pH Dependence of Chitosan Enzymolysis
Abstract
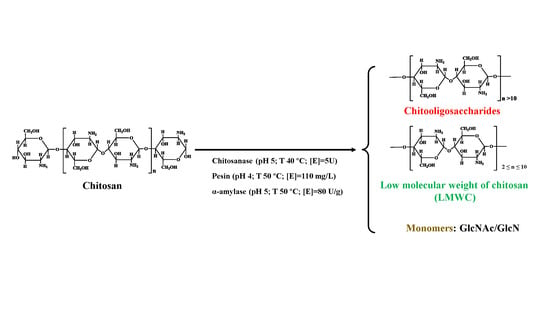
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Hydrolysis Experiments
2.3. Experimental Design
2.4. Determination of pKa Values
2.5. Characterizations of Chitosan before and after Hydrolysis
3. Results and Discussion
3.1. Influence of the Main Factors on the Hydrolysis Process
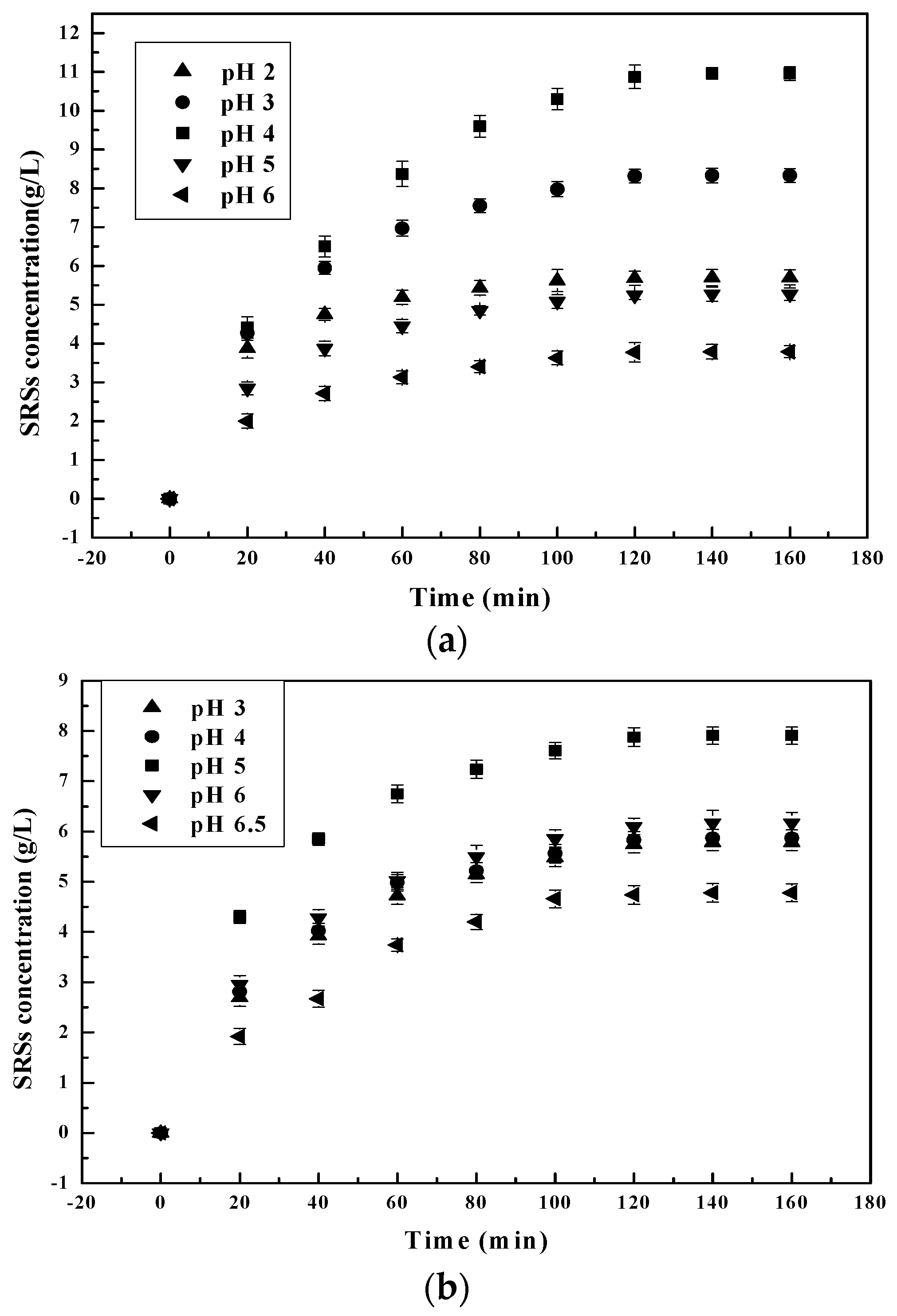
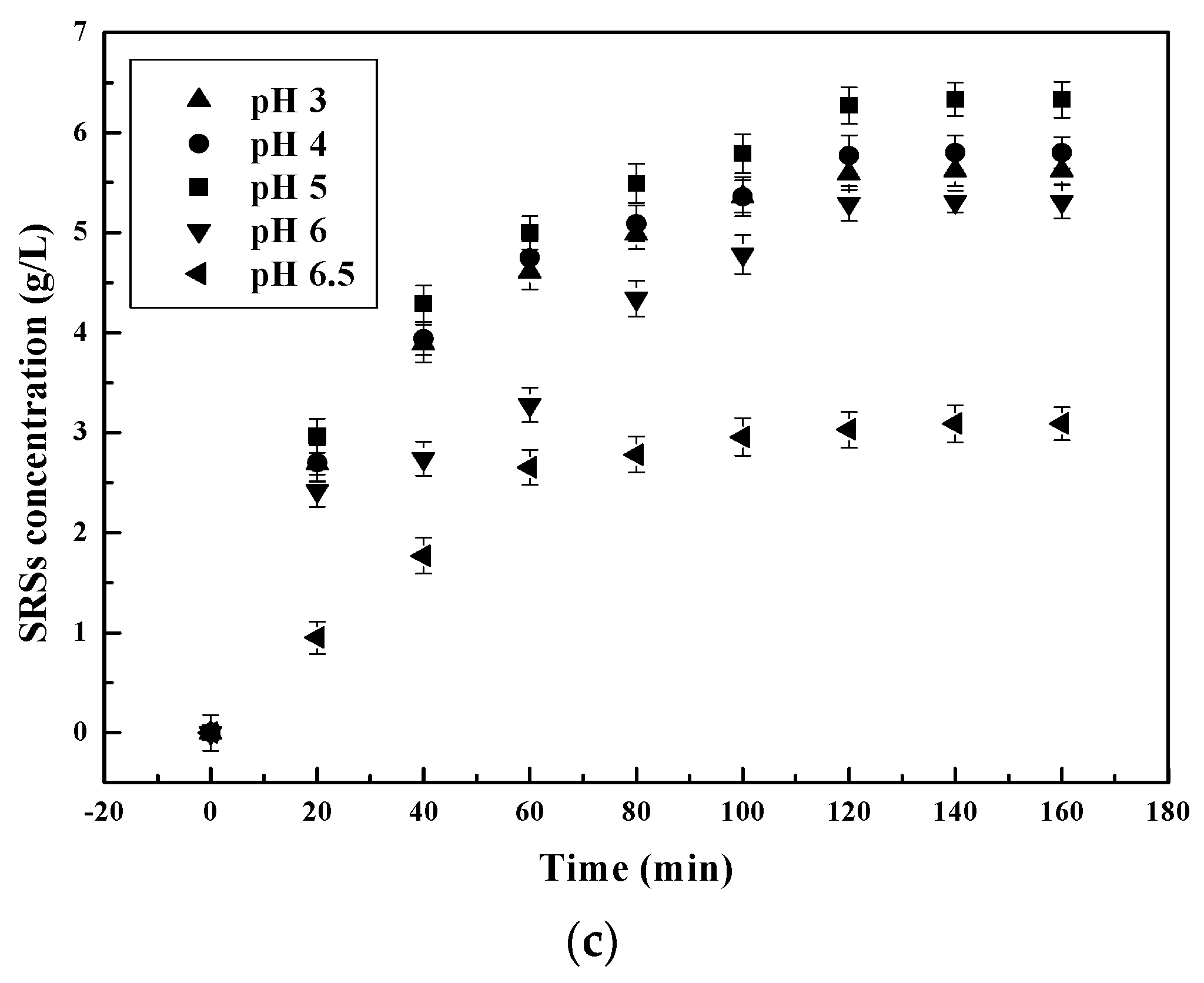
3.1.1. Effect of pH on the Chitosan Hydrolysis Process
3.1.2. Effect of Temperature on the Chitosan Hydrolysis Process
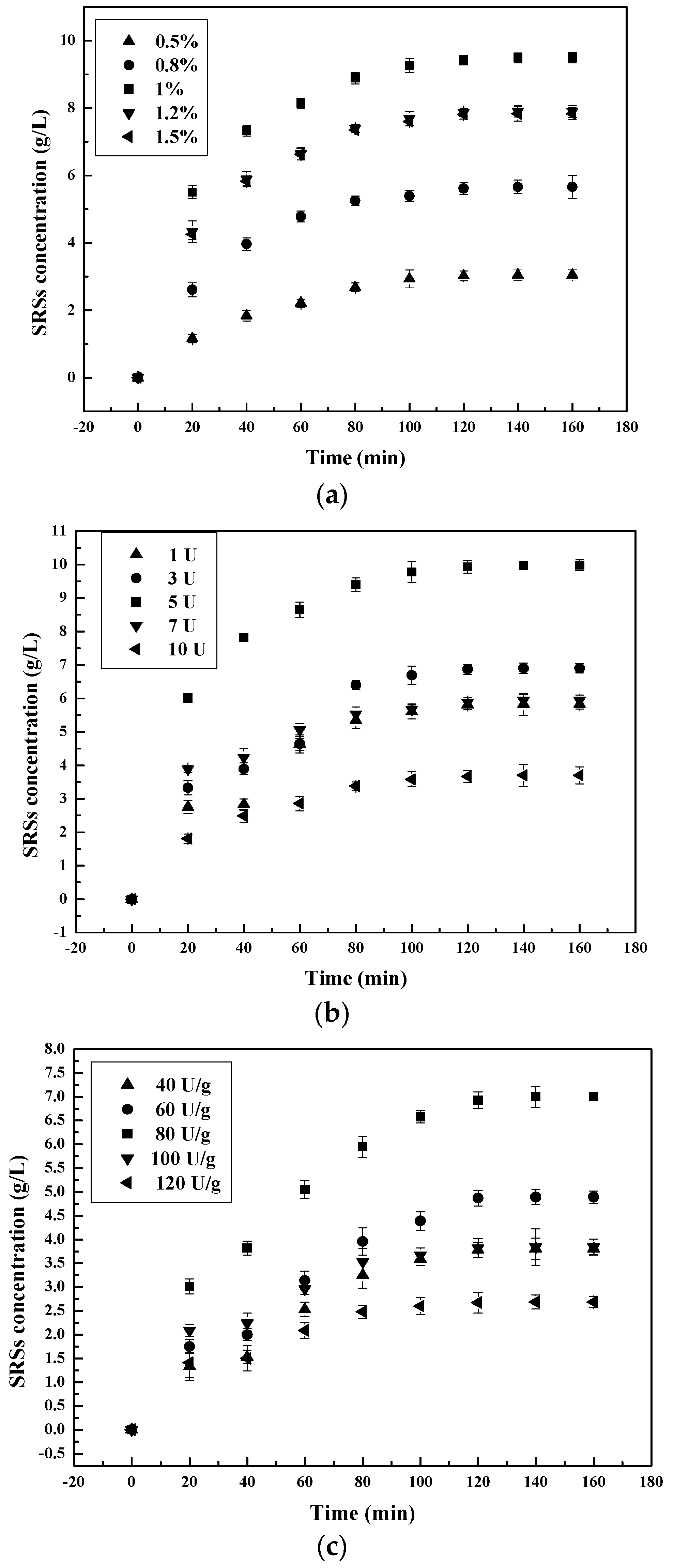
3.1.3. Effect of Pepsin Concentration on the Chitosan Hydrolysis Process
3.2. Enzymolysis Kinetics
3.2.1. pH Dependent Kinetic Parameters
3.2.2. Temperature Dependence of Kinetic Parameters
3.2.3. Hydrolysis Process Dependence on Enzyme Concentration
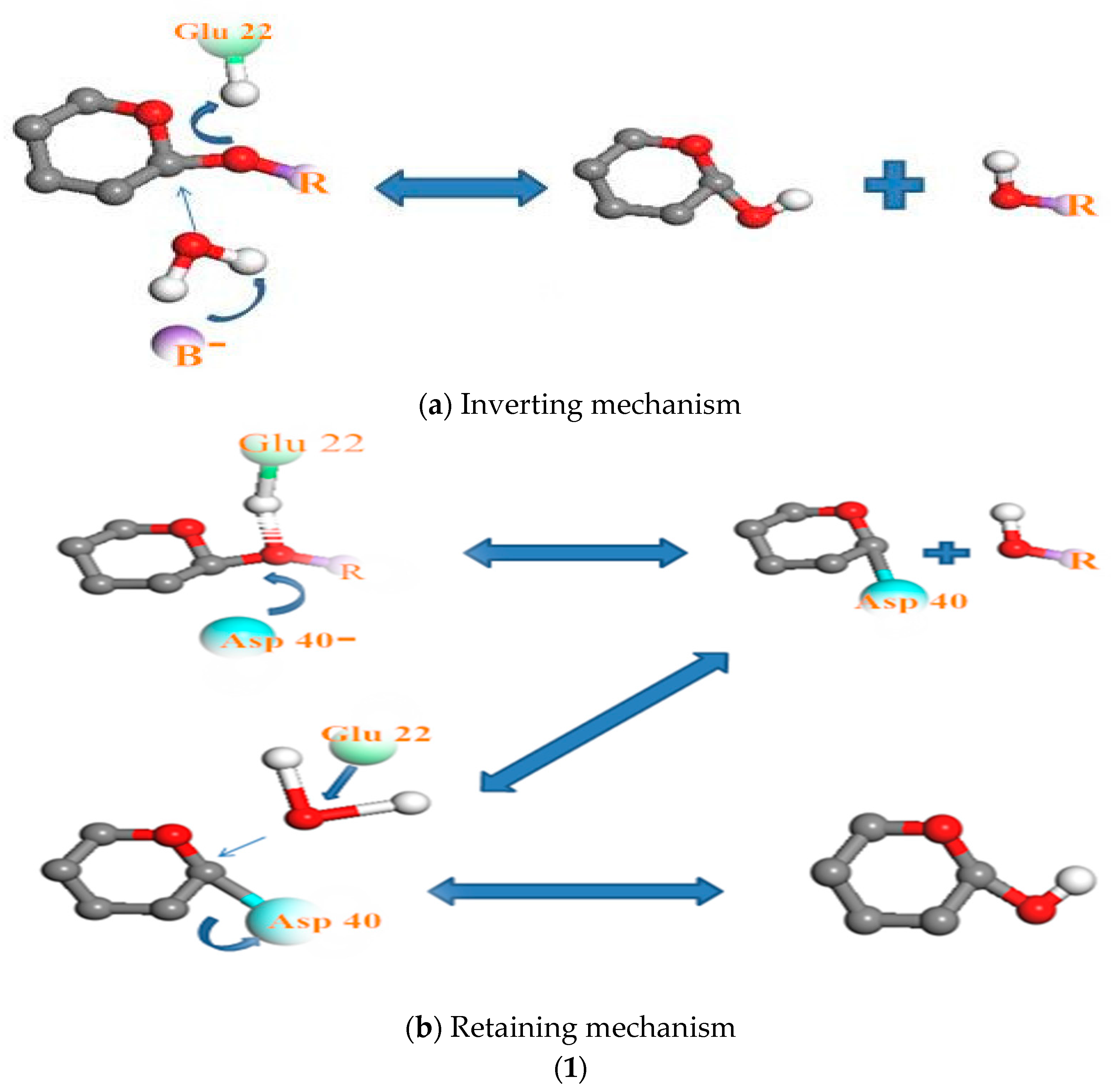
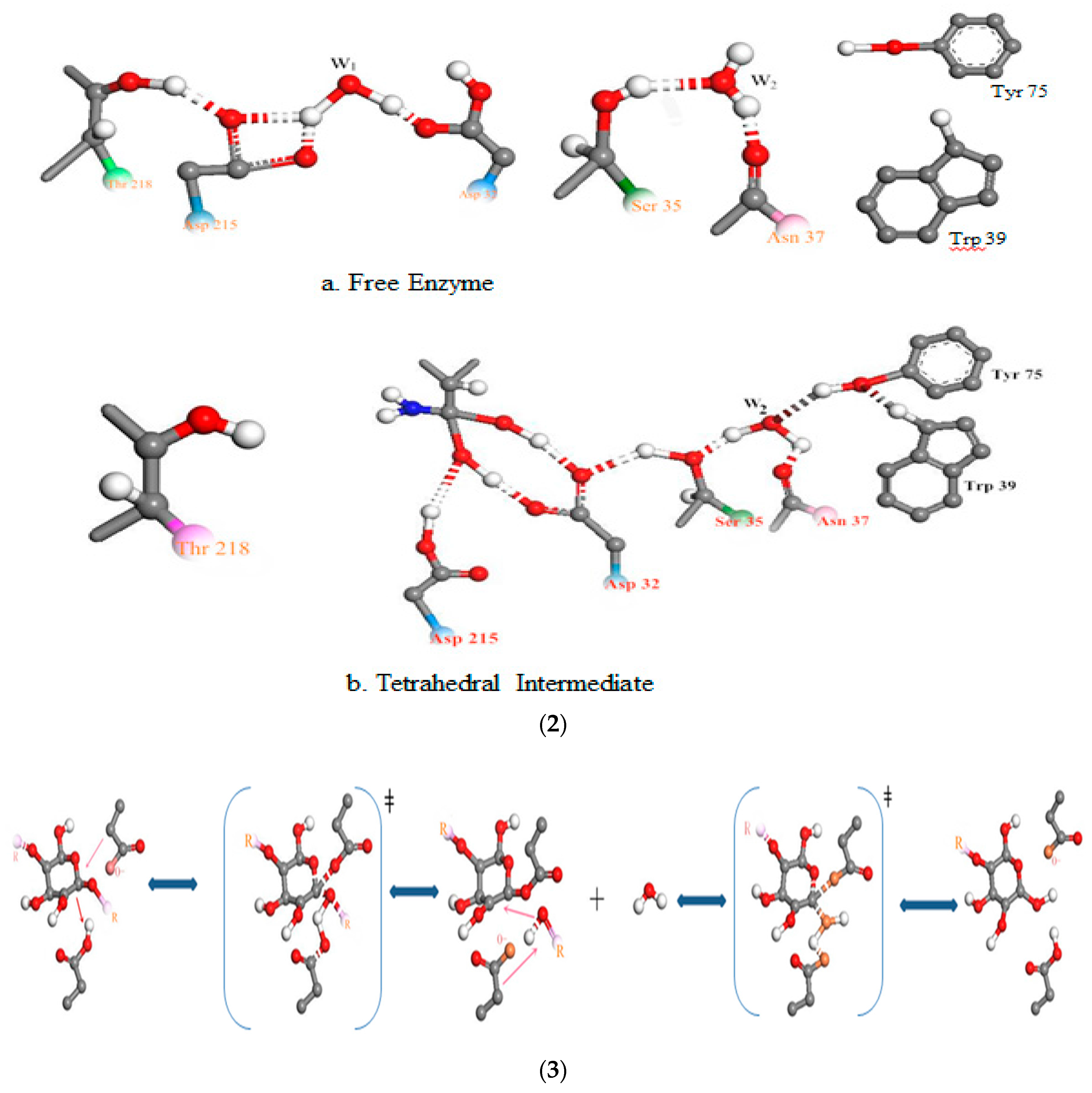
3.3. Relationship Between pH and Degradation Mechanisms of Each Enzyme
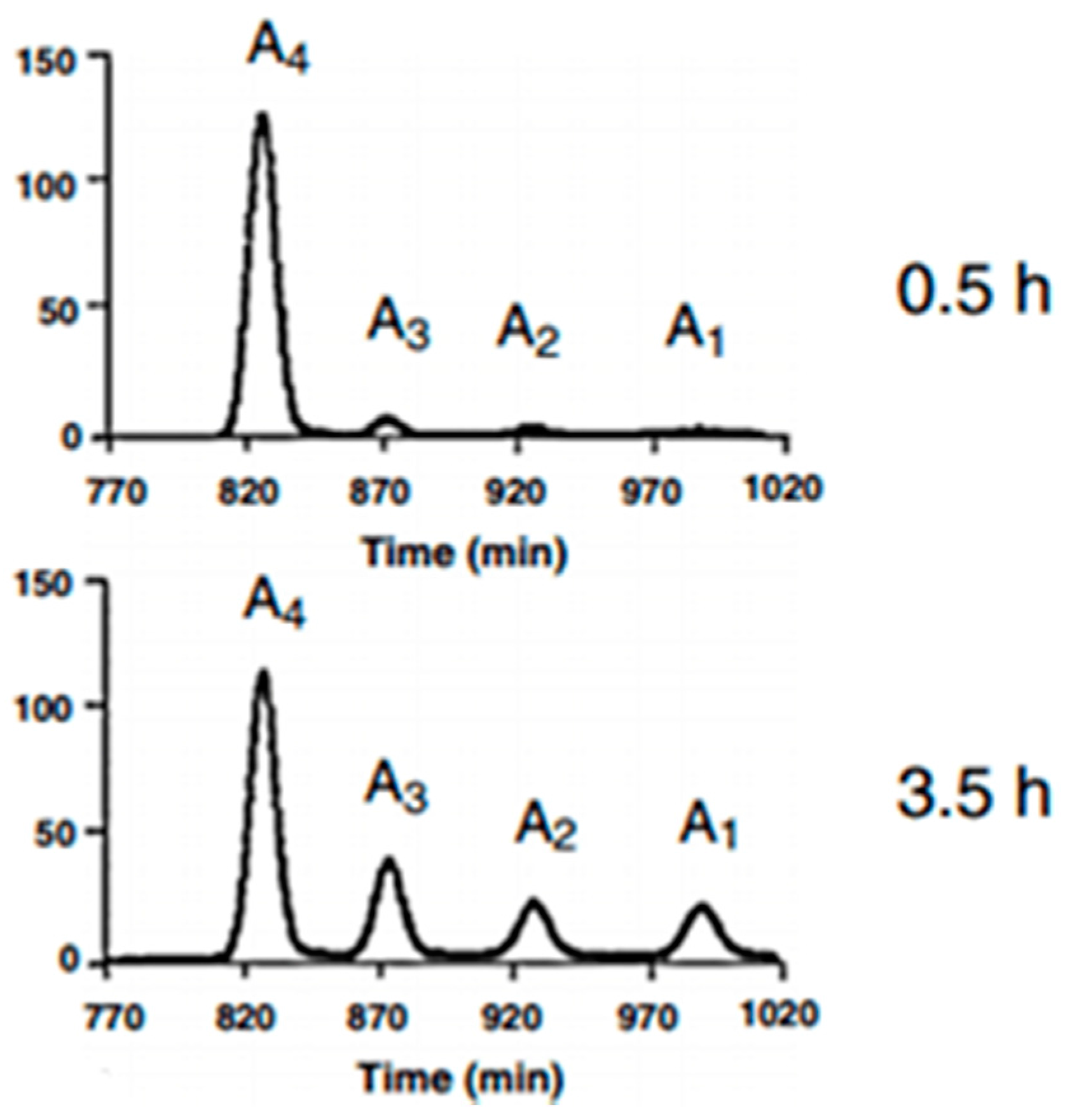
3.4. Characterizations of the Enzymolysis Products
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kerton, F.M.; Liu, Y.; Omari, K.W.; Hawboldt, K. Green chemistry and the ocean-based biorefinery. Green Chem. 2013, 15, 860–871. [Google Scholar] [CrossRef]
- Chen, X.; Yang, H.Y.; Yan, N. Shell Biorefinery: Dream or Reality? Chem. Eur. J. 2016, 22, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Yan, N.; Chen, X. Don’t waste seafood waste: Turning cast-off shells into nitrogen-rich chemicals would benefit economies and the environment. Nature 2015, 524, 155–157. [Google Scholar] [CrossRef] [PubMed]
- Muzzarelli, R.A.A.; Muzzarelli, C. Chitosan Chemistry: Relevance to the Biomedical Sciences, book Polysaccharides I. Adv. Polym. Sci. 2005, 186, 151–209. [Google Scholar]
- Li, J.; Du, Y.; Yang, J.; Feng, T.; Li, A.; Chen, P. Preparation and characterization of low molecular weight chitosan and chito-oligomers by a commercial enzyme. Polym. Degrad. Stabil. 2005, 87, 441–448. [Google Scholar] [CrossRef]
