DNA Compaction and Charge Inversion Induced by Organic Monovalent Ions
Abstract
:1. Introduction
2. Experimental Procedures
2.1. Materials
2.2. AFM Imaging
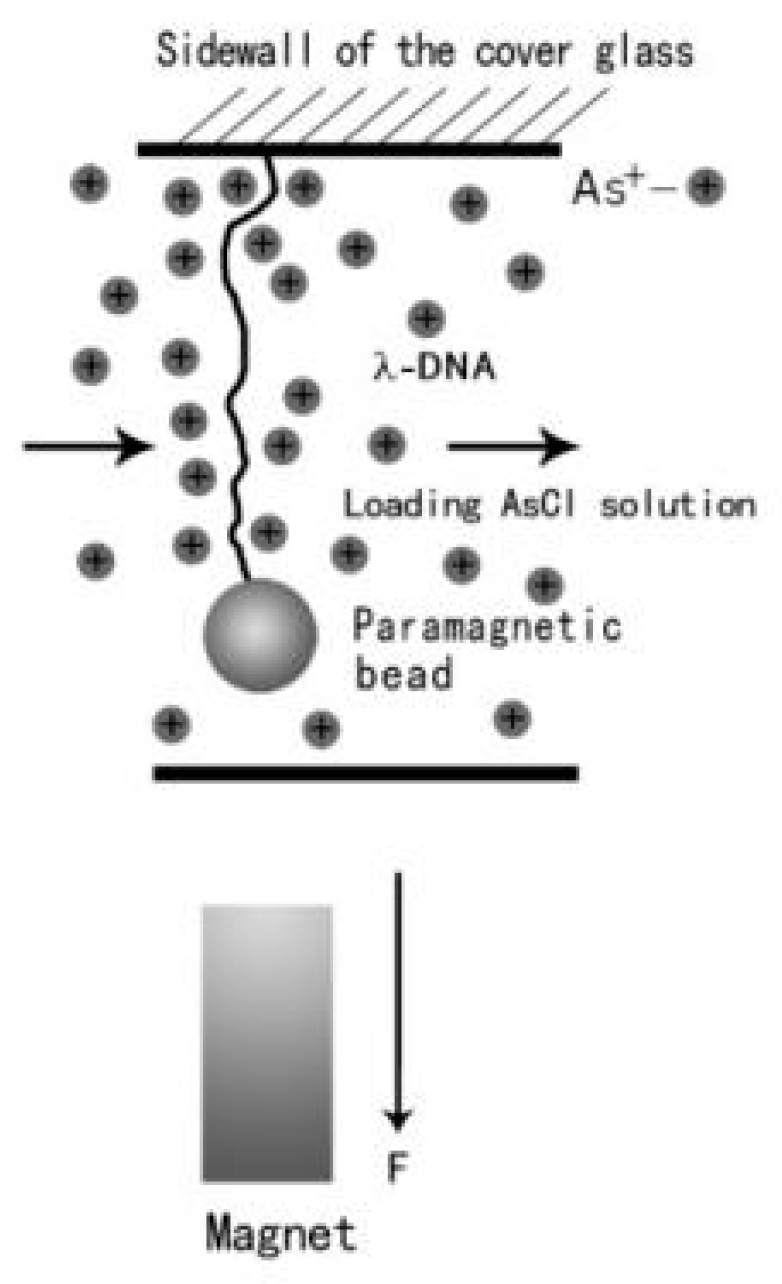
2.3. Magnetic Tweezers Experiment
2.4. Electrophoretic Mobility Measurement by Dynamic Light Scattering (DLS)
3. Results and Discussion
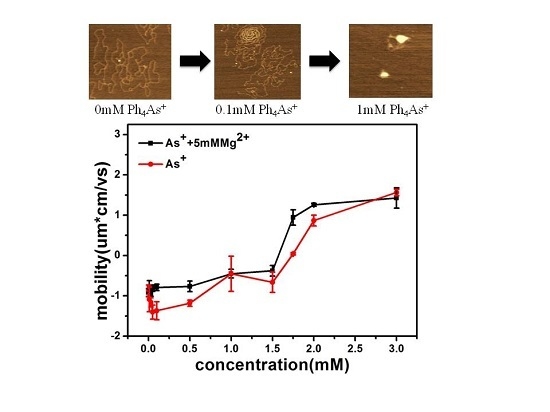
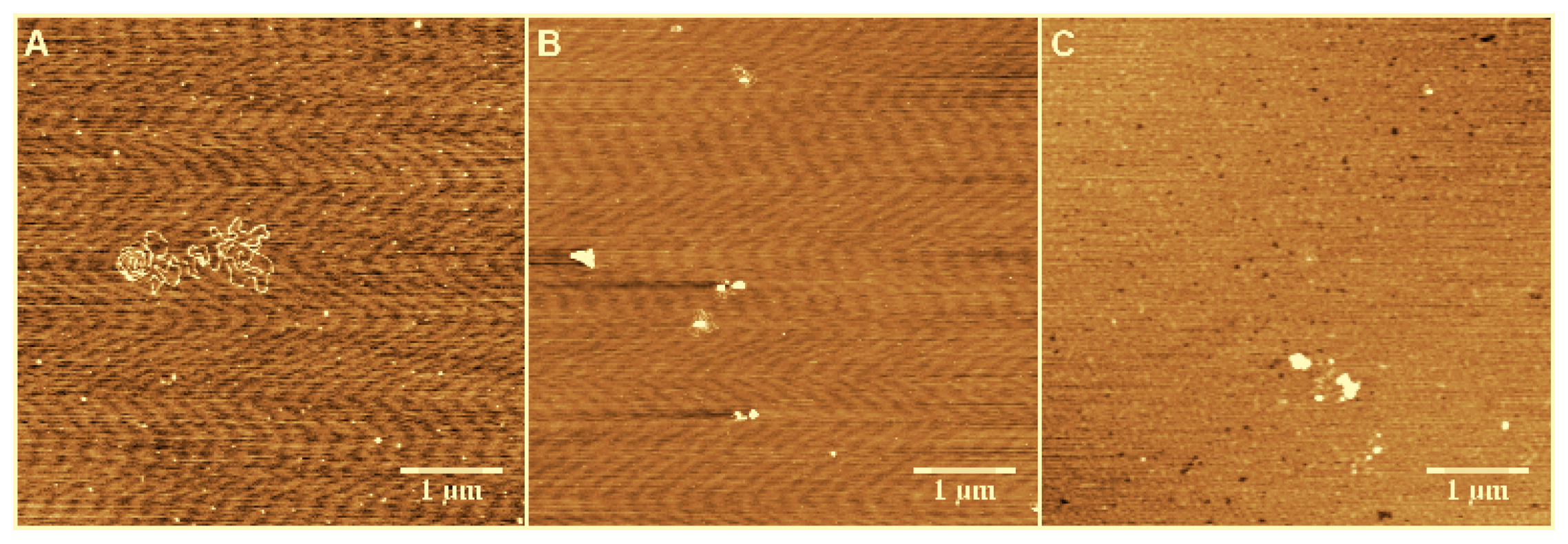
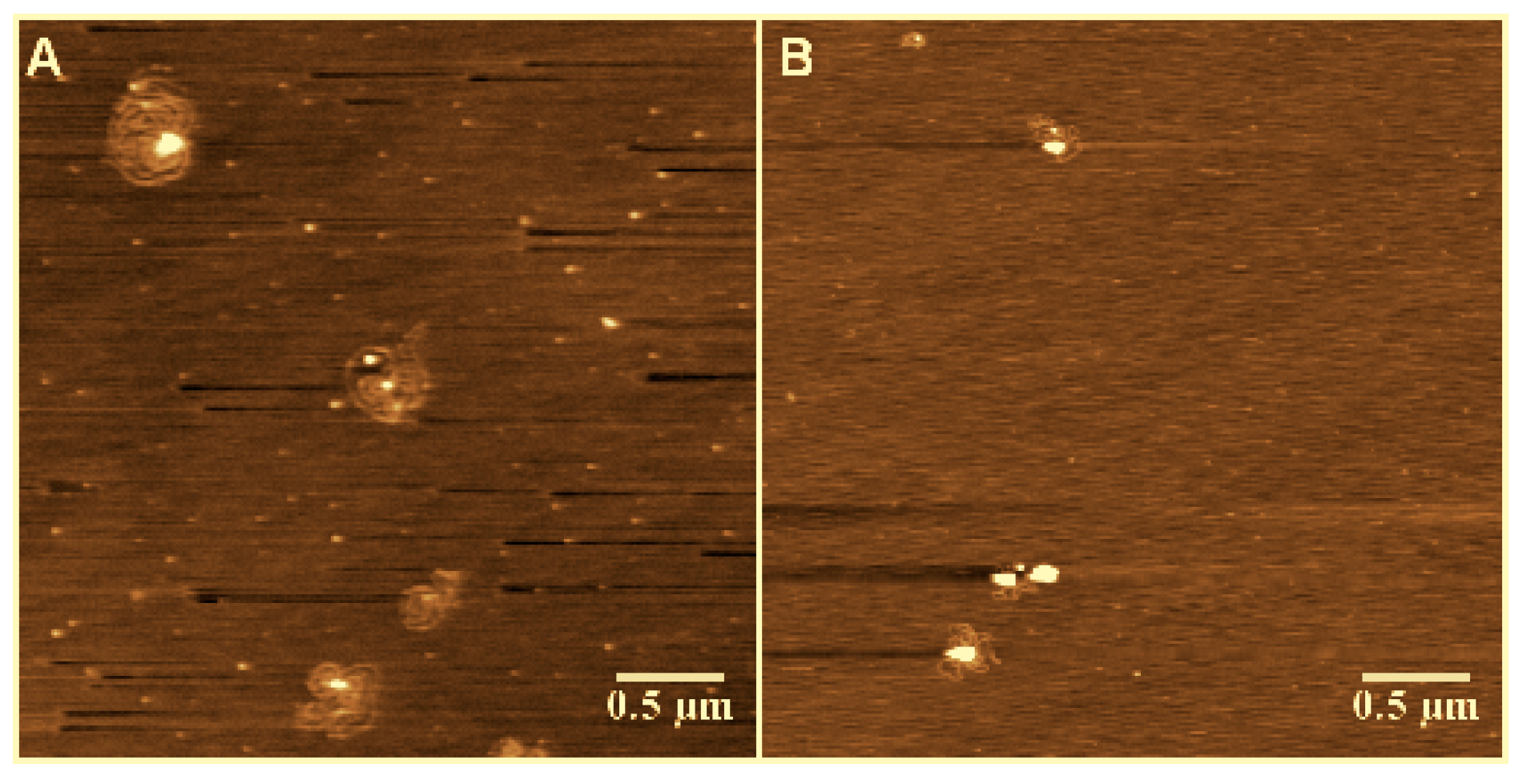
3.1. Observation by AFM
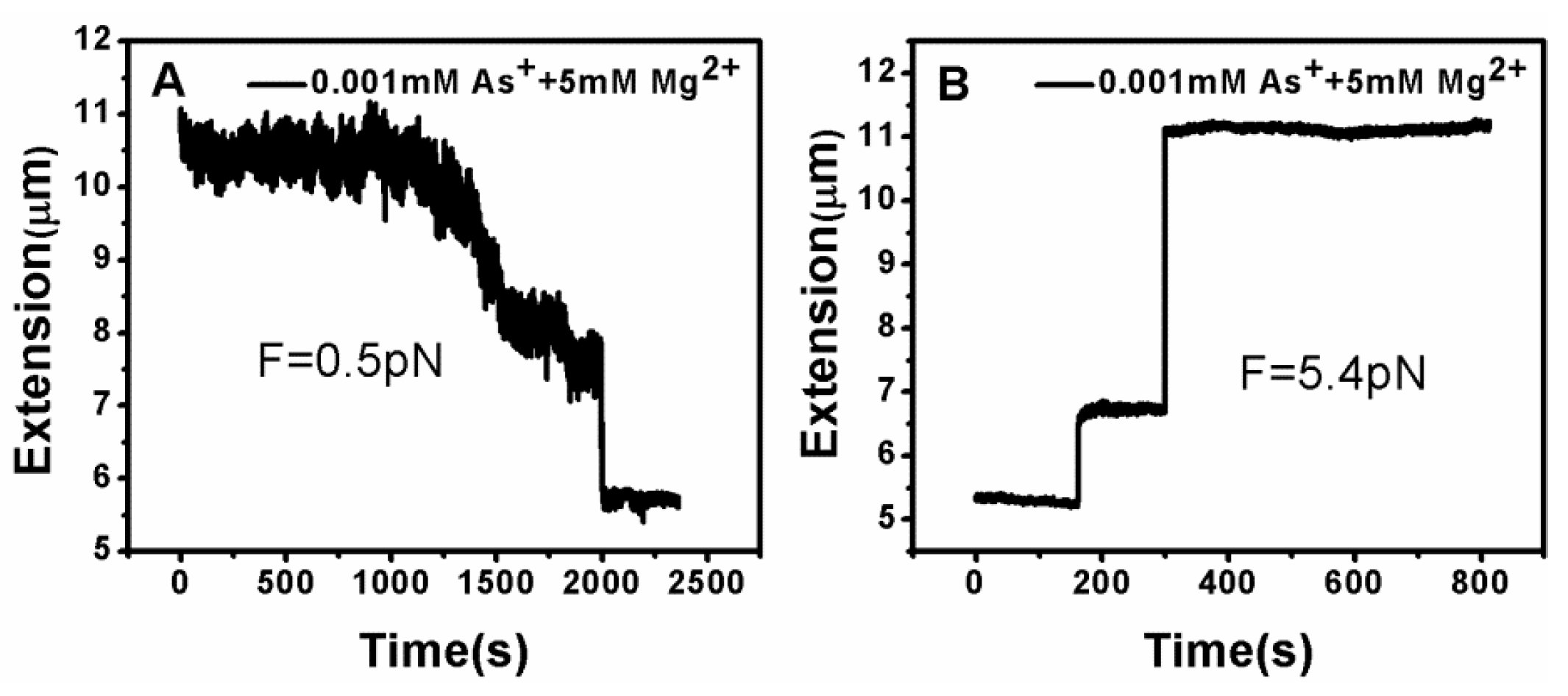
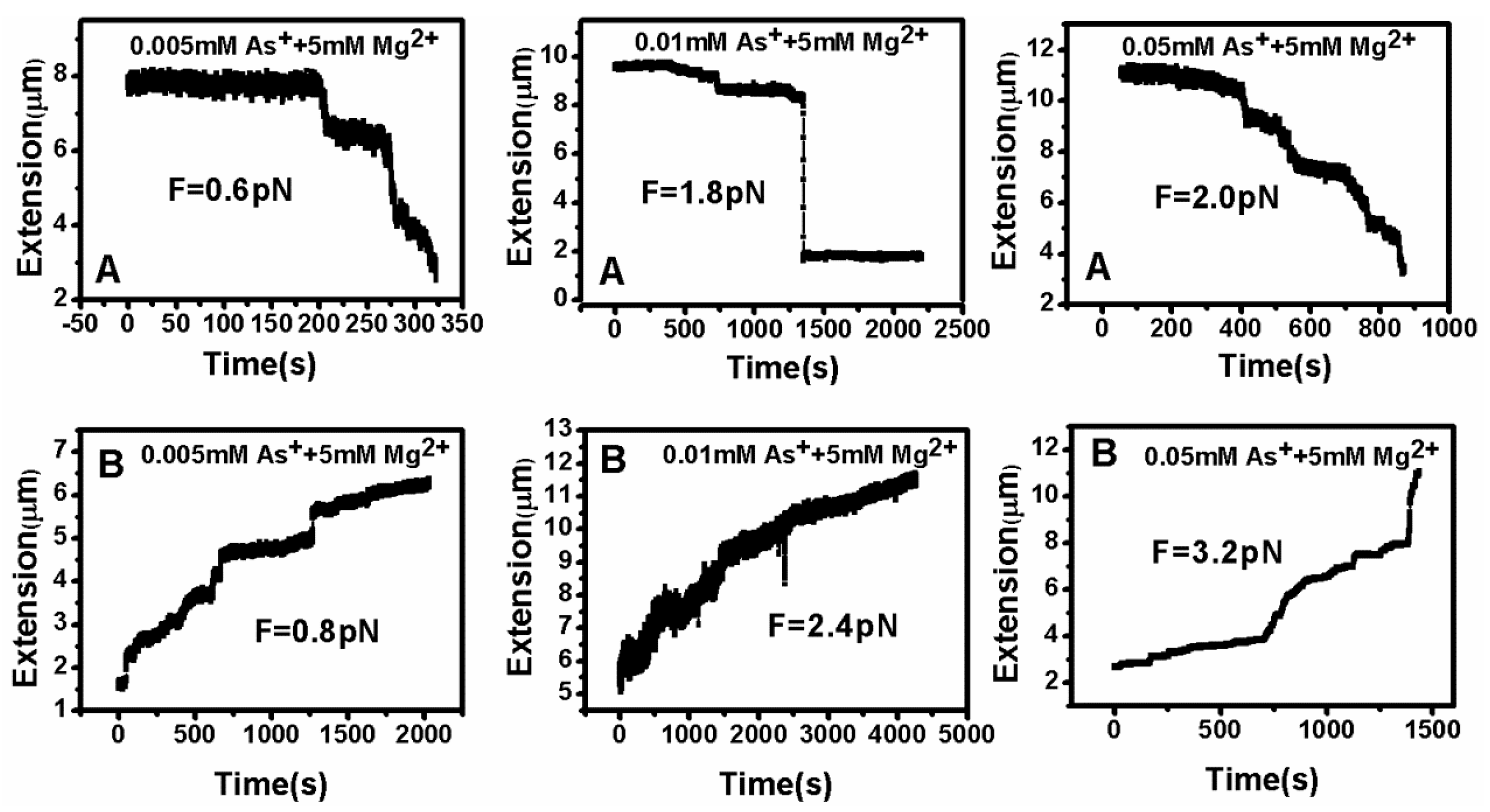
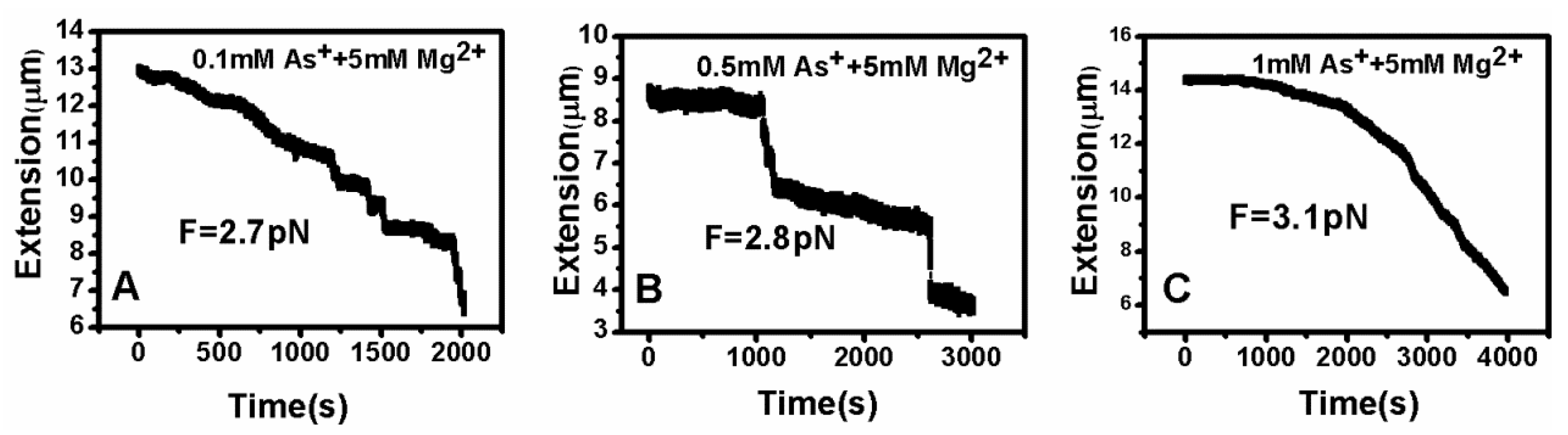
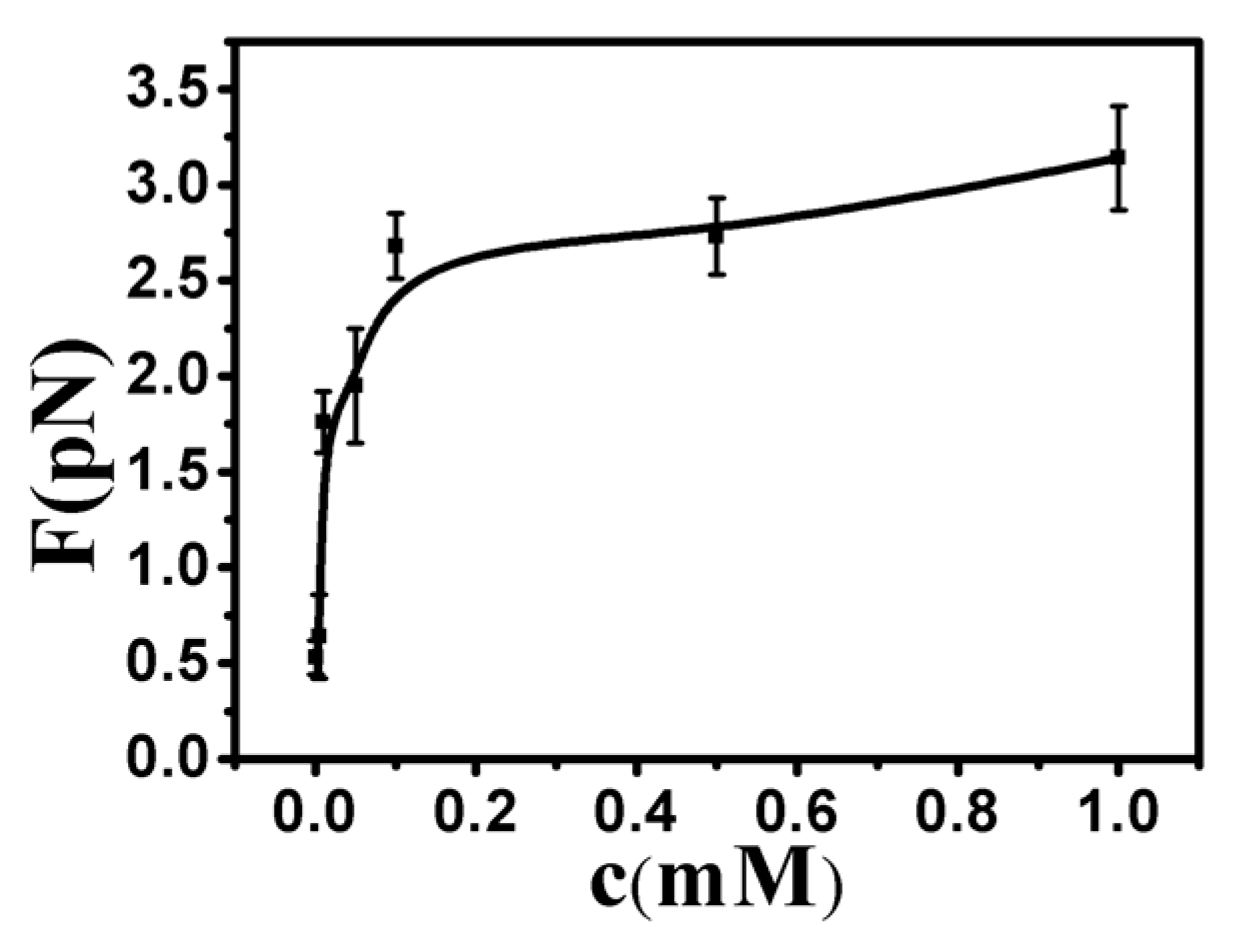
3.2. Tethering of Single DNA Molecules
3.3. Electrophoretic Mobility of DNA
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bloomfield, V.A. DNA condensation by multivalent cations. Biopolymers 1997, 44, 269–282. [Google Scholar] [CrossRef]
- Teif, V.B.; Bohinc, K. Condensed DNA: Condensing the concepts. Prog. Biophys. Mol. Biol. 2011, 105, 208–222. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Rouzina, I.; Shklovskii, B. Reentrant condensation of DNA induced by multivalent counterions. J. Chem. Phys. 2000, 112, 2562–2568. [Google Scholar] [CrossRef]
