Optimization of 3D Surfaces of Dextran with Different Molecule Weights for Real-Time Detection of Biomolecular Interactions by a QCM Biosensor
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
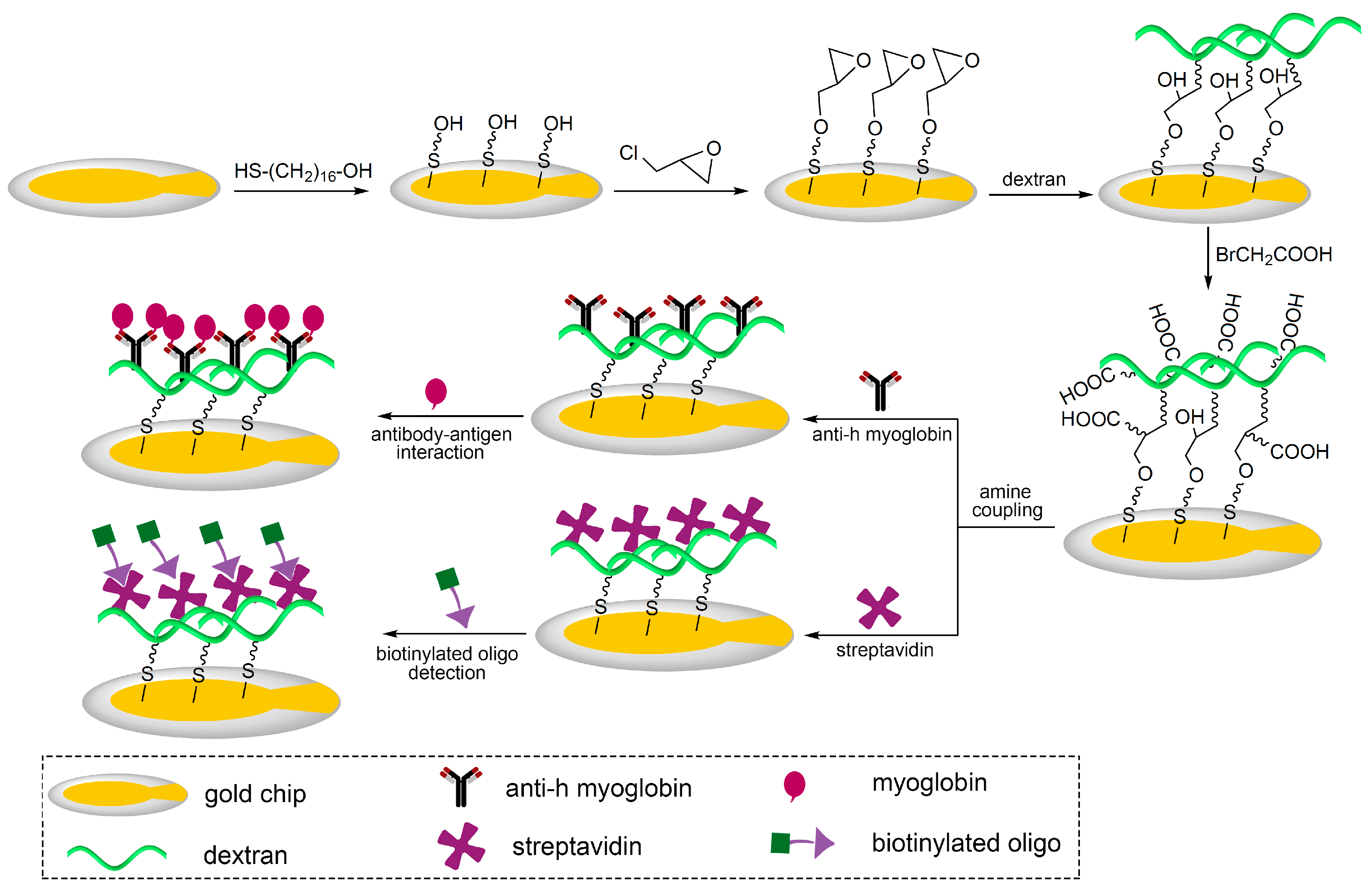
2.2. Fabrication of the 3D Carboxymethyl Dextran Sensor Chip with Different Molecular Weight Dextran
2.3. Immobilization of Protein on the Sensor Surface
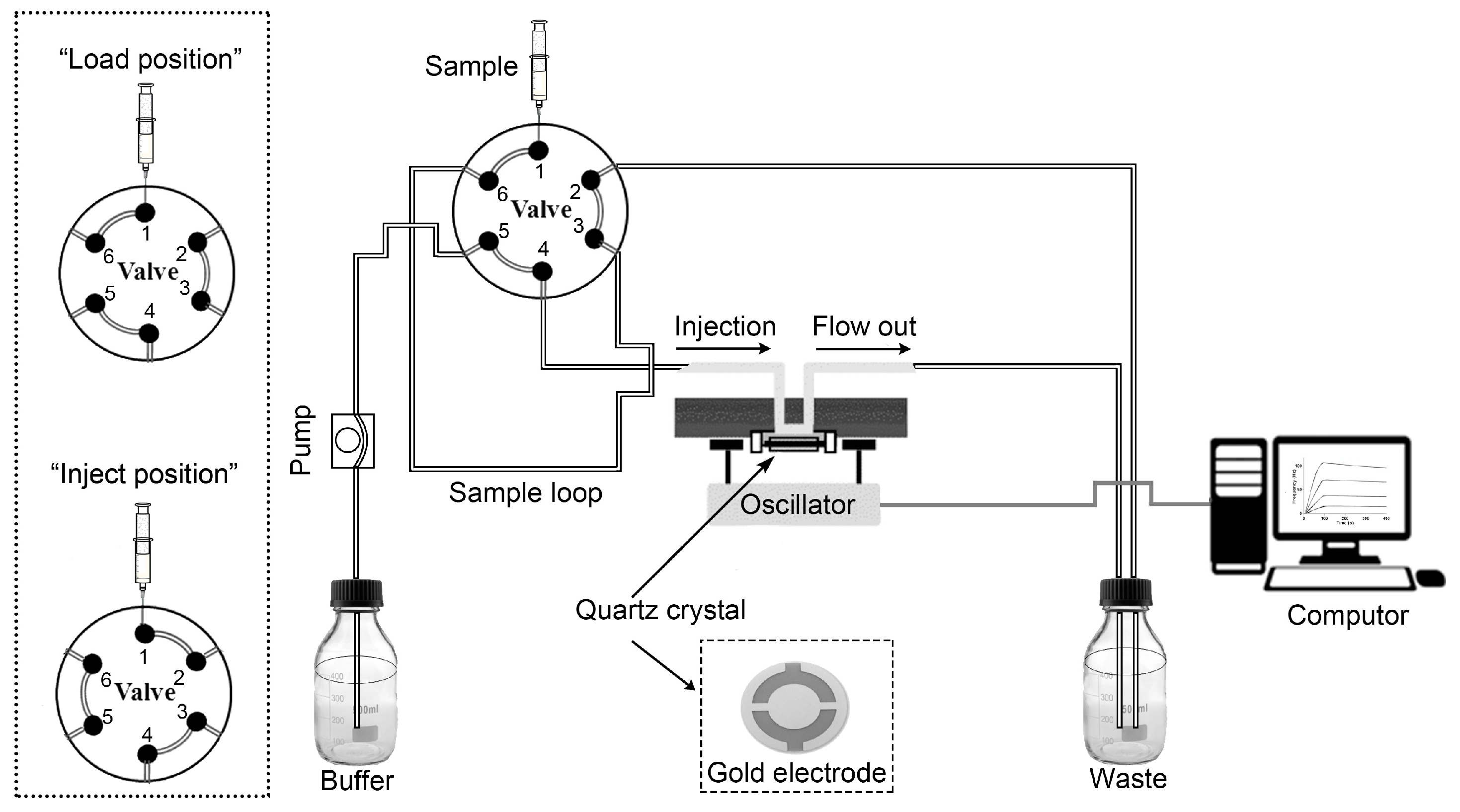
2.4. Protein–Protein Interactions and Kinetic Characterization with QCM
2.5. Detection of Low Molecular Weight Oligo by QCM
3. Results and Discussion
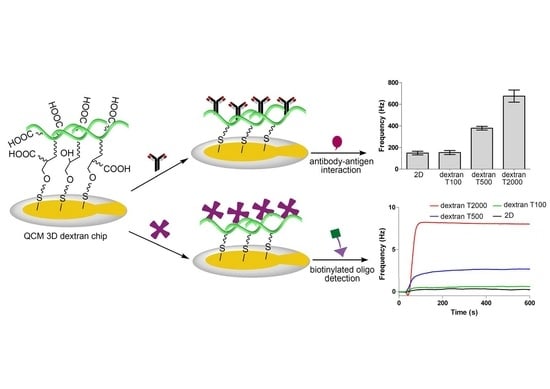
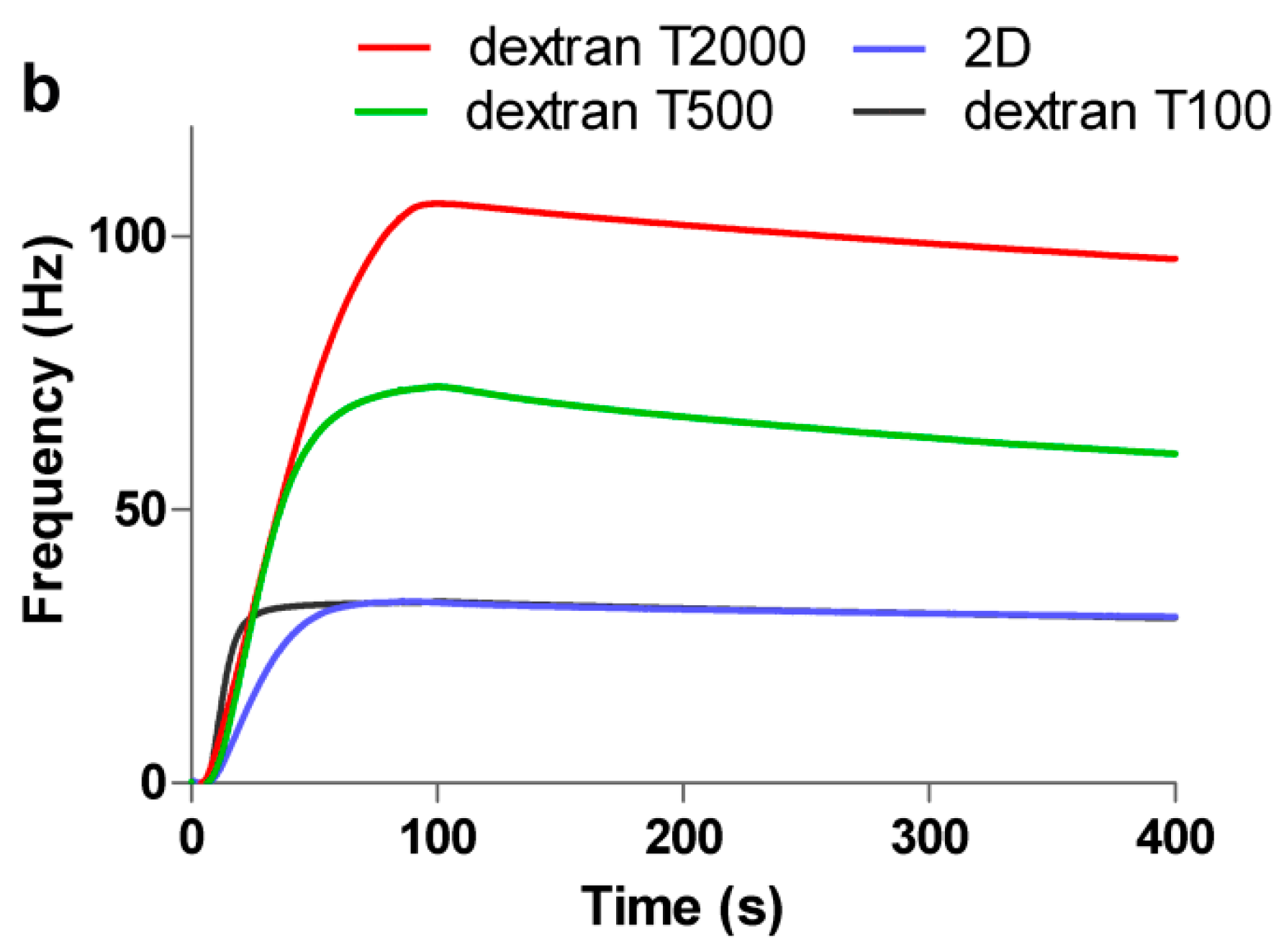
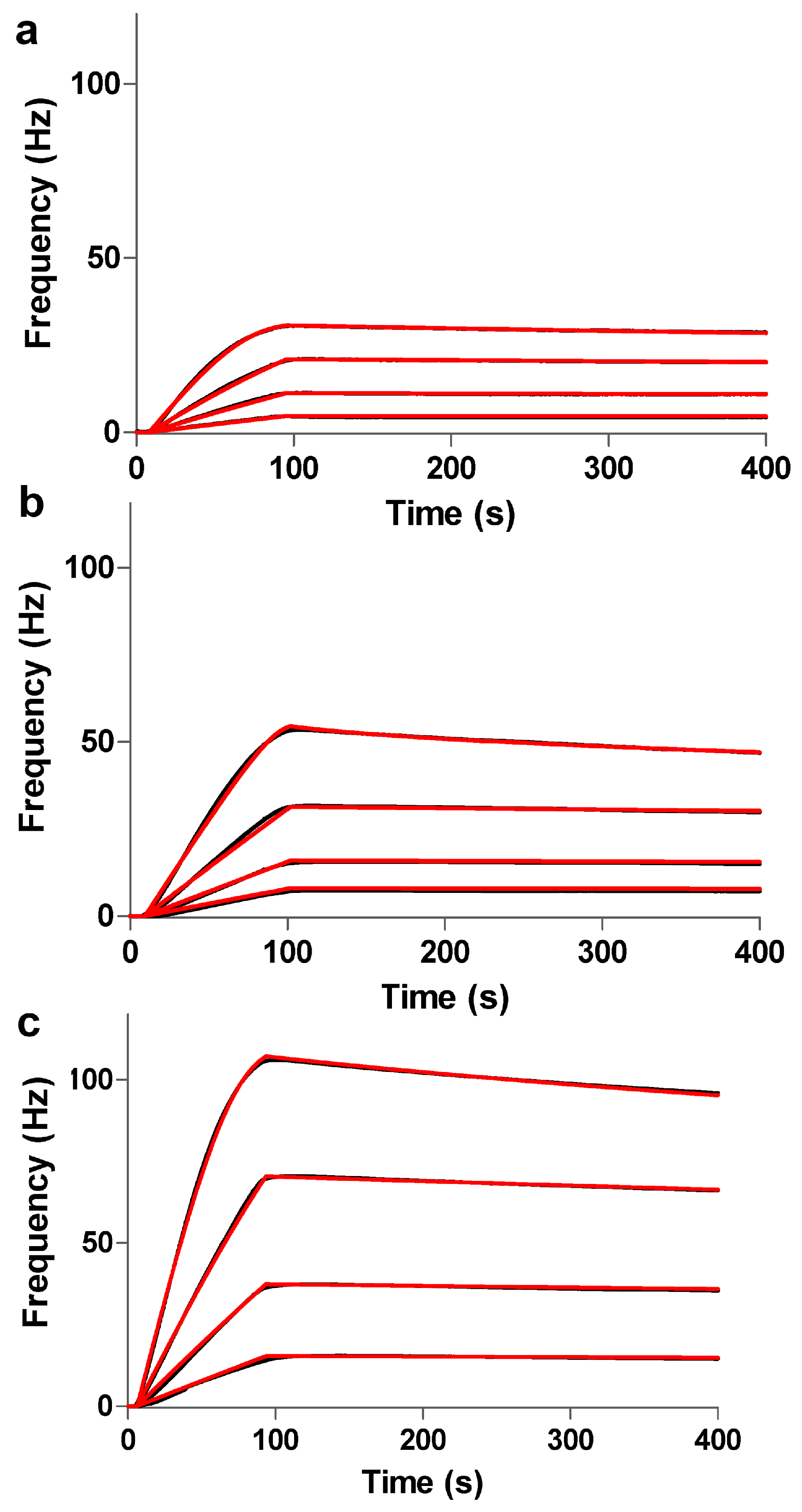
3.1. Comparison of Immobilization and Binding Capacity on 2D and 3D Surfaces
3.2. Interaction and Kinetic Studies
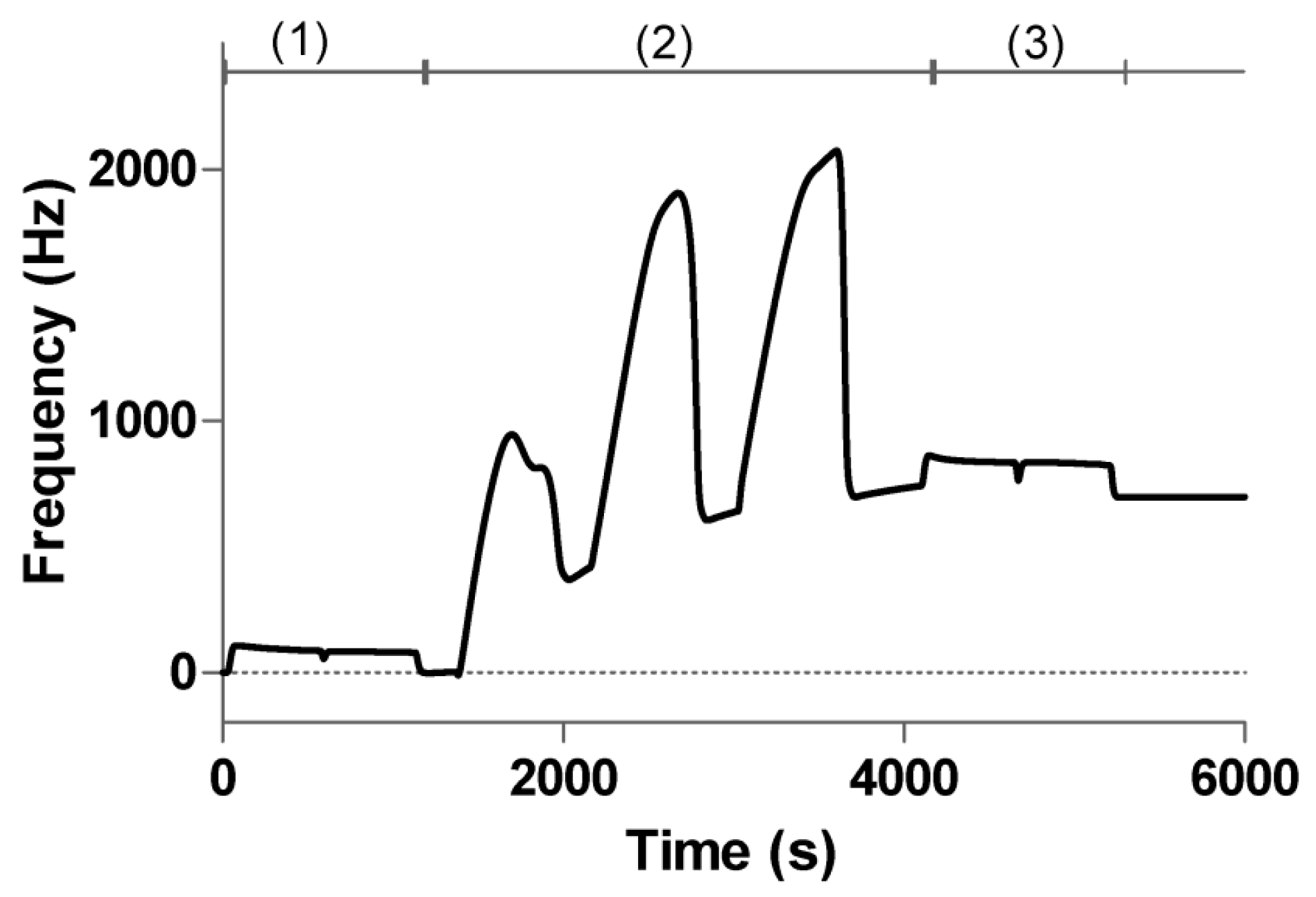
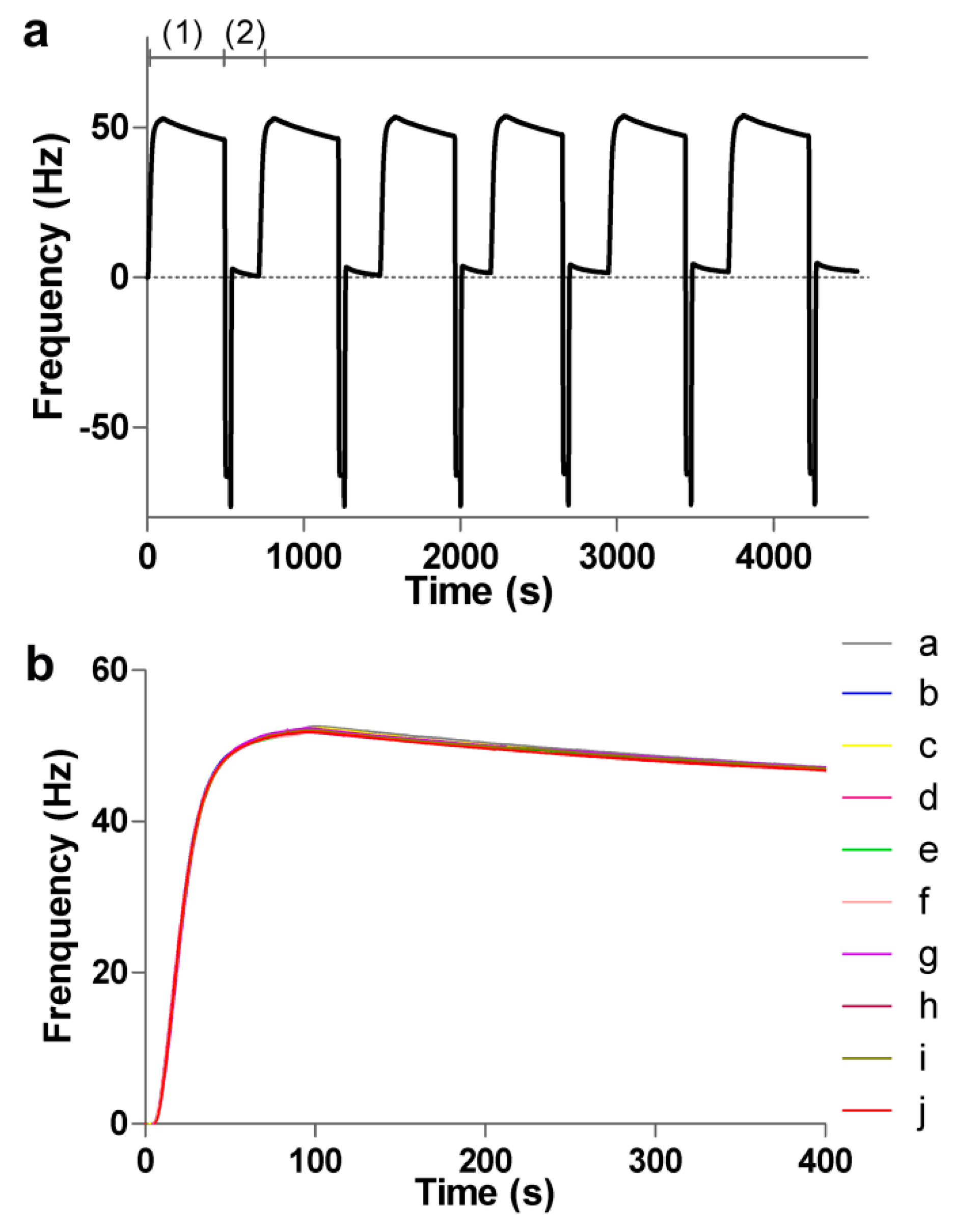
3.3. Regeneration and Stability of the Sensing Surface
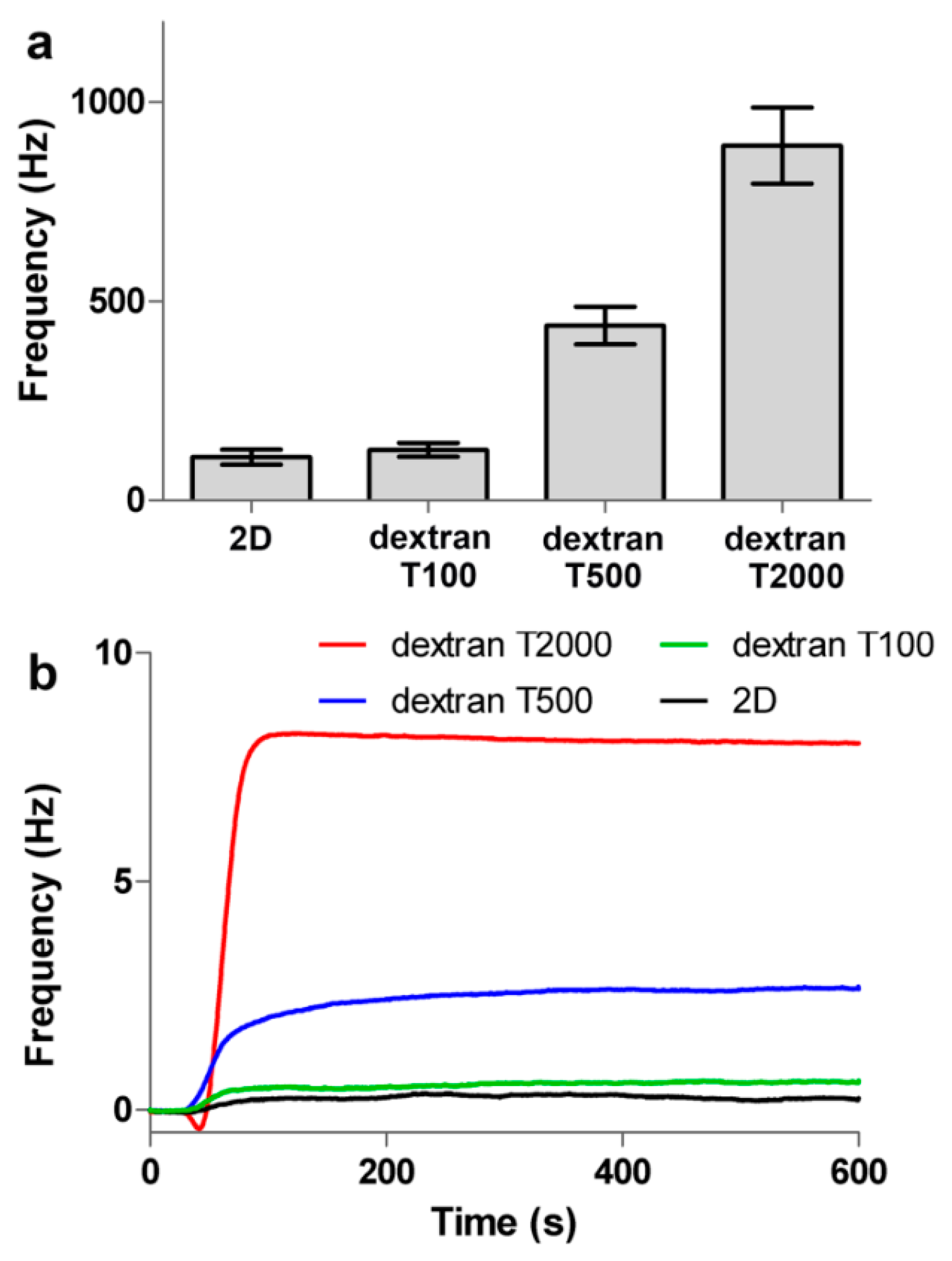
3.4. Evaluating Sensitivity of the Surface
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Takahashi, H.; Nakanishi, T.; Kami, K.; Arata, Y.; Shimada, I. A novel nmr method for determining the interfaces of large protein-protein complexes. Nat. Struct. Biol. 2000, 7, 220–223. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, K.; Wagner, G. Nmr studies of protein interactions. Curr. Opin. Struct. Biol. 2006, 16, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Carlomagno, T.; Blommers, M.J.J.; Meiler, J.; Jahnke, W.; Schupp, T.; Petersen, F.; Schinzer, D.; Altmann, K.H.; Griesinger, C. The high-resolution solution structure of epothilone a bound to tubulin: An understanding of the structure-activity relationships for a powerful class of antitumor agents. Angew. Chem. Int. Ed. 2003, 42, 2511–2515. [Google Scholar] [CrossRef] [PubMed]