- Choi, W.; Ahn, K.; Lee, D.; Byun, M.; Park, H. Preparation of chitosan oligomers by irradiation. Polym. Degrad. Stabil. 2002, 78, 533–538. [Google Scholar] [CrossRef]
- Mourya, V.K.; Inamdar, N.N.; Choudhari, Y.M. Chitooligosaccharides: Synthesis, characterization and applications. Polym. Sci. Ser. A 2011, 53, 583–612. [Google Scholar] [CrossRef]
- Chen, Q.; Xiao, W.J.; Zhou, L.L.; Wu, T.H.; Wu, Y. Hydrolysis of chitosan under microwave irradiation in ionic liquids promoted by sulfonic acid-functionalized ionic liquids. Polym. Degrad. Stabil. 2012, 97, 49–53. [Google Scholar] [CrossRef]
- Philippova, O.E.; Korchagina, E.V. Chitosan and its hydrophobic derivatives: Preparation and aggregation in dilute aqueous solutions. Polym. Sci. Ser. A 2012, 54, 552–572. [Google Scholar] [CrossRef]
- Zhang, J.G.; Yan, N. Formic acid-mediated liquefaction of chitin. Green Chem. 2016, 18, 5050–5058. [Google Scholar] [CrossRef]
- Pierson, Y.; Chen, X.; Bobbink, F.D.; Zhang, J.G.; Yan, N. Acid-Catalyzed Chitin Liquefaction in Ethylene Glycol. ACS Sustain. Chem. Eng. 2014, 2, 2081–2089. [Google Scholar] [CrossRef]
- Muzzarelli, R.A.; Terbojevich, M.; Muzzarelli, C.; Francescangeli, O. Chitosans depolymerized with the aid of papain and stabilized asglycosylamines. Carbohydr. Polym. 2002, 50, 69–78. [Google Scholar] [CrossRef]
- Eide, K.B.; Norberg, A.L.; Heggset, E.B.; Lindbom, A.R.; Vårum, K.M.; Eijsink, V.G.H.; Sørlie, M. Human Chitotriosidase Catalyzed Hydrolysis of Chitosan. ACS Biochem. 2012, 51, 487–495. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, K.; Chrispinas, O.A.; Yoshida, N.; Inoue, J.; Kariya, K. Chitosanase, its manufacture, and manufacture of chitooligosaccharides. In J. T.K Koho. (Ed.) Jpn. 2001, 0069975. [Google Scholar]
- Kim, S.K.; Rajapakse, N. Enzymatic production and biological activities of chitosan oligosaccharides (COS): A review. Carbohydr. Polym. 2005, 62, 357–368. [Google Scholar] [CrossRef]
