Synthesis and Nanoprecipitation of HEMA-CLn Based Polymers for the Production of Biodegradable Nanoparticles
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
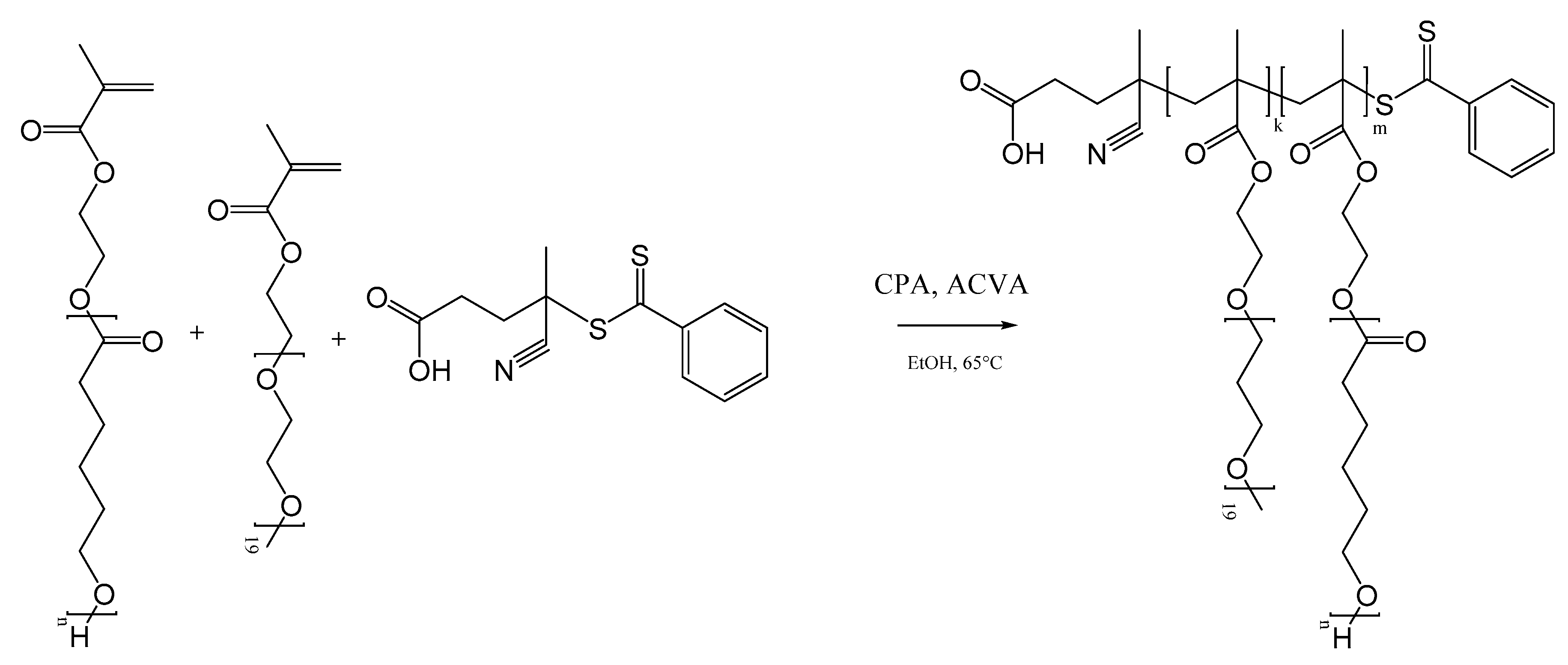
2.2. Polymer Synthesis
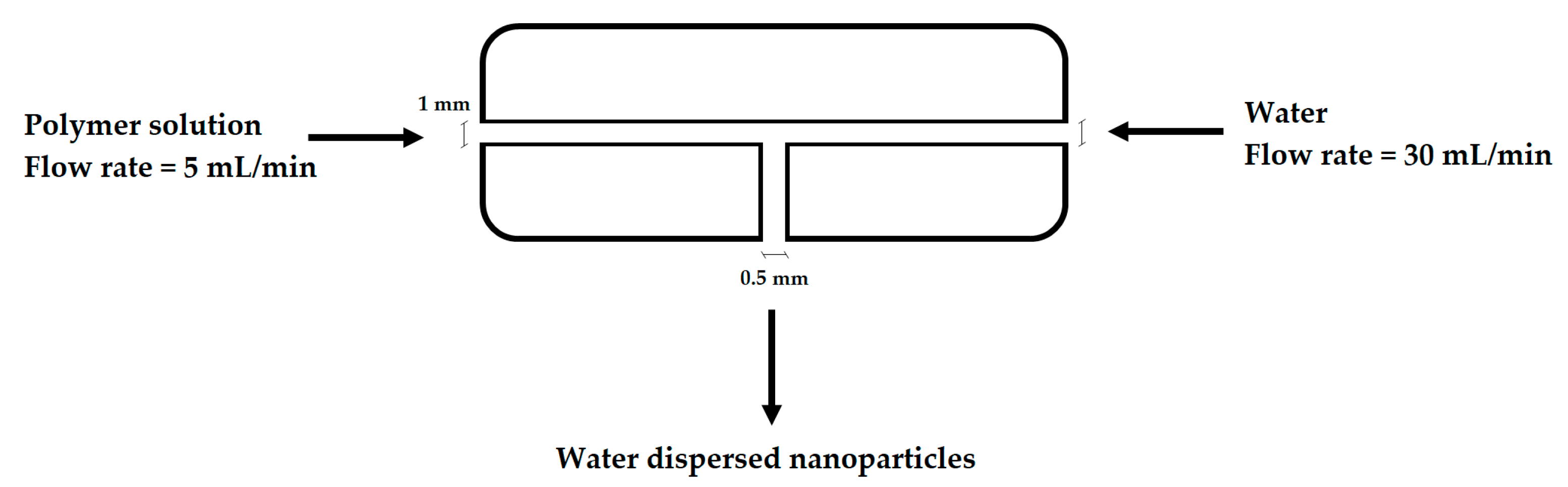
2.3. Nanoparticle Production, Stability and Degradation Studies
3. Results
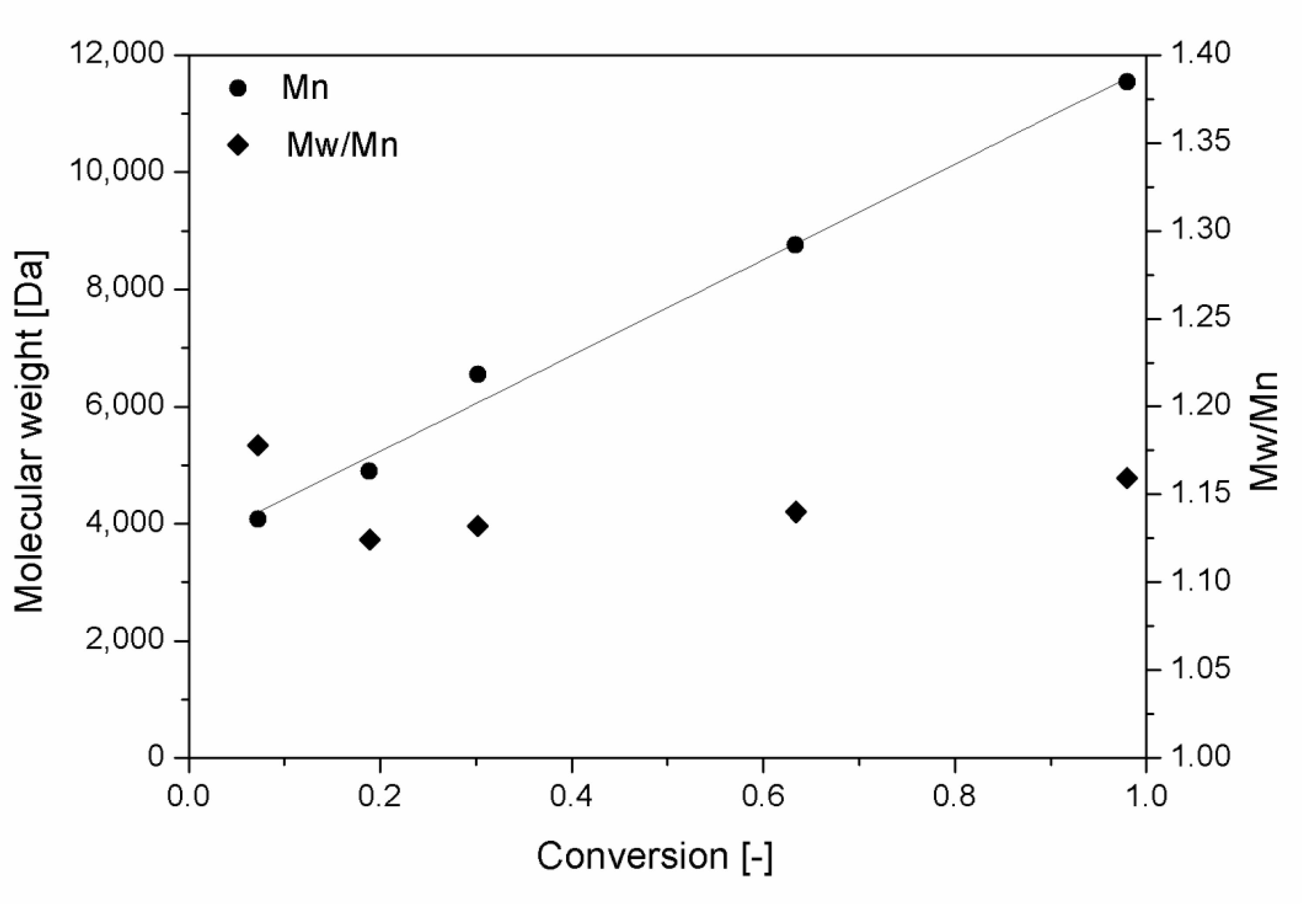
3.1. Polymer Production and Characterization
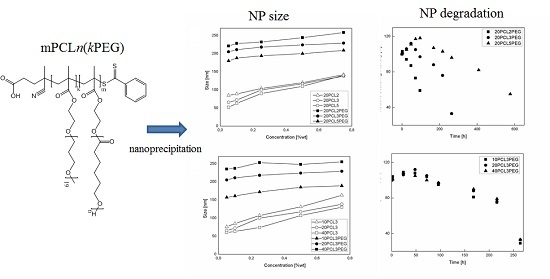
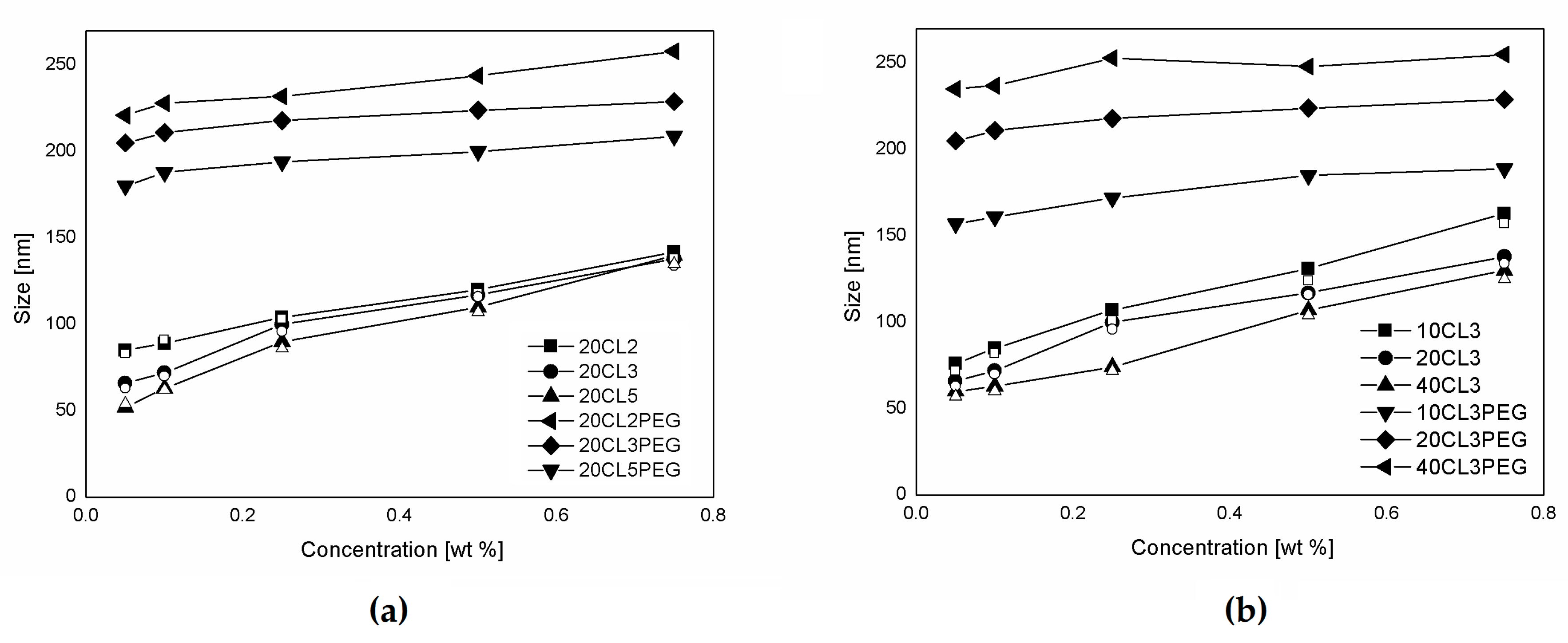
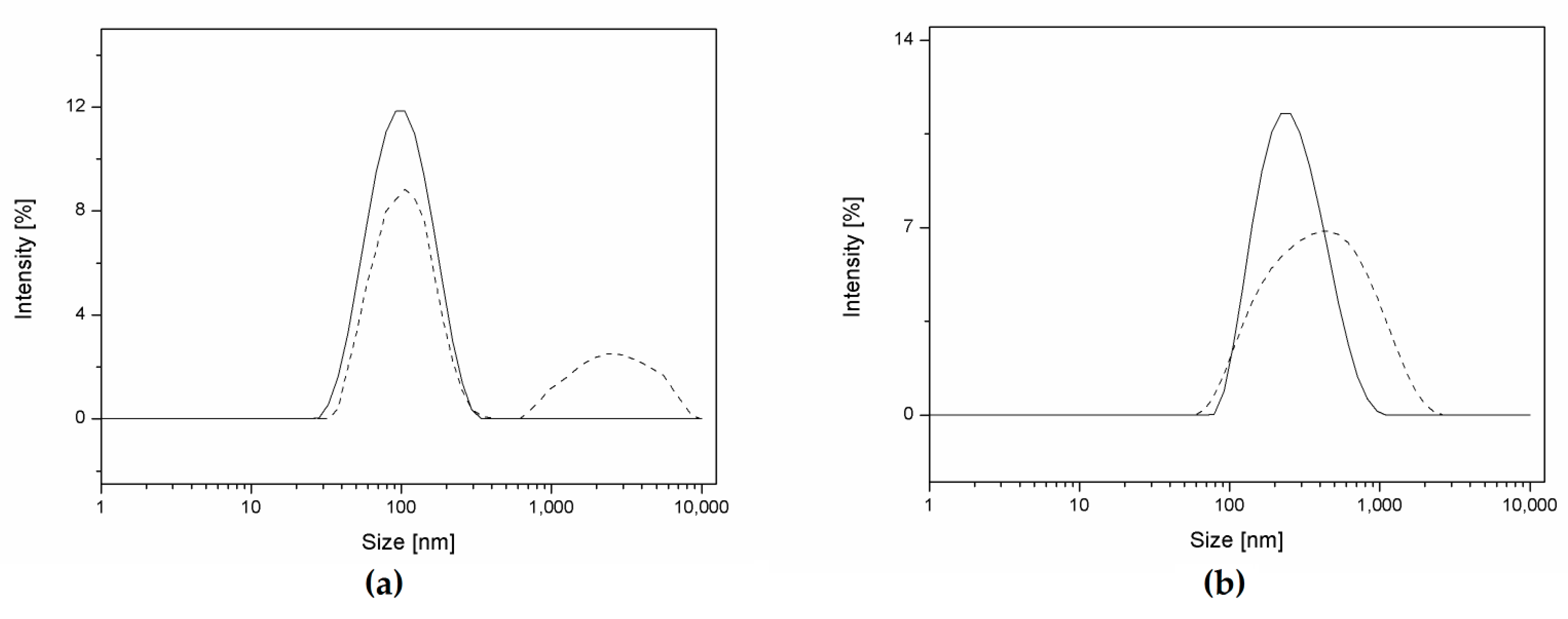
3.2. NP Production and Characterization
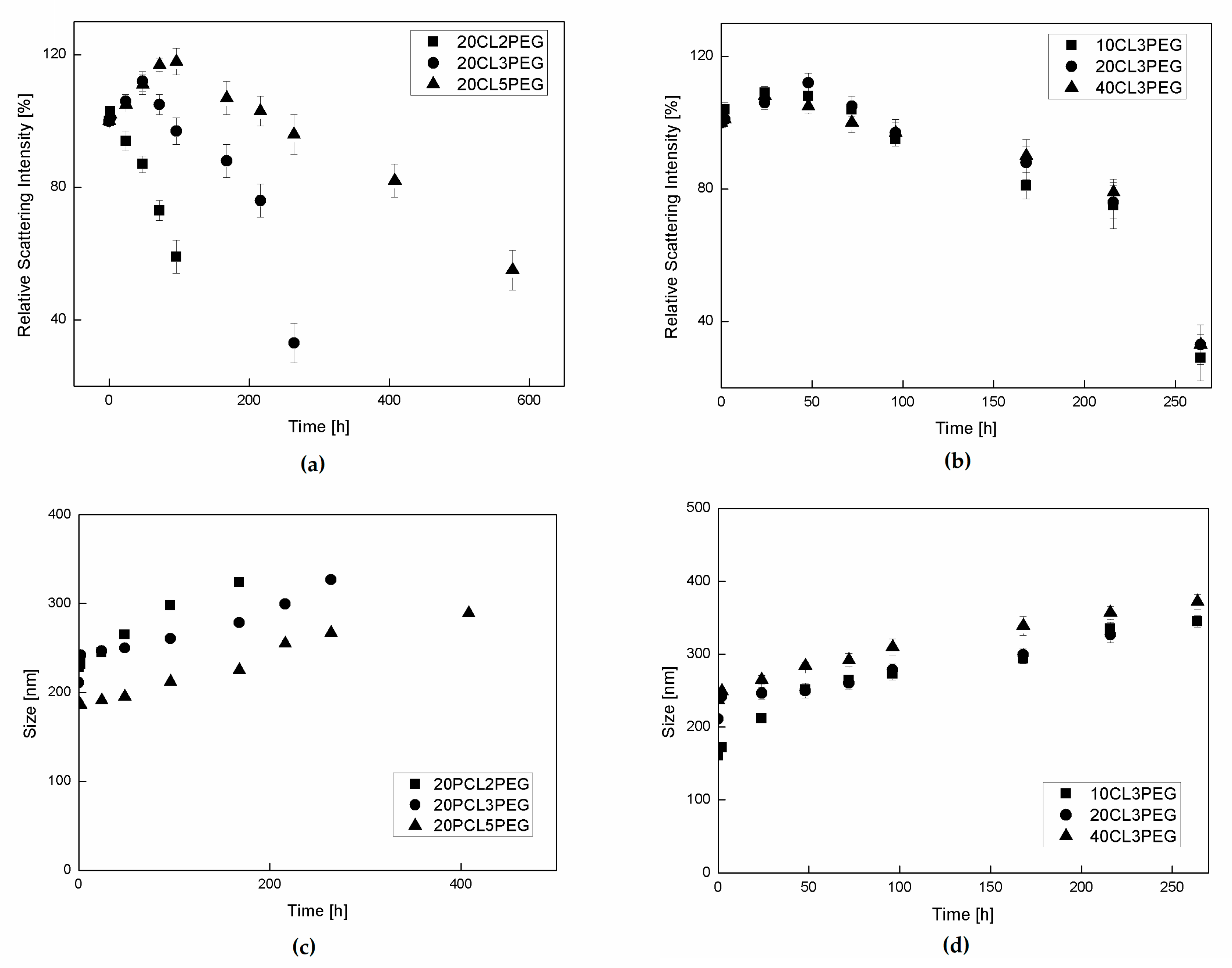
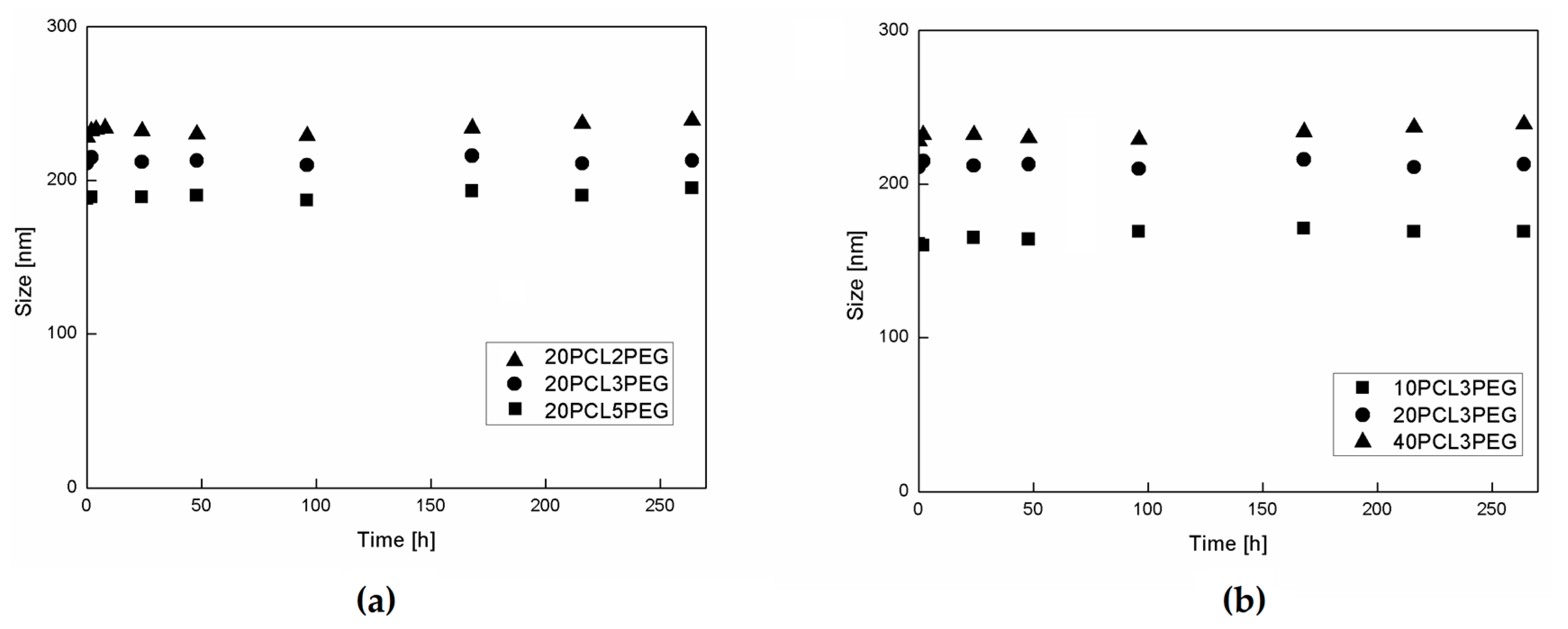
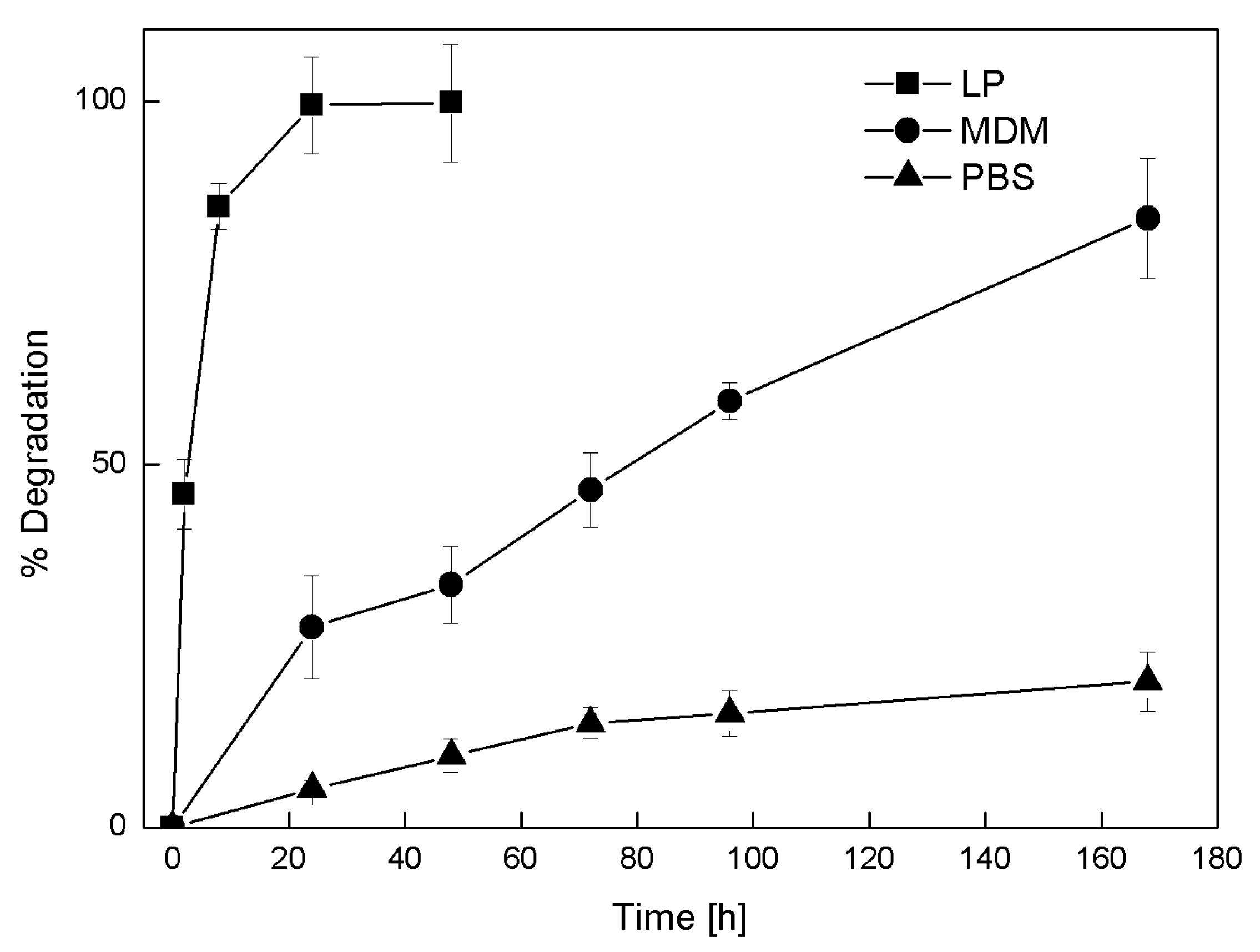
3.3. NP Stability and Degradation Behavior
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kumari, A.; Yadav, S.K.; Yadav, S.C. Biodegradable polymeric nanoparticles based drug delivery systems. Colloid Surf. B 2010, 75, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Votruba, A.R.; Farokhzad, O.C.; Langer, R. Nanotechnology in drug delivery and tissue engineering: From discovery to applications. Nano Lett. 2010, 10, 3223–3230. [Google Scholar] [CrossRef] [PubMed]
- Couvreur, P. Nanoparticles in drug delivery: Past, present and future. Adv. Drug Deliv. Rev. 2013, 65, 21–23. [Google Scholar] [CrossRef] [PubMed]
- Musumeci, T.; Ventura, C.A.; Giannone, I.; Ruozi, B.; Montenegro, L.; Pignatello, R.; Puglisi, G. Pla/plga nanoparticles for sustained release of docetaxel. Int. J. Pharm. 2006, 325, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Kedar, U.; Phutane, P.; Shidhaye, S.; Kadam, V. Advances in polymeric micelles for drug delivery and tumor targeting. Nanomed. Nanotechnol. Biol. Med. 2010, 6, 714–729. [Google Scholar] [CrossRef] [PubMed]
