To Tie or Not to Tie? That Is the Question
Abstract
:1. Introduction
2. Entanglement in Proteins
3. Advantages and Disadvantages of a Complex Topology
3.1. Folding
Misfolding and Aggregation
3.2. Stability
3.3. Function
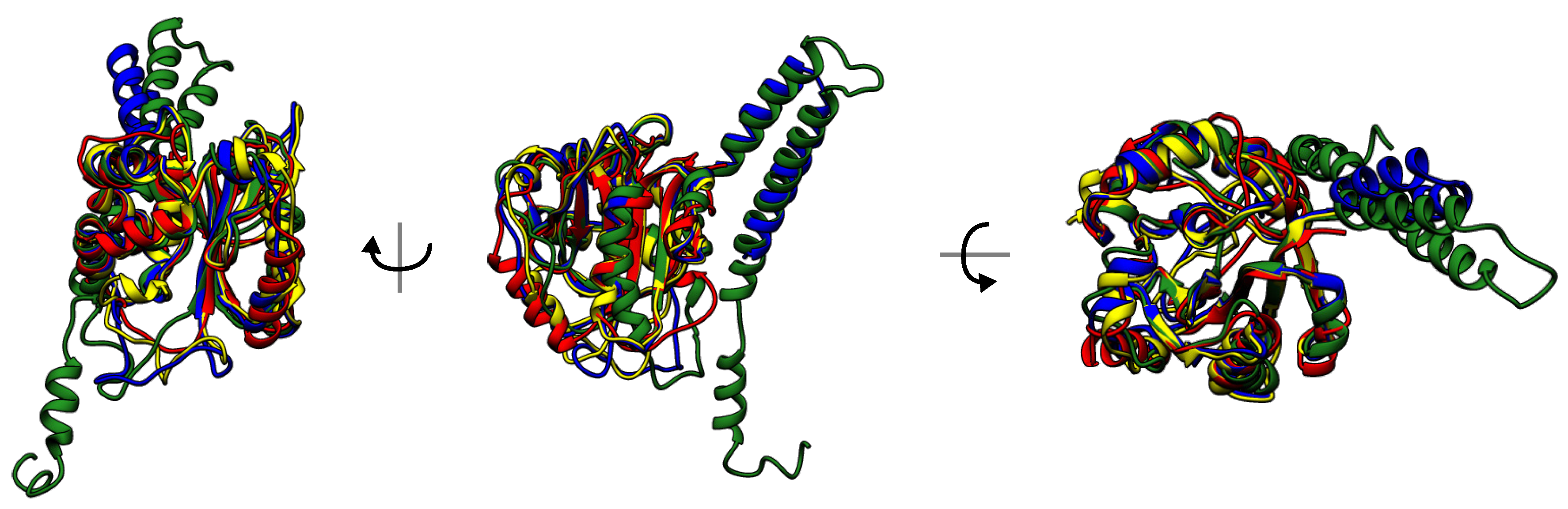
3.4. Evolutionary Conservation
4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sogo, J.M.; Stasiak, A.; Martıńez-Robles, M.L.; Krimer, D.B.; Hernández, P.; Schvartzman, J.B. Formation of knots in partially replicated DNA molecules. J. Mol. Biol. 1999, 286, 637–643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mueller, J.E.; Du, S.M.; Seeman, N.C. Design and synthesis of a knot from single-stranded DNA. J. Am. Chem. Soc. 1991, 113, 6306–6308. [Google Scholar] [CrossRef]
- Arsuaga, J.; Vazquez, M.; McGuirk, P.; Trigueros, S.; Roca, J.; Sumners, D.W. DNA knots reveal a chiral organization of DNA in phage capsids. Proc. Natl. Acad. Sci. USA 2005, 102, 9165–9169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.F.; Liu, C.C.; Alberts, B.M. Type II DNA topoisomerases: Enzymes that can unknot a topologically knotted DNA molecule via a reversible double-strand break. Cell 1980, 19, 697–707. [Google Scholar] [CrossRef]
- Rawdon, E.J.; Dorier, J.; Racko, D.; Millett, K.C.; Stasiak, A. How topoisomerase IV can efficiently unknot and decatenate negatively supercoiled DNA molecules without causing their torsional relaxation. Nucleic Acids Res. 2016, 44, 4528–4538. [Google Scholar] [CrossRef] [PubMed]
- Racko, D.; Benedetti, F.; Dorier, J.; Burnier, Y.; Stasiak, A. Generation of supercoils in nicked and gapped DNA drives DNA unknotting and postreplicative decatenation. Nucleic Acids Res. 2015, 43, 7229–7236. [Google Scholar] [CrossRef] [PubMed]
- Mansfield, M.L. Are there knots in proteins? Nat. Struct. Mol. Biol. 1994, 1, 213–214. [Google Scholar] [CrossRef]
- Mansfield, M.L. Fit to be tied. Nat. Struct. Mol. Biol. 1997, 4, 166–167. [Google Scholar] [CrossRef]
- Taylor, W.R. A deeply knotted protein structure and how it might fold. Nature 2000, 406, 916–919. [Google Scholar] [CrossRef] [PubMed]
- Taylor, W.R.; Lin, K. Protein knots: A tangled problem. Nature 2003, 421, 25. [Google Scholar] [CrossRef] [PubMed]
- Nureki, O.; Shirouzu, M.; Hashimoto, K.; Ishitani, R.; Terada, T.; Tamakoshi, M.; Oshima, T.; Chijimatsu, M.; Takio, K.; Vassylyev, D.G.; et al. An enzyme with a deep trefoil knot for the active-site architecture. Acta Crystallogr. Sect. D Biol. Crystallogr. 2002, 58, 1129–1137. [Google Scholar] [CrossRef]
- Nureki, O.; Watanabe, K.; Fukai, S.; Ishii, R.; Endo, Y.; Hori, H.; Yokoyama, S. Deep knot structure for construction of active site and cofactor binding site of tRNA modification enzyme. Structure 2004, 12, 593–602. [Google Scholar] [CrossRef] [PubMed]
- King, N.P.; Yeates, E.O.; Yeates, T.O. Identification of rare slipknots in proteins and their implications for stability and folding. J. Mol. Biol. 2007, 373, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Jamroz, M.; Niemyska, W.; Rawdon, E.J.; Stasiak, A.; Millett, K.C.; Sułkowski, P.; Sulkowska, J.I. KnotProt: A database of proteins with knots and slipknots. Nucleic Acids Res. 2014, 43, D306–D314. [Google Scholar] [CrossRef] [PubMed]
- Sułkowska, J.I.; Rawdon, E.J.; Millett, K.C.; Onuchic, J.N.; Stasiak, A. Conservation of complex knotting and slipknotting patterns in proteins. Proc. Natl. Acad. Sci. USA 2012, 109, E1715–E1723. [Google Scholar] [CrossRef] [PubMed]