- Kumar, A.V.; Varadaraj, M.C.; Gowda, L.R.; Tharanathan, R.N. Low molecular weight chitosan-preparation with the aid of pronase, characterization and their bactericidal activity towards Bacillus cereus and Escherichia coli. Biochim. Biophys. Acta 2007, 1771, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.Q.; Du, Y.M.; Xiao, L.; Li, Z.; Gao, X.H. Enzyme preparation of water-soluble chitosan oligomers and their antitumour activity. Int. J. Biol. Macromol. 2002, 31, 111–117. [Google Scholar] [CrossRef]
- Kumar, A.V.; Varadaraj, M.C.; Gowda, L.R.; Tharanathan, R.N. Low molecular weight chitosan-preparation with aid of pepsin, characterization, and its bactericidal activity. Biomacromolecules 2007, 8, 566–572. [Google Scholar] [CrossRef] [PubMed]
- Xia, W.S.; Liu, P.; Liu, J. Advance in chitosan hydrolysis by non-specific cellulases. Bioresour. Technol. 2008, 99, 6751–6762. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.J. Preparation of water soluble chitosan by hydrolysis with commercial α-amylase containing chitosanase activity. Food Chem. 2011, 128, 769–772. [Google Scholar] [CrossRef]
- Yan, X.; Evenocheck, H.M. Chitosananalysis using acid hydrolysis and HPLC/UV. Carbohydr. Polym. 2012, 87, 1774–1778. [Google Scholar] [CrossRef]
- Xia, Z.Q.; Wu, S.J.; Chen, J.H. Preparation of water soluble chitosan by hydrolysis using hydrogen peroxide. Int. J. Biol. Macromol. 2013, 59, 242–245. [Google Scholar] [CrossRef] [PubMed]
- Dzirila, M.; Gribb, H.; Laribi-Habchi, H.; Drouicheb, N.; Abdib, N.; Lounicib, H.; Paussd, A.; Mamerid, N. Chitin oligomers and monomers production by coupling g radiation and enzymatic hydrolysis. J. Ind. Eng. Chem. 2015, 26, 396–401. [Google Scholar] [CrossRef]
- Pan, A.D.; Zeng, H.Y.; Gohi, B.F.C.A.; Li, Y.Q. Enzymolysis of Chitosan by Papain and Its Kinetics. Carbohydr. Polym. 2016, 135, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Gohi, B.F.A.C.; Zeng, H.Y.; Pan, A.D. Optimization and Characterization of Chitosan Enzymolysis by Pepsin. Bioingineering 2016, 3, 17. [Google Scholar] [CrossRef]
- Roncal, T.; Oviedo, A.; de Armentia, I.L.; Fernández, L.; Villarán, M.C. High yield production of monomer-free chitosan oligosaccharides by pepsin catalyzed hydrolysis of a high deacetylation degree chitosan. Carbohydr. Res. 2007, 342, 2750–2756. [Google Scholar] [CrossRef] [PubMed]