- Elsabahy, M.; Wooley, K.L. Design of polymeric nanoparticles for biomedical delivery applications. Chem. Soc. Rev. 2012, 41, 2545–2561. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, J.; Mura, S.; Brambilla, D.; Mackiewicz, N.; Couvreur, P. Design, functionalization strategies and biomedical applications of targeted biodegradable/biocompatible polymer-based nanocarriers for drug delivery. Chem. Soc. Rev. 2013, 42, 1147–1235. [Google Scholar] [CrossRef] [PubMed]
- Braunecker, W.A.; Matyjaszewski, K. Controlled/living radical polymerization: Features, developments, and perspectives. Prog. Polym. Sci. 2007, 32, 93–146. [Google Scholar] [CrossRef]
- Rocha, N.; Mendonça, P.; Góis, J.R.; Cordeiro, R.; Fonseca, A.; Ferreira, P.; Guliashvili, T.; Matyjaszewski, K.; Serra, A.; Coelho, J. The importance of controlled/living radical polymerization techniques in the design of tailor made nanoparticles for drug delivery systems. In Drug Delivery Systems: Advanced Technologies Potentially Applicable in Personalised Treatment; Springer: Dordrecht, The Netherlands, 2013; pp. 315–357. [Google Scholar]
- Georges, M.K.; Veregin, R.P.N.; Kazmaier, P.M.; Hamer, G.K. Narrow molecular-weight resins by a free-radical polymerization process. Macromolecules 1993, 26, 2987–2988. [Google Scholar] [CrossRef]