- Vijayanathan, V.; Thomas, T.; Thomas, T.J. DNA nanoparticals and development of DNA delivery vehicles for gene therapy. Biochemistry 2002, 41, 14085–14094. [Google Scholar] [CrossRef] [PubMed]
- Thomas, T.J.; Tajmir-Riahi, H.; Thomas, T. Polyamine-DNA interactions and development of gene delivery vehicles. Amino Acids 2016, 48, 2423–2431. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Lu, X.; Jia, F.; Liu, X.; Sun, Y.; Logan, J.K.; Zhang, K. Blurring the role of oligonucleotides: Spherical nucleic acids as a drug delivery vehicle. J. Am. Chem. Soc. 2016, 138, 10834–10837. [Google Scholar] [CrossRef] [PubMed]
- Besteman, K.; van Eijk, K.; Lemay, S. Charge inversion accompanies DNA condensation by multivalent ions. Nat. Phys. 2007, 3, 641–644. [Google Scholar] [CrossRef]
- Grosberg, A.Y.; Nguyen, T.T.; Shklovskii, B.I. Colloquium: The physics of charge inversion in chemical and biological systems. Rev. Mod. Phys. 2002, 74, 329–345. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Shklovskii, B.I. Model of inversion of DNA charge by a positive polymer: Fractionalization of the polymer charge. Phys. Rev. Lett. 2002, 89, 018101. [Google Scholar] [CrossRef] [PubMed]