- Einbu, A.; Grasdalen, H.; Varum, K.M. Kinetics of hydrolysis of chitin/chitosan oligomers in concentrated hydrochloric acid. Carbohydr. Res. 2007, 342, 1055–1062. [Google Scholar] [CrossRef] [PubMed]
- Sørbotten, A.; Horn, S.J.; Eijsink, V.G.H.; Vårum, K.M. Degradation of chitosans with Chitinase B from Serratia marcescens; production of chito-oligosaccharides and insight into enzyme processivity. FEBS J. 2005, 272, 538–549. [Google Scholar] [CrossRef] [PubMed]
- Radeva, G.; Valchev, I.; Petrin, S.; Valcheva, E.; Tsekova, P. Kinetic model of enzymatic hydrolysis of steam-exploded wheat straw. Carbohydr. Polym. 2012, 87, 1280–1285. [Google Scholar] [CrossRef]
- Boeris, V.; Micheletto, Y.; Lionzo, M.; Nadya, P.S.S.; Pico, G. Interaction behavior between chitosan and pepsin. Carbohydr. Polym. 2011, 84, 459–464. [Google Scholar] [CrossRef]
- Wu, C.; Zhou, S.Q.; Wang, W. A dynamic laser light-scattering study of chitosan in aqueous-solution. Biopolymers 1995, 35, 385–392. [Google Scholar] [CrossRef]
- Andras, F.; Sinkovits, B.C.; Bryksa, T.T.; Rickey, Y.Y. Understanding the structural-functional role of specific catalytic residues in a model food related enzyme: Pepsin. Enzyme Microb. Technol. 2007, 40, 1175–1180. [Google Scholar]
- Choi, Y.J.; Kim, E.J.; Piao, Z.; Yun, Y.C.; Shin, Y.C. Purification and Characterization of Chitosanase from Bacillus sp. Strain KCTC 0377BP and Its Application for the Production of Chitosan Oligosaccharides. Appl. Environ. Microbiol. 2004, 70, 4522–4531. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.K.; Wu, S.J.; Kim, J.M. Preparation of glucosamine by hydrolysis of chitosan with commercial α-amylase and glucoamylase. J. Zhejiang Univ. Sci. B (Biomed. Biotechnol.) 2011, 12, 931–934. [Google Scholar]
- Kamatari, Y.O.; Dobson, C.M.; Konno, T. Structural dissection of alkaline-denatured pepsin. Protein Sci. 2003, 12, 717–724. [Google Scholar] [CrossRef] [PubMed]
- Fruton, J.S. Pepsin. In The Enzymes; Boyer, P.D., Ed.; Academic Press: New York, NY, USA, 1971; pp. 119–164. [Google Scholar]