- Harrisson, S.; Nicolas, J.; Maksimenko, A.; Bui, D.T.; Mougin, J.; Couvreur, P. Nanoparticles with in vivo anticancer activity from polymer prodrug amphiphiles prepared by living radical polymerization. Angew. Chem. Int. Ed. 2013, 52, 1678–1682. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.S.; Matyjaszewski, K. Controlled living radical polymerization-atom-transfer radical polymerization in the presence of transition-metal complexes. J. Am. Chem. Soc. 1995, 117, 5614–5615. [Google Scholar] [CrossRef]
- Simakova, A.; Mackenzie, M.; Averick, S.E.; Park, S.; Matyjaszewski, K. Bioinspired iron-based catalyst for atom transfer radical polymerization. Angew. Chem. Int. Ed. 2013, 52, 12148–12151. [Google Scholar] [CrossRef] [PubMed]
- Chiefari, J.; Chong, Y.K.; Ercole, F.; Krstina, J.; Jeffery, J.; Le, T.P.T.; Mayadunne, R.T.A.; Meijs, G.F.; Moad, C.L.; Moad, G.; et al. Living free-radical polymerization by reversible addition-fragmentation chain transfer: The raft process. Macromolecules 1998, 31, 5559–5562. [Google Scholar] [CrossRef]
- Moad, G.; Rizzardo, E.; Thang, S.H. Raft polymerization and some of its applications. Chem. Asian J. 2013, 8, 1634–1644. [Google Scholar] [CrossRef] [PubMed]
- De Rybel, N.; Van Steenberge, P.H.; Reyniers, M.F.; Barner-Kowollik, C.; D’hooge, D.R.; Marin, G. An update on the pivotal role of kinetic modeling for the mechanistic understanding and design of bulk and solution raft polymerization. Macromol. Theory Simul. 2017, 26. [Google Scholar] [CrossRef]
- Boyer, C.; Bulmus, V.; Davis, T.P.; Ladmiral, V.; Liu, J.; Perrier, S.B. Bioapplications of raft polymerization. Chem. Rev. 2009, 109, 5402–5436. [Google Scholar] [CrossRef] [PubMed]