- Harada, S.; Hiromori, Y.; Nakamura, S.; Kawahara, K.; Fukakusa, S.; Maruno, T.; Noda, M.; Uchiyama, S.; Fukui, K.; Nishikawa, J.; et al. Structural basis for ppar gamma transactivation by endocrine-disrupting organotin compounds. Sci. Rep. 2015, 5, 8520. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, C.; Boelens, R.; Bonvin, A. Haddock: A protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003, 125, 1731–1737. [Google Scholar] [CrossRef] [PubMed]
- Arroyo-Maya, I.J.; Campos-Teran, J.; Hernandez-Arana, A.; McClements, D.J. Characterization of flavonoid-protein interactions using fluorescence spectroscopy: Binding of pelargonidin to dairy proteins. Food Chem. 2016, 213, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Z.; Zhou, B.; Liu, Y.X.; Zhou, C.X.; Ding, X.L.; Liu, Y. Fluorescence study on the interaction of bovine serum albumin with p-aminoazobenzene. J. Fluoresc. 2008, 18, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Glish, G.L.; Vachet, R.W. The basics of mass spectrometry in the twenty-first century. Nat. Rev. Drug Discov. 2003, 2, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Li, R.L.; Lin, C.W.; Shao, Y.L.; Chang, C.W.; Yao, F.K.; Kowal, M.D.; Wang, H.S.; Yeung, M.T.; Huang, S.C.; Kaner, R.B. Characterization of aniline tetramer by maldi tof mass spectrometry upon oxidative and reductive cycling. Polymers 2016, 8, 401. [Google Scholar] [CrossRef]