- Besteman, K.; Zevenbergen, M.A.; Heering, H.A.; Lemay, S.G. Direct observation of charge inversion by multivalent ions as a universal electrostatic phenomenon. Phys. Rev. Lett. 2004, 93, 170802. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Guang, Y.; Yan, W. A dynamic light scattering study of counter-ions condensation on DNA. Acta Phys. Sin. 2013, 62, 118702. [Google Scholar]
- Luan, B.; Aksimentiev, A. Electric and electrophoretic inversion of the DNA charge in multivalent electrolytes. Soft Matter 2010, 6, 243–246. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, R.; Cao, B.; Guo, Z.; Yang, G. Single molecular demonstration of modulating charge inversion of DNA. Sci. Rep. 2016, 6, 38628. [Google Scholar] [CrossRef] [PubMed]
- Qiu, S.; Wang, Y.; Cao, B.; Guo, Z.; Chen, Y.; Yang, G. The suppression and promotion of DNA charge inversion by mixing counterions. Soft Matter 2015, 11, 4099–4105. [Google Scholar] [CrossRef] [PubMed]
- Raspaud, E.; Pelta, J.; de Frutos, M.; Livolant, F. Solubility and charge inversion of complexes of DNA and basic protein. Phys. Rev. Lett. 2006, 97, 068103. [Google Scholar] [CrossRef] [PubMed]
- Kabanov, A.V.; Kabanov, V. DNA complexes with polycations for the delivery of gene material into cells. Bioconjug. Chem. 1995, 6, 7–20. [Google Scholar] [PubMed]
- Petzold, G.; Nebel, A.; Buchhammer, H.-M.; Lunkwitz, K. Preparation and characterization of different polyelectrolyte complexes and their application as flocculants. Colloid Polym. Sci. 1998, 276, 125–130. [Google Scholar] [CrossRef]
- Buchhammer, H.-M.; Petzold, G.; Lunkwitz, K. Salt effect on formation and properties of interpolyelectrolyte complexes and their interactions with silica particles. Langmuir 1999, 15, 4306–4310. [Google Scholar] [CrossRef]
- Kekkonen, J.; Lattu, H.; Stenius, P. Adsorption kinetics of complexes formed by oppositely charged polyelectrolytes. J. Colloid Interface Sci. 2001, 234, 384–392. [Google Scholar] [CrossRef] [PubMed]
- Choosakoonkriang, S.; Lobo, B.A.; Koe, G.S.; Koe, J.G.; Middaugh, C.R. Biophysical characterization of PEI/DNA complexes. J. Pharm. Sci. 2003, 92, 1710–1722. [Google Scholar] [CrossRef] [PubMed]