- Barner-Kowollik, C.; Perrier, S. The future of reversible addition fragmentation chain transfer polymerization. J. Polym. Sci. Polym. Chem. 2008, 46, 5715–5723. [Google Scholar] [CrossRef]
- Ferrari, R.; Yu, Y.C.; Morbidelli, M.; Hutchinson, R.A.; Moscatelli, D. Epsilon-caprolactone-based macromonomers suitable for biodegradable nanoparticles synthesis through free radical polymerization. Macromolecules 2011, 44, 9205–9212. [Google Scholar] [CrossRef]
- Ferrari, R.; Rooney, T.R.; Lupi, M.; Ubezio, P.; Hutchinson, R.A.; Moscatelli, D. A methyl methacrylate–hema-cln copolymerization investigation: From kinetics to bioapplications. Macromol. Biosci. 2013, 13, 1347–1357. [Google Scholar] [CrossRef] [PubMed]
- Storey, R.F.; Sherman, J.W. Kinetics and mechanism of the stannous octoate-catalyzed bulk polymerization of epsilon-caprolactone. Macromolecules 2002, 35, 1504–1512. [Google Scholar] [CrossRef]
- Nuyken, O.; Pask, S.D. Ring-opening polymerization—An introductory review. Polymers 2013, 5, 361–403. [Google Scholar] [CrossRef]
- Yu, Y.; Ferrari, R.; Lattuada, M.; Storti, G.; Morbidelli, M.; Moscatelli, D. Pla-based nanoparticles with tunable hydrophobicity and degradation kinetics. J. Polym. Sci. A 2012, 50, 5191–5200. [Google Scholar] [CrossRef]
- Ferrari, R.; Colombo, C.; Dossi, M.; Moscatelli, D. Tunable plga-based nanoparticles synthesized through free-radical polymerization. Macromol. Mater. Eng. 2013, 298, 730–739. [Google Scholar] [CrossRef]