- Li, B.; Zhang, J.L.; Dai, F.T.; Xia, W.S. Purification of chitosan by using sol-gel immobilized pepsin deproteinization. Carbohydr. Polym. 2012, 88, 206–212. [Google Scholar] [CrossRef]
- Thadathil, N.; Velappan, S.P. Recent developments in chitosanase research and its biotechnologicalapplications: A review. Food Chem. 2014, 150, 392–399. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.G.; Pan, S.K.; Xia, Z.Q.; Wu, S.J. A new assay of chitosan in the presence of protein by hydrolysis with commercial α-amylase. Eur. Food Res. Technol. 2011, 233, 717–719. [Google Scholar] [CrossRef]
- Shee, F.L.T.; Arul, J.; Brunet, S.; Bazinet, L. Effect of bipolar membrane electrobasification on chitosanase activity during chitosan hydrolysis. J. Biotechnol. 2008, 134, 305–311. [Google Scholar]
- Peller, L.; Alberty, R.A. Multiple Intermediates in Steady State Enzyme Kinetics.1,2 I. The Mechanism Involving a Single Substrate and Product. J. Am. chem. Soc. 1959, 81, 5907–5914. [Google Scholar] [CrossRef]
- Nathan, S.M.; Christine, M.L.; Michael, R.L. Characterization of Acid catalytic Domains for Cellulose hydrolysis and glucose degradation. Biotechnol. Bioeng. 2002, 79, 610–618. [Google Scholar]
- Malester, I.A.; Green, M.; Shelef, G. kinetics of dilute acid hydrolysis of cellulose originating from municipal solide waste. Ind. Eng. Chem. 1992, 31, 1998–2003. [Google Scholar] [CrossRef]
- Richard, J.P. The Enhancement of enzyme rate acceleration by bronsted acid-base catalysis. Biochemistry 1998, 37, 4305–4309. [Google Scholar] [CrossRef] [PubMed]
- Alireza, S.; Azardokht, A.; Alireza, F.; Steven, R.K. Effect of Hydrogen bonds on pKa values: Importance of Networking. J. Am. Chem. Soc. 2012, 134, 10646–10650. [Google Scholar]
- Cleland, W.W.; Frey, P.A.; Gerlt, J.A. The low barrier hydrogen bond in enzymic catalysis. J. Biol. Chem. 1998, 273, 25529–25532. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, J.L.; Xu, H.J.; Yuan, Z.H.; Guo, Y. Cellulase deactivation based kinetic modeling of enzymatic hydrolysis of steam-exploded wheat straw. Bioresour. Technol. 2010, 101, 8261–8266. [Google Scholar] [CrossRef] [PubMed]