- Mengarelli, V.; Auvray, L.; Zeghal, M. Phase behaviour and structure of stable complexes of oppositely charged polyelectrolytes. Europhys. Lett. 2009, 85, 58001. [Google Scholar] [CrossRef]
- Mengarelli, V.; Zeghal, M.; Auvray, L.; Clemens, D. Phase behavior and structure of stable complexes between a long polyanion and a branched polycation. Phys. Rev. E 2011, 84, 021805. [Google Scholar] [CrossRef] [PubMed]
- Störkle, D.; Duschner, S.; Heimann, N.; Maskos, M.; Schmidt, M. Complex formation of DNA with oppositely charged polyelectrolytes of different chain topology: Cylindrical brushes and dendrimers. Macromolecules 2007, 40, 7998–8006. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, X.; Yang, G. Single molecular analysis of the interaction between DNA and chitosan. RSC Adv. 2015, 5, 29594–29600. [Google Scholar] [CrossRef]
- Kabanov, V.; Sergeyev, V.; Pyshkina, O.; Zinchenko, A.; Zezin, A.; Joosten, J.; Brackman, J.; Yoshikawa, K. Interpolyelectrolyte complexes formed by DNA and astramol poly(propylene imine) dendrimers. Macromolecules 2000, 33, 9587–9593. [Google Scholar] [CrossRef]