- Lee, C.S.; Muthusamy, A.; Abdul-Rahman, P.S.; Bhavanandan, V.P.; Hashim, O.H. An improved lectin-based method for the detection of mucin-type O-glycans in biological samples. Analyst 2013, 138, 3522–3529. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.R.; Liu, R.T.; Qin, P.F.; Wang, L.; Zhao, X.C. Spectroscopic studies on the interaction of acid yellow with bovine serum albumin. J. Lumin. 2010, 130, 611–617. [Google Scholar] [CrossRef]
- Citartan, M.; Gopinath, S.C.B.; Tominaga, J.; Chen, Y.; Tang, T.H. Use of UV–Vis–NIR spectroscopy to monitor label-free interaction between molecular recognition elements and erythropoietin on a gold-coated polycarbonate platform. Talanta 2014, 126, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Christau, S.; Thurandt, S.; Yenice, Z.; von Klitzing, R. Stimuli-responsive polyelectrolyte brushes as a matrix for the attachment of gold nanoparticles: The effect of brush thickness on particle distribution. Polymers 2014, 6, 1877–1896. [Google Scholar] [CrossRef]
- Homola, J. Surface plasmon resonance sensors for detection of chemical and biological species. Chem. Rev. 2008, 108, 462–493. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.A.; Thomas, W.D.; Kiessling, L.L.; Corn, R.M. Surface plasmon resonance imaging studies of protein–carbohydrate interactions. J. Am. Chem. Soc. 2003, 125, 6140–6148. [Google Scholar] [CrossRef] [PubMed]
- Green, R.J.; Frazier, R.A.; Shakesheff, K.M.; Davies, M.C.; Roberts, C.J.; Tendler, S.J.B. Surface plasmon resonance analysis of dynamic biological interactions with biomaterials. Biomaterials 2000, 21, 1823–1835. [Google Scholar] [CrossRef]