- Niemyska, W.; Dabrowski-Tumanski, P.; Kadlof, M.; Haglund, E.; Sułkowski, P.; Sulkowska, J.I. Complex lasso: New entangled motifs in proteins. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Dabrowski-Tumanski, P.; Sulkowska, J.I. Topological knots and links in proteins. Proc. Natl. Acad. Sci. USA 2017, 114, 3415–3420. [Google Scholar] [CrossRef] [PubMed]
- Dabrowski-Tumanski, P.; Jarmolinska, A.I.; Niemyska, W.; Rawdon, E.J.; Millett, K.C.; Sulkowska, J.I. LinkProt: A database collecting information about biological links. Nucleic Acids Res. 2017, 45, D243–D249. [Google Scholar] [CrossRef] [PubMed]
- Dabrowski-Tumanski, P.; Niemyska, W.; Pasznik, P.; Sulkowska, J.I. LassoProt: Server to analyze biopolymers with lassos. Nucleic Acids Res. 2016, 44, W383–W389. [Google Scholar] [CrossRef] [PubMed]
- Virnau, P.; Mirny, L.A.; Kardar, M. Intricate knots in proteins: Function and evolution. PLoS Comput. Biol. 2006, 2, e122. [Google Scholar] [CrossRef] [PubMed]
- Yeates, T.O.; Norcross, T.S.; King, N.P. Knotted and topologically complex proteins as models for studying folding and stability. Current Opin. Chem. Biol. 2007, 11, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Taylor, W.R. Protein knots and fold complexity: Some new twists. Comput. Biol. Chem. 2007, 31, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Virnau, P.; Mallam, A.; Jackson, S. Structures and folding pathways of topologically knotted proteins. J. Phys. Condens. Matter 2010, 23, 033101. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.E.; Suma, A.; Micheletti, C. How to fold intricately: Using theory and experiments to unravel the properties of knotted proteins. Curr. Opin. Struct. Biol. 2017, 42, 6–14. [Google Scholar] [CrossRef] [PubMed]
- Mallam, A.L. How does a knotted protein fold? FEBS J. 2009, 276, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Faísca, P.F. Knotted proteins: A tangled tale of structural biology. Comput. Struct. Biotechnol. J. 2015, 13, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Lim, N.C.; Jackson, S.E. Molecular knots in biology and chemistry. J. Phys. Condens. Matter 2015, 27, 354101. [Google Scholar] [CrossRef] [PubMed]
- Jarmolinska, A.I.; Perlinska, A.P.; Runkel, R.; Trefz, B.; Virnau, P.; Sulkowska, J.I. Proteins a knotty problems. Under Rev. 2017. under review. [Google Scholar]
- Kolesov, G.; Virnau, P.; Kardar, M.; Mirny, L.A. Protein knot server: Detection of knots in protein structures. Nucleic Acids Res. 2007, 35, W425–W428. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.L.; Yen, S.C.; Yu, S.H.; Hwang, J.K. pKNOT: The protein KNOT web server. Nucleic Acids Res. 2007, 35, W420–W424. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.L.; Chen, C.C.; Hwang, J.K. pKNOT v. 2: The protein KNOT web server. Nucleic Acids Res. 2012, 40, W228–W231. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.J.; Zhou, M.; Fang, Z.P.; Wang, M.; Zhou, X.L.; Wang, E.D. The tRNA recognition mechanism of the minimalist SPOUT methyltransferase, TrmL. Nucleic Acids Res. 2013, 41, 7828–7842. [Google Scholar] [CrossRef] [PubMed]
- Bölinger, D.; Sułkowska, J.I.; Hsu, H.P.; Mirny, L.A.; Kardar, M.; Onuchic, J.N.; Virnau, P. A Stevedore’s protein knot. PLoS Comput. Biol. 2010, 6, e1000731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Millett, K.C.; Rawdon, E.J.; Stasiak, A.; Sułkowska, J.I. Identifying knots in proteins. Biochem. Soc. Trans. 2013, 41, 533–537. [Google Scholar] [CrossRef] [PubMed]