- Ferrari, R.; Yu, Y.; Lattuada, M.; Storti, G.; Morbidelli, M.; Moscatelli, D. Controlled pegylation of pla-based nanoparticles. Macromol. Chem. Phys. 2012, 213, 2012–2018. [Google Scholar] [CrossRef]
- Papa, S.; Rossi, F.; Ferrari, R.; Mariani, A.; De Paola, M.; Caron, I.; Fiordaliso, F.; Bisighini, C.; Sammali, E.; Colombo, C. Selective nanovector mediated treatment of activated proinflammatory microglia/macrophages in spinal cord injury. ACS Nano 2013, 7, 9881–9895. [Google Scholar] [CrossRef] [PubMed]
- Colombo, C.; Morosi, L.; Bello, E.; Ferrari, R.; Licandro, S.A.; Lupi, M.; Ubezio, P.; Morbidelli, M.; Zucchetti, M.; D’Incalci, M. Pegylated nanoparticles obtained through emulsion polymerization as paclitaxel carriers. Mol. Pharm. 2015, 13, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Colombo, C.; Dragoni, L.; Gatti, S.; Pesce, R.M.; Rooney, T.R.; Mavroudakis, E.; Ferrari, R.; Moscatelli, D. Tunable degradation behavior of pegylated polyester-based nanoparticles obtained through emulsion free radical polymerization. Ind. Eng. Chem. Res. 2014, 53, 9128–9135. [Google Scholar] [CrossRef]
- Moad, G.; Chong, Y.; Postma, A.; Rizzardo, E.; Thang, S.H. Advances in raft polymerization: The synthesis of polymers with defined end-groups. Polymer 2005, 46, 8458–8468. [Google Scholar] [CrossRef]
- York, A.W.; Kirkland, S.E.; McCormick, C.L. Advances in the synthesis of amphiphilic block copolymers via raft polymerization: Stimuli-responsive drug and gene delivery. Adv. Drug Deliv. Rev. 2008, 60, 1018–1036. [Google Scholar] [CrossRef] [PubMed]