- Cooper, M.A.; Singleton, V.T. A survey of the 2001 to 2005 quartz crystal microbalance biosensor literature: Applications of acoustic physics to the analysis of biomolecular interactions. J. Mol. Recognit. 2007, 20, 154–184. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.I.; Chang, Y.P.; Chu, Y.H. Biomolecular interactions and tools for their recognition: Focus on the quartz crystal microbalance and its diverse surface chemistries and applications. Chem. Soc. Rev. 2012, 41, 1947–1971. [Google Scholar] [CrossRef] [PubMed]
- Li, X.M.; Pei, Y.X.; Zhang, R.N.; Shuai, Q.; Wang, F.; Aastrup, T.; Pei, Z.C. A suspension-cell biosensor for real-time determination of binding kinetics of protein-carbohydrate interactions on cancer cell surfaces. Chem. Commun. 2013, 49, 9908–9910. [Google Scholar] [CrossRef] [PubMed]
- Becker, B.; Cooper, M.A. A survey of the 2006–2009 quartz crystal microbalance biosensor literature. J. Mol. Recognit. 2011, 24, 754–787. [Google Scholar] [CrossRef] [PubMed]
- Homola, J.; Yee, S.S.; Gauglitz, G. Surface plasmon resonance sensors: Review. Sens. Actuators B 1999, 54, 3–15. [Google Scholar] [CrossRef]
- Pei, Y.X.; Yu, H.; Pei, Z.C.; Theurer, M.; Ammer, C.; Andre, S.; Gabius, H.J.; Yan, M.D.; Ramstrom, O. Photoderivatized polymer thin films at quartz crystal microbalance surfaces: Sensors for carbohydrate-protein interactions. Anal. Chem. 2007, 79, 6897–6902. [Google Scholar] [CrossRef] [PubMed]
- Kanoh, N.; Kyo, M.; Inamori, K.; Ando, A.; Asami, A.; Nakao, A.; Osada, H. Spr imaging of photo-cross-linked small-molecule arrays on gold. Anal. Chem. 2006, 78, 2226–2230. [Google Scholar] [CrossRef] [PubMed]