- Walker, H.W.; Grant, S.B. Factors Influencing the flocculation of colloidal particles by a model anionic polyelectrolyte. Colloids Surf. 1996, 119, 229–239. [Google Scholar] [CrossRef]
- Keren, K.; Soen, Y.; Yoseph, G.B.; Gilad, R.; Braun, E.; Sivan, U.; Talmon, Y. Microscopics of complexation between long DNA molecules and positively charged colloids. Phys. Rev. Lett. 2002, 89, 088103. [Google Scholar] [CrossRef] [PubMed]
- Gössl, I.; Shu, L.; Schlüter, A.D.; Rabe, J.P. Molecular structure of single DNA complexes with positively charged dendronized polymers. J. Am. Chem. Soc. 2002, 124, 6860–6865. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kimura, K.; Huang, Q.; Dubin, P.L.; Jaeger, W. Effect of salt on polyelectrolyte-micelle coacervation. Macromolecules 1999, 32, 7128–7134. [Google Scholar] [CrossRef]
- Fu, W.-B.; Wang, X.-L.; Zhang, X.-H.; Ran, S.-Y.; Yan, J.; Li, M. Compaction dynamics of single DNA molecules under tension. J. Am. Chem. Soc. 2006, 128, 15040–15041. [Google Scholar] [CrossRef] [PubMed]
- Raspaud, E.; de la Cruz, M.O.; Sikorav, J.-L.; Livolant, F. Precipitation of DNA by polyamines: A polyelectrolyte behavior. Biophys. J. 1998, 74, 381–393. [Google Scholar] [CrossRef]