- Colombo, C.; Gatti, S.; Ferrari, R.; Casalini, T.; Cuccato, D.; Morosi, L.; Zucchetti, M.; Moscatelli, D. Self-assembling amphiphilic pegylated block copolymers obtained through raft polymerization for drug-delivery applications. J. Appl. Polym. Sci. 2015, 133. [Google Scholar] [CrossRef]
- Xia, Y.; Tang, S.; Olsen, B.D. Site-specific conjugation of raft polymers to proteins via expressed protein ligation. Chem. Commun. 2013, 49, 2566–2568. [Google Scholar] [CrossRef] [PubMed]
- Perrier, S.; Barner-Kowollik, C.; Quinn, J.F.; Vana, P.; Davis, T.P. Origin of inhibition effects in the reversible addition fragmentation chain transfer (RAFT) polymerization of methyl acrylate. Macromolecules 2002, 35, 8300–8306. [Google Scholar] [CrossRef]
- Ferrari, R.; Colombo, C.; Casali, C.; Lupi, M.; Ubezio, P.; Falcetta, F.; D’Incalci, M.; Morbidelli, M.; Moscatelli, D. Synthesis of surfactant free PCL–PEG brushed nanoparticles with tunable degradation kinetics. Int. J. Pharm. 2013, 453, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Azzi, J.; Tang, L.; Moore, R.; Tong, R.; El Haddad, N.; Akiyoshi, T.; Mfarrej, B.; Yang, S.; Jurewicz, M.; Ichimura, T. Polylactide-cyclosporin a nanoparticles for targeted immunosuppression. FASEB J. 2010, 24, 3927–3938. [Google Scholar] [CrossRef] [PubMed]
- Valente, I.; Celasco, E.; Marchisio, D.; Barresi, A. Nanoprecipitation in confined impinging jets mixers: Production, characterization and scale-up of pegylated nanospheres and nanocapsules for pharmaceutical use. Chem. Eng. Sci. 2012, 77, 217–227. [Google Scholar] [CrossRef]