- Pei, Z.; Saint-Guirons, J.; Kack, C.; Ingemarsson, B.; Aastrup, T. Real-time analysis of the carbohydrates on cell surfaces using a qcm biosensor: A lectin-based approach. Biosens. Bioelectron. 2012, 35, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Frasconi, M.; Mazzarino, M.; Botre, F.; Mazzei, F. Surface plasmon resonance immunosensor for cortisol and cortisone determination. Anal. Bioanal. Chem. 2009, 394, 2151–2159. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Zhao, Z.J.; Cheng, P.; He, Z.H.; Cheng, Z.Q.; Peng, J.X.; Wang, H.Y.; Wang, C.; Yang, Y.L.; Hu, Z.Y. Antibody-mimetic peptoid nanosheet for label-free serum-based diagnosis of alzheimer’s disease. Adv. Mater. 2017, 29, 1700057. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Yang, M.; Zhou, W.; Zhang, B.; Cheng, Z. Kinetic and high-throughput profiling of epigenetic interactions by 3D-carbene chip-based surface plasmon resonance imaging technology. Proc. Natl. Acad. Sci. USA 2017, 114, E7245–E7254. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.M.; Wang, C.X.; Cheng, Z.Q.; Zhang, D.D.; Li, S.P.; Song, L.H.; Zhou, W.F.; Yang, M.; Wang, Z.Y.; Zheng, Z.; et al. Spri determination of inter-peptide interaction by using 3D supramolecular co-assembly polyrotaxane film. Biosens. Bioelectron. 2015, 66, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.L.; Yang, L.J.; Su, X.L.; Li, Y.B. A nanoparticle amplification based quartz crystal microbalance DNA sensor for detection of escherichia coli O157: H7. Biosens. Bioelectron. 2006, 21, 1178–1185. [Google Scholar] [CrossRef] [PubMed]