- Pelta, J.; Livolant, F.; Sikorav, J.-L. DNA aggregation induced by polyamines and cobalthexamine. J. Biol. Chem. 1996, 271, 5656–5662. [Google Scholar] [CrossRef] [PubMed]
- Todd, B.A.; Rau, D.C. Interplay of ion binding and attraction in DNA condensed by multivalent cations. Nucleic Acids Res. 2008, 36, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Cherstvy, A.G. Electrostatic interactions in biological DNA-related systems. Phys. Chem. Chem. Phys. 2011, 13, 9942–9968. [Google Scholar] [CrossRef] [PubMed]
- Cortini, R.; Caré, B.R.; Victor, J.-M.; Barbi, M. Theory and simulations of toroidal and rod-like structures in single-molecule DNA condensation. J. Chem. Phys. 2015, 142, 105102. [Google Scholar] [CrossRef] [PubMed]
- Martín-Molina, A.; Calero, C.; Faraudo, J.; Quesada-Pérez, M.; Travesset, A.; Hidalgo-Álvarez, R. The hydrophobic effect as a driving force for charge inversion in colloids. Soft Matter 2009, 5, 1350–1353. [Google Scholar] [CrossRef]
- Wang, Y.; Ran, S.; Man, B.; Yang, G. Ethanol induces condensation of single DNA molecules. Soft Matter 2011, 7, 4425–4434. [Google Scholar] [CrossRef]