- Palmiero, U.C.; Agostini, A.; Gatti, S.; Sponchioni, M.; Valenti, V.; Brunel, L.; Moscatelli, D. Raft macro-surfmers and their use in the ab initio raft emulsion polymerization to decouple nanoparticle size and polymer molecular weight. Macromolecules 2016, 49, 8387–8396. [Google Scholar] [CrossRef]
- Palmiero, U.C.; Chovancová, A.; Cuccato, D.; Storti, G.; Lacík, I.; Moscatelli, D. The raft copolymerization of acrylic acid and acrylamide. Polymer 2016, 98, 156–164. [Google Scholar] [CrossRef]
- Mehan, S.; Chinchalikar, A.J.; Kumar, S.; Aswal, V.K.; Schweins, R. Small-angle neutron scattering study of structure and interaction of nanoparticle, protein, and surfactant complexes. Langmuir 2013, 29, 11290–11299. [Google Scholar] [CrossRef] [PubMed]
- Colombo, C.; Li, M.; Watanabe, S.; Messa, P.; Edefonti, A.; Montini, G.; Moscatelli, D.; Rastaldi, M.P.; Cellesi, F. Polymer nanoparticle engineering for podocyte repair: From in vitro models to new nanotherapeutics in kidney diseases. ACS Omega 2017, 2, 599–610. [Google Scholar] [CrossRef]
- Cretu, A.; Gattin, R.; Brachais, L.; Barbier-Baudry, D. Synthesis and degradation of poly (2-hydroxyethyl methacrylate)-graft-poly (ε-caprolactone) copolymers. Polym. Degrad. Stab. 2004, 83, 399–404. [Google Scholar] [CrossRef]
- Lee, H.J.; Bae, Y. Brushed block copolymer micelles with pH-sensitive pendant groups for controlled drug delivery. Pharma. Res. 2013, 30, 2077–2086. [Google Scholar] [CrossRef] [PubMed]

| Sample Name | m | n | k | χ | Mn th [Da] | Mn GPC [Da] | Mw/Mn |
|---|---|---|---|---|---|---|---|
| 20CL2 | 20 | 2 | - | 0.956 | 7440 | 6862 | 1.14 |