- Mahon, E.; Aastrup, T.; Barboiu, M. Dynamic glycovesicle systems for amplified qcm detection of carbohydrate-lectin multivalent biorecognition. Chem. Commun. 2010, 46, 2441–2443. [Google Scholar] [CrossRef] [PubMed]
- Li, S.P.; Yang, M.; Zhou, W.F.; Johnston, T.G.; Wang, R.; Zhu, J.S. Dextran hydrogel coated surface plasmon resonance imaging (spri) sensor for sensitive and label-free detection of small molecule drugs. Appl. Surf. Sci. 2015, 355, 570–576. [Google Scholar] [CrossRef]
- Löfås, S.; Johnsson, B. A novel hydrogel matrix on gold surfaces in surface plasmon resonance sensors for fast and efficient covalent immobilization of ligands. J. Chem. Soc. Chem. Commun. 1990, 1526–1528. [Google Scholar] [CrossRef]
- Johnsson, B.; Lofas, S.; Lindquist, G. Immobilization of proteins to a carboxymethyldextran-modified gold surface for biospecific interaction analysis in surface plasmon resonance sensors. Anal. Biochem. 1991, 198, 268–277. [Google Scholar] [CrossRef]
- Lofas, S.; Johnsson, B.; Edstrom, A.; Hansson, A.; Lindquist, G.; Hillgren, R.M.M.; Stigh, L. Methods for site controlled coupling to carboxymethyldextran surfaces in surface-plasmon resonance sensors. Biosens. Bioelectron. 1995, 10, 813–822. [Google Scholar] [CrossRef]
- Mohseni, S.; Moghadam, T.T.; Dabirmanesh, B.; Jabbari, S.; Khajeh, K. Development of a label-free spr sensor for detection of matrixmetalloproteinase-9 by antibody immobilization on carboxymethyldextran chip. Biosens. Bioelectron. 2016, 81, 510–516. [Google Scholar] [CrossRef] [PubMed]