- Rajesh, G.; Lee, Y.Y. Mechanism of cellulase reaction on pure cellulosic substrates. Biotechnol. Bioeng. 2009, 102, 1570–1581. [Google Scholar]
- Bardhan, K.D.; Strugala, V.; Dettmar, P.W. Reflux Revisited: Advancing the Role of Pepsin. Int. J. Otolaryngol. 2012, 2012, 646901. [Google Scholar] [CrossRef] [PubMed]
- Fukamizo, T.; Juffer, A.H.; Vogel, H.J.; Honda, Y.; Tremblay, H.; Boucher, I. Theoretical calculation of pKa reveals an important role of Arg205 in the activity and stability of Streptomyces sp. N174 chitosanase. J. Biol. Chem. 2000, 275, 25633–25640. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Hart, C.A.; Sales, D.; Roberts, N.B. Bacterial killing in gastric juice-effect of pH and pepsin on Escherichia coli and Helicobacter pylori. J. Med. Microbiol. 2006, 55, 1265–1270. [Google Scholar] [CrossRef] [PubMed]
- Savarino, V.; di Mario, F.; Scarpignato, C. Proton pump inhibitors in GORD. An overview of their pharmacology, efficacy and safety. Pharmacol. Res. 2009, 59, 135–153. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, E.P.; Matthews, B.W. Core-packing constraints, hydrophobicity and protein design. Curr. Opin. Biotechnol. 1994, 5, 396–402. [Google Scholar] [CrossRef]
- MacGregor, E.A.; Janecek, S.I.; Svensson, B. Relationship of sequence and structure to specificity in the K-amylase family of enzymes. Biochim. Biophys. Acta 2001, 1546, 1–20. [Google Scholar] [CrossRef]
- Davies, G.; Sinnott, M.L.; Withers, S.G. Glycosyl transfer. In Comprehensive Biological Catalysis; Sinnott, M., Ed.; Academic Press: New York, NY, USA, 1998; pp. 119–208. [Google Scholar]
- Uitdehaag, J.C.M.; Mosi, R.; Kalk, K.H.; van der Veen, B.A.; Dijkhuizen, L.; Withers, S.G.; Dijkstra, B.W. X-ray structures along the reaction pathway of cyclodextrin glycosyltransferase elucidate catalysis in the K-amylase family. Nat. Struct. Biol. 1999, 6, 432–436. [Google Scholar] [CrossRef] [PubMed]