| 10CL3 | 10 | 3 | - | 0.921 | 5000 | 7453 | 1.09 |
| 20CL3 | 20 | 3 | - | 0.983 | 9720 | 11,539 | 1.16 |
| 40CL3 | 40 | 3 | - | 0.952 | 19,160 | 18,641 | 1.12 |
| 20CL5 | 20 | 5 | - | 0.967 | 14,280 | 15,549 | 1.14 |
| 20CL2PEG | 20 | 2 | 2 | 0.981 | 9440 | 8810 | 1.11 |
| 10CL3PEG | 10 | 3 | 1 | 0.965 | 6000 | 7511 | 1.08 |
| 20CL3PEG | 20 | 3 | 2 | 0.977 | 11,720 | 12,301 | 1.1 |
| 40CL3PEG | 40 | 3 | 4 | 0.96 | 23,160 | 20,509 | 1.13 |
| 20CL5PEG | 20 | 5 | 2 | 0.941 | 16,280 | 16527 | 1.09 |
| Polymer Sample | NP Size [nm] | PDI |
|---|---|---|
| 10CL3 | 85 | 0.114 |
| 20CL2 | 89 | 0.121 |
| 20CL3 | 72 | 0.106 |
| 20CL5 | 63 | 0.073 |
| 40CL3 | 63 | 0.090 |
| 20CL2PEG | 228 | 0.142 |
| 10CL3PEG | 161 | 0.101 |
| 20CL3PEG | 211 | 0.122 |
| 40CL3PEG | 237 | 0.145 |
| 20CL5PEG | 188 | 0.089 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gatti, S.; Agostini, A.; Ferrari, R.; Moscatelli, D. Synthesis and Nanoprecipitation of HEMA-CLn Based Polymers for the Production of Biodegradable Nanoparticles. Polymers 2017, 9, 389. https://doi.org/10.3390/polym9090389
Gatti S, Agostini A, Ferrari R, Moscatelli D. Synthesis and Nanoprecipitation of HEMA-CLn Based Polymers for the Production of Biodegradable Nanoparticles. Polymers. 2017; 9(9):389. https://doi.org/10.3390/polym9090389
Chicago/Turabian StyleGatti, Simone, Azzurra Agostini, Raffaele Ferrari, and Davide Moscatelli. 2017. "Synthesis and Nanoprecipitation of HEMA-CLn Based Polymers for the Production of Biodegradable Nanoparticles" Polymers 9, no. 9: 389. https://doi.org/10.3390/polym9090389