- Jabbari, S.; Dabirmanesh, B.; Arab, S.S.; Amanlou, M.; Daneshjou, S.; Gholami, S.; Khajeh, K. A novel enzyme based spr-biosensor to detect bromocriptine as an ergoline derivative drug. Sens. Actuators B 2017, 240, 519–527. [Google Scholar] [CrossRef]
- Yang, N.; Su, X.; Tjong, V.; Knoll, W. Evaluation of two- and three-dimensional streptavidin binding platforms for surface plasmon resonance spectroscopy studies of DNA hybridization and protein-DNA binding. Biosens. Bioelectron. 2007, 22, 2700–2706. [Google Scholar] [CrossRef] [PubMed]
- Marx, K.A. Quartz crystal microbalance: A useful tool for studying thin polymer films and complex biomolecular systems at the solution-surface interface. Biomacromolecules 2003, 4, 1099–1120. [Google Scholar] [CrossRef] [PubMed]
- Pei, Z.; Anderson, H.; Aastrup, T.; Ramstrom, O. Study of real-time lectin-carbohydrate interactions on the surface of a quartz crystal microbalance. Biosens. Bioelectron. 2005, 21, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Dubiel, E.A.; Martin, B.; Vigier, S.; Vermette, P. Real-time label-free detection and kinetic analysis of etanercept-protein a interactions using quartz crystal microbalance. Colloids Surf. B 2017, 149, 312–321. [Google Scholar] [CrossRef] [PubMed]
- Pei, Z.; Anderson, H.; Myrskog, A.; Duner, G.; Ingemarsson, B.; Aastrup, T. Optimizing immobilization on two-dimensional carboxyl surface: Ph dependence of antibody orientation and antigen binding capacity. Anal. Biochem. 2010, 398, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Li, X.M.; Song, S.Y.; Shuai, Q.; Pei, Y.H.; Aastrup, T.; Pei, Y.X.; Pei, Z.C. Real-time and label-free analysis of binding thermodynamics of carbohydrate-protein interactions on unfixed cancer cell surfaces using a qcm biosensor. Sci. Rep. 2015, 5, 14066. [Google Scholar] [CrossRef] [PubMed]
