/808/484 | 6910/0/382 |
| Quality-of-fit indicator c | 1.076 | 1.062 | 1.037 | 1.067 | 1.053 |
| Final R indices [I > 2σ(I)] a,b | R1 = 0.0329 | R1 = 0.0472 | R1 = 0.0401 | R1 = 0.0666 | R1 = 0.0353 |
| wR2 = 0.0731 | wR2 = 0.1282 | wR2 = 0.1018 | wR2 = 0.1864 | wR2 = 0.0925 | |
| R indices (all data) | R1 = 0.0473 | R1 = 0.0756 | R1 = 0.0533 | R1 = 0.0955 | R1 = 0.0426 |
| wR2 = 0.0793 | wR2 = 0.1460 | wR2 = 0.1096 | wR2 = 0.2044 | wR2 = 0.0962 | |
| Complex | 6 | 7 | 8 | 9 | 10 |
| Formula | C72H88Co3N12O26 | C28H21CuN5O7 | C19H20CuN3O7 | C28H25ZnN5O9 | C26H29ZnN5O9 |
| Formula weight | 1714.33 | 603.04 | 465.92 | 640.90 | 620.91 |
| Crystal system | Triclinic | Triclinic | Orthorhombic | Monoclinic | Monoclinic |
| Space group | Pī | Pī | Pbca | C2/c | P21/c |
| a, Å | 10.0136(2) | 9.7425(2) | 12.8022(2) | 26.4979(10) | 12.5193(1) |
| b, Å | 11.9834(3) | 9.9983(2) | 14.5368(2) | 12.7299(5) | 12.8079(2) |
| c, Å | 17.2930(4) | 15.1361(2) | 20.5531(2) | 17.1560(6) | 16.8353(2) |
| α, ° | 98.375(2) | 81.021(1) | 90 | 90 | 90 |
| β, ° | 90.027(1) | 84.597(1) | 90 | 108.868(2) | 91.488(1) |
| γ, ° | 95.361(1) | 61.683(1) | 90 | 90 | 90 |
| V, Å3 | 2043.79(8) | 1281.71(4) | 3824.99(9) | 5476.0(4) | 2698.56(6) |
| Z | 1 | 2 | 8 | 8 | 4 |
| dcalc, mg/m3 | 1.393 | 1.563 | 1.618 | 1.555 | 1.528 |
| F(000) | 893 | 618 | 1920 | 2640 | 1288 |
| µ(Mo Kα), mm−1 | 0.685 | 0.911 | 1.192 | 0.962 | 0.973 |
| Range(2θ) for data collection | 3.89 to 52.00 | 4.66 to 56.64 | 4.68 to 56.64 | 3.25 to 56.66 | 3.26 to 56.58 |
| Independent reflections | 8041 | 6365 | 4760 | 6783 | 6539 |
| [R(int) = 0.0411] | [R(int) = 0.0551] | [R(int) = 0.0486] | [R(int) = 0.0609] | [R(int) = 0.0193] | |
| Data/restraints/parameters | 8041/0/568 | 6265/0/374 | 4760/0/283 | 6783/0/397 | 6539/0/370 |
| Quality-of-fit indicator c | 1.053 | 1.056 | 1.021 | 1.024 | 1.023 |
| Final R indices [I > 2σ(I)] a,b | R1 = 0.0660, | R1 = 0.0453, | R1 = 0.0438, | R1 = 0.0521, | R1 = 0.0399, |
| wR2 = 0.1807 | wR2 = 0.1040 | wR2 = 0.0997 | wR2 = 0.0930 | wR2 = 0.1144 | |
| R indices (all data) | R1 = 0.0907, | R1 = 0.0714, | R1 = 0.0724, | R1 = 0.1033, | R1 = 0.0510, |
| wR2 = 0.1980 | wR2 = 0.1150 | wR2 = 0.1129 | wR2 = 0.1090 | wR2 = 0.1219 |
| Complex | Ligand conformation | Bonding mode |
|---|---|---|
| {[Ni(L1)(3,5-PDA)(H2O)3]·2H2O}n, 1 | AAAAA trans syn-syn | μ1-κ1,κ0,κ0,κ0 |
| {[Ni2(L1)2(1,3,5-HBTC)2(H2O)4]·H2O}n, 2 | AAAAA trans anti-syn AGAGA trans syn-anti | μ2-κ1,κ0,κ1,κ0,κ0,κ0 |
| {[Ni(L2)(5-tert-IPA)(H2O)2]·2H2O}n, 3 | AAA trans anti-anti GAG trans syn-anti | μ2-κ1,κ0,κ1,κ0 |
| [Ni(L3)1.5(5-tert-IPA)]n, 4 | AAA trans GAG trans | μ2-κ1,κ0,κ1,κ0 |
| [Co(L1)(1,3,5-HBTC)(H2O)]n, 5 | GAAAA cis anti-syn | μ2-κ1,κ0,κ1,κ1,κ0,κ0 |
| {[Co3(L1)3(1,3,5-BTC)2(H2O)2]·6H2O}n, 6 | AAGGA cis syn-syn | μ3-κ1,κ0,κ1,κ0,κ1,κ1 |
| AGAGA trans syn-syn | ||
| [Cu(L4)(AIPA)]n, 7 | trans syn-syn | μ3-κ0,κ1,κ1,κ1 |
| {[Cu(L2)0.5(AIPA)]·MeOH}n, 8 | GAG trans anti-anti | μ4-κ1,κ1,κ1,κ1 |
| {[Zn(L4)(AIPA)]·2H2O}n, 9 | trans anti-anti | μ3-κ0,κ1,κ1,κ1 |
| {[Zn(L2)(AIPA)]·2H2O}n, 10 | GAG trans anti-anti | μ3-κ0,κ1,κ1,κ1 |
| Complex | λabs (nm) | λex (nm) | λem (nm) |
|---|---|---|---|
| L2 | 300 | 370 | 415 |
| L4 | 341 | 330/365 | 420 |
| H2AIPA | 323 | 336 | 425 |
| 9 | 340 | 340/400 | 460 |
| 10 | 322 | 325/350 | 415 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, M.-N.; Yang, X.-K.; Chhetri, P.M.; Chen, J.-D. Metal and Ligand Effects on the Construction of Divalent Coordination Polymers Based on bis-Pyridyl-bis-amide and Polycarboxylate Ligands. Polymers 2017, 9, 691. https://doi.org/10.3390/polym9120691
Chang M-N, Yang X-K, Chhetri PM, Chen J-D. Metal and Ligand Effects on the Construction of Divalent Coordination Polymers Based on bis-Pyridyl-bis-amide and Polycarboxylate Ligands. Polymers. 2017; 9(12):691. https://doi.org/10.3390/polym9120691
Chicago/Turabian StyleChang, Miao-Ning, Xiang-Kai Yang, Pradhumna Mahat Chhetri, and Jhy-Der Chen. 2017. "Metal and Ligand Effects on the Construction of Divalent Coordination Polymers Based on bis-Pyridyl-bis-amide and Polycarboxylate Ligands" Polymers 9, no. 12: 691. https://doi.org/10.3390/polym9120691