- Uitdehaag, J.C.M.; Kalk, K.H.; van der Veen, B.A.; Dijkhuizen, L.; Dijkstra, B.W. The cyclization mechanism of cyclodextrin glycosyltransferase (CGTase) as revealed by a Q-cyclodextrin-CGTase complex at 1.8-A resolution. J. Biol. Chem. 1999, 274, 34868–34876. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Du, Y.; Liang, H. Low Molecular Weight Water-Soluble Chitosans: Preparation with the Aid of Cellulase, Characterization, and Solubility. J. Appl. Polym. Sci. 2006, 102, 1098–1105. [Google Scholar]
- Kumar, A.B.V.; Tharanathan, R.N. A comparative study on depolymerization of chitosan by proteolytic enzymes. Carbohydr. Polym. 2004, 58, 275–283. [Google Scholar]
- Sun, Y.Y.; Zhang, J.Q.; Wu, S.J.; Wang, S.J. Preparation of d-glucosamine by hydrolysis of chitosan with chitosanase and β-D-glucosaminidase. Int. J. Biol. Macromol. 2013, 61, 160–163. [Google Scholar] [CrossRef] [PubMed]
- Horn, S.J.; Sørbotten, A.; Synstad, B.; Sikorski, P.; Sørlie, M.; Vårum, K.M.; Eijsink, V.G. Endo/exo mechanism and processivity of family 18 chitinases produced by Serratia marcescens. FEBS J. 2006, 273, 491–503. [Google Scholar] [CrossRef] [PubMed]
- Andreeva, N.S.; Rumsh, L.D. Analysis of crystal structures of aspartic proteinases: On the role of amino acid residues adjacent to the catalytic site of pepsin-like enzymes. Protein Sci. 2001, 10, 2439–2450. [Google Scholar] [CrossRef] [PubMed]

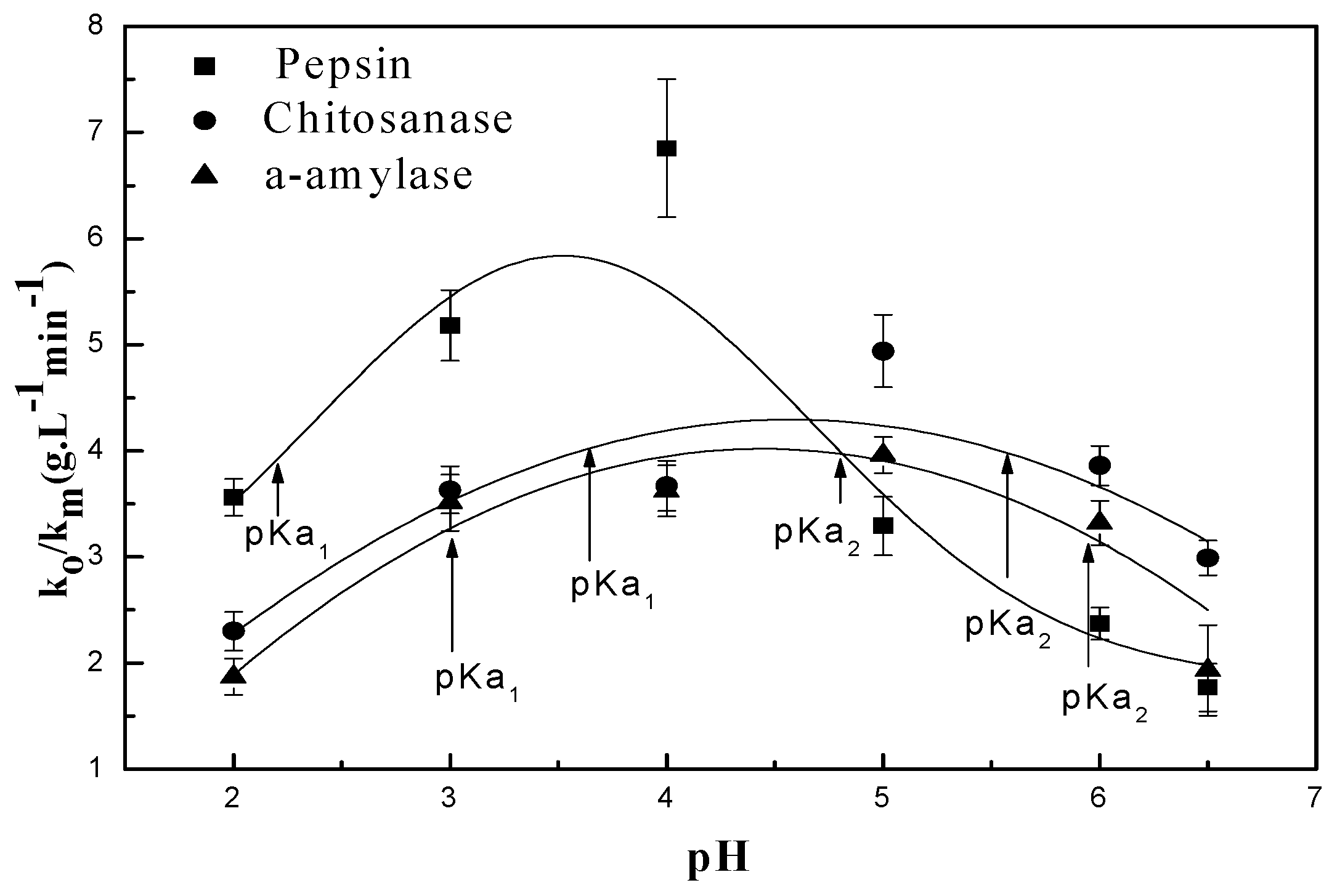
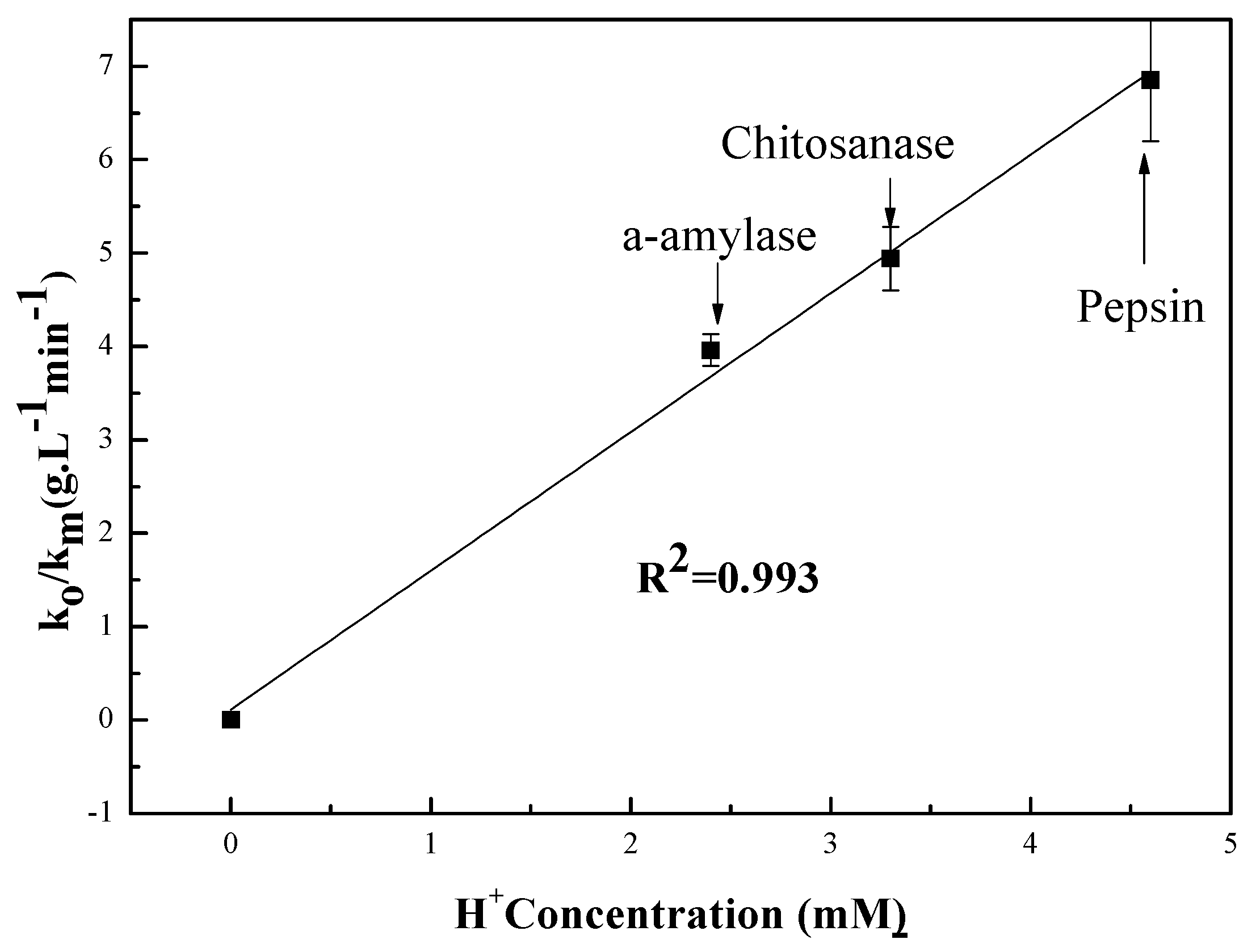

| Enzymes | Mw (kDa) | (gL−1 min−1) | pKa | ||
|---|---|---|---|---|---|
| 1 | 2 | 3 | |||
| α-amylase | 55 | 3.96 | 3 | 6 | - |
| Chitosanase | 47 | 4.94 | 3.59 | 5.54 | - |
| Pepsin | 34.64 | 6.85 | 2.22 | 4.81 | - |
| Enzyme | A/min−1 | Ea/kJ·mol−1 | R2 |
|---|---|---|---|
| Pepsin | 5.8 × 107 | 15.028 | 0.986 |
| Chitosanase | 1.02 × 105 | 12.822 | 0.982 |
| α-amylase | 66.282 | 5.439 | 0.956 |
| Pepsin | |||||
| Ratio Sub-Enzyme | First-Order Qt = Qe(1 − exp( − k1t)) * | Second-Order t/Qt = 1/(k2Qe2) + t/Qe * | |||
| k1 (1/min) | R2 | k2 (L/(g·min)) | h (g/(L·min)) | R2 | |
| 0.5% | 0.030 | 0.999 | 0.00488 | 4.168 | 0.992 |
| 0.8% | 0.039 | 0.995 | 0.00484 | 6.943 | 0.996 |
| 1% | 0.036 | 0.995 | 0.00483 | 10.840 | 0.999 |
| 1.2% | 0.035 | 0.996 | 0.00487 | 9.226 | 0.998 |