- Myszka, D.G.; Morton, T.A. Clamp: A biosensor kinetic data analysis program. Trends Biochem. Sci. 1998, 23, 149–150. [Google Scholar] [CrossRef]
- Li, X.M.; Song, S.Y.; Pei, Y.X.; Dong, H.; Aastrup, T.; Pei, Z.C. Oriented and reversible immobilization of his-tagged proteins on two- and three-dimensional surfaces for study of protein-protein interactions by a qcm biosensor. Sens. Actuators B 2016, 224, 814–822. [Google Scholar] [CrossRef]
- Cui, X.Q.; Yang, F.; Sha, Y.F.; Yang, X.R. Real-time immunoassay of ferritin using surface plasmon resonance biosensor. Talanta 2003, 60, 53–61. [Google Scholar] [CrossRef]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, S.; Lu, Y.; Li, X.; Cao, S.; Pei, Y.; Aastrup, T.; Pei, Z. Optimization of 3D Surfaces of Dextran with Different Molecule Weights for Real-Time Detection of Biomolecular Interactions by a QCM Biosensor. Polymers 2017, 9, 409. https://doi.org/10.3390/polym9090409
Song S, Lu Y, Li X, Cao S, Pei Y, Aastrup T, Pei Z. Optimization of 3D Surfaces of Dextran with Different Molecule Weights for Real-Time Detection of Biomolecular Interactions by a QCM Biosensor. Polymers. 2017; 9(9):409. https://doi.org/10.3390/polym9090409
Chicago/Turabian StyleSong, Siyu, Yuchao Lu, Xueming Li, Shoupeng Cao, Yuxin Pei, Teodor Aastrup, and Zhichao Pei. 2017. "Optimization of 3D Surfaces of Dextran with Different Molecule Weights for Real-Time Detection of Biomolecular Interactions by a QCM Biosensor" Polymers 9, no. 9: 409. https://doi.org/10.3390/polym9090409