- Todd, B.A.; Parsegian, V.A.; Shirahata, A.; Thomas, T.; Rau, D.C. Attractive forces between cation condensed DNA double helices. Biophys. J. 2008, 94, 4775–4782. [Google Scholar] [CrossRef] [PubMed]
- Strick, T.; Allemand, J.-F.; Bensimon, D.; Croquette, V. Behavior of supercoiled DNA. Biophys. J. 1998, 74, 2016–2028. [Google Scholar] [CrossRef]
- Estévez-Torres, A.; Baigl, D. DNA Compaction: Fundamentals and applications. Soft Matter 2011, 7, 6746. [Google Scholar] [CrossRef]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, W.; Wang, Y.; Yang, A.; Yang, G. DNA Compaction and Charge Inversion Induced by Organic Monovalent Ions. Polymers 2017, 9, 128. https://doi.org/10.3390/polym9040128
Xia W, Wang Y, Yang A, Yang G. DNA Compaction and Charge Inversion Induced by Organic Monovalent Ions. Polymers. 2017; 9(4):128. https://doi.org/10.3390/polym9040128
Chicago/Turabian StyleXia, Wenyan, Yanwei Wang, Anthony Yang, and Guangcan Yang. 2017. "DNA Compaction and Charge Inversion Induced by Organic Monovalent Ions" Polymers 9, no. 4: 128. https://doi.org/10.3390/polym9040128