| 1.5% | 0.021 | 0.996 | 0.00486 | 9.163 | 0.998 |
| Chitosanase | |||||
| Enzyme Concentration | First-Order Qt = Qe(1 − exp( − k1t)) * | Second-Order t/Qt = 1/(k2Qe2) + t/Qe * | |||
| k1 (1/min) | R2 | k2 (L/(g·min)) | H (g/(L·min)) | R2 | |
| 1 U | 0.022 | 0.964 | 0.0028 | 7.841 | 0.961 |
| 3 U | 0.042 | 0.994 | 0.00247 | 9.157 | 0.966 |
| 5 U | 0.042 | 0.972 | 0.00508 | 11.282 | 0.999 |
| 7 U | 0.028 | 0.992 | 0.00913 | 6.585 | 0.989 |
| 10 U | 0.022 | 0.964 | 0.00695 | 4.560 | 0.994 |
| α-Amylase | |||||
| Ratio Sub-Enzyme | First-Order Qt = Qe(1 − exp( − k1t)) * | Second-Order t/Qt = 1/(k2Qe2) + t/Qe * | |||
| k1 (1/min) | R2 | k2 (L/(g·min)) | h (g/(L·min)) | R2 | |
| 40 U/g | 0.014 | 0.97851 | 0.00188 | 6.218 | 0.968 |
| 60 U/g | 0.021 | 0.98822 | 0.00137 | 8.078 | 0.976 |
| 80 U/g | 0.026 | 0.96801 | 0.00209 | 9.532 | 0.990 |
| 100 U/g | 0.026 | 0.96857 | 0.00614 | 4.812 | 0.980 |
| 120 U/g | 0.015 | 0.97291 | 0.00826 | 3.400 | 0.972 |
| Source | Mw (×103) | DD (%) | Viscosity Decrease (%) | Yield (%) |
|---|---|---|---|---|
| Native | 300 | - | - | - |
| CH2 | 186–126 | 65.50 a | 55 a | - |
| 78.9 b | 75 b | - | ||
| CH3 | 110–80 | 86.00 a | 77 a | - |
| 90 b | 88 b | - | ||
| COS 2 | 90–85 | - | - | 86.34 a |
| 55.00 b | ||||
| COS 3 | 64–47 | - | - | 77.30 a |
| 42.40 b | ||||
| LMWC 2 | 20–15 | - | - | 13.66 a |
| 41.80 b | ||||
| LMWC 3 | 10–1.2 | - | - | 19.70 a |
| 53.70 b | ||||
| Monomers 2 | 0.004–0.01 | - | - | 1.0 a |
| 2.6 b | ||||
| Monomers 3 | 0.004–0.01 | - | 3.0 a | |
| 4.10 b |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gohi, B.F.C.A.; Zeng, H.-Y.; Pan, A.D.; Han, J.; Yuan, J. pH Dependence of Chitosan Enzymolysis. Polymers 2017, 9, 174. https://doi.org/10.3390/polym9050174
Gohi BFCA, Zeng H-Y, Pan AD, Han J, Yuan J. pH Dependence of Chitosan Enzymolysis. Polymers. 2017; 9(5):174. https://doi.org/10.3390/polym9050174
Chicago/Turabian StyleGohi, Bi Foua Claude Alain, Hong-Yan Zeng, A Dan Pan, Jing Han, and Jian Yuan. 2017. "pH Dependence of Chitosan Enzymolysis" Polymers 9, no. 5: 174. https://doi.org/10.3390/polym9050174