- Rawdon, E.J.; Millett, K.C.; Sułkowska, J.I.; Stasiak, A. Knot localization in proteins. Biochem. Soc. Trans. 2013, 41, 538–541. [Google Scholar] [CrossRef] [PubMed]
- Khatib, F.; Weirauch, M.T.; Rohl, C.A. Rapid knot detection and application to protein structure prediction. Bioinformatics 2006, 22, e252–e259. [Google Scholar] [CrossRef] [PubMed]
- Tubiana, L.; Orlandini, E.; Micheletti, C. Probing the entanglement and locating knots in ring polymers: A comparative study of different arc closure schemes. Prog. Theor. Phys. Suppl. 2011, 191, 192–204. [Google Scholar] [CrossRef]
- Rawdon, E.J.; Millett, K.C.; Stasiak, A. Subknots in ideal knots, random knots, and knotted proteins. Sci. Rep. 2015, 5, 8928. [Google Scholar] [CrossRef] [PubMed]
- Hyde, D.A.; Henrich, J.; Rawdon, E.J.; Millett, K.C. Knotting fingerprints resolve knot complexity and knotting pathways in ideal knots. J. Phys. Condens. Matter 2015, 27, 354112. [Google Scholar] [CrossRef] [PubMed]
- Alexander, K.; Taylor, A.J.; Dennis, M.R. Proteins analysed as virtual knots. Sci. Rep. 2017, 7. [Google Scholar] [PubMed]
- Goundaroulis, D.; Dorier, J.; Benedetti, F.; Stasiak, A. Studies of global and local entanglements of individual protein chains using the concept of knotoids. arXiv, 2017; arXiv:1705.07849. [Google Scholar]
- Caraglio, M.; Micheletti, C.; Orlandini, E. Physical Links: Defining and detecting inter-chain entanglement. Sci. Rep. 2017, 7, 1156. [Google Scholar] [CrossRef] [PubMed]
- Baiesi, M.; Orlandini, E.; Trovato, A.; Seno, F. Linking in domain-swapped protein dimers. Sci. Rep. 2016, 6, 33872. [Google Scholar] [CrossRef] [PubMed]
- Boutz, D.R.; Cascio, D.; Whitelegge, J.; Perry, L.J.; Yeates, T.O. Discovery of a thermophilic protein complex stabilized by topologically interlinked chains. J. Mol. Biol. 2007, 368, 1332–1344. [Google Scholar] [CrossRef] [PubMed]
- Blankenship, J.W.; Dawson, P.E. Thermodynamics of a designed protein catenane. J. Mol. Biol. 2003, 327, 537–548. [Google Scholar] [CrossRef]
- Yan, L.Z.; Dawson, P.E. Design and synthesis of a protein catenane. Angew. Chem. Int. Ed. 2001, 40, 3625–3627. [Google Scholar] [CrossRef]
- Salomon, R.; Farías, R.N. Microcin 25, a novel antimicrobial peptide produced by Escherichia coli. J. Bacteriol. 1992, 174, 7428–7435. [Google Scholar] [CrossRef] [PubMed]
- Bryngelson, J.D.; Onuchic, J.N.; Socci, N.D.; Wolynes, P.G. Funnels, pathways, and the energy landscape of protein folding: A synthesis. Proteins Struct. Funct. Bioinform. 1995, 21, 167–195. [Google Scholar] [CrossRef] [PubMed]
- Mallam, A.L.; Jackson, S.E. Knot formation in newly translated proteins is spontaneous and accelerated by chaperonins. Nat. Chem. Biol. 2012, 8, 147–153. [Google Scholar] [CrossRef] [PubMed]
- King, N.P.; Jacobitz, A.W.; Sawaya, M.R.; Goldschmidt, L.; Yeates, T.O. Structure and folding of a designed knotted protein. Proc. Natl. Acad. Sci. USA 2010, 107, 20732–20737. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, F.; Lim, N.C.; Mandal, S.S.; Pelz, B.; Ng, W.P.; Schlierf, M.; Jackson, S.E.; Rief, M. Knotting and unknotting of a protein in single molecule experiments. Proc. Natl. Acad. Sci. USA 2016, 113, 7533–7538. [Google Scholar] [CrossRef] [PubMed]
- Chien, C.T.H.; Wang, L.W.; Liu, Y.N.; Hsu, B.D.; Lyu, P.C.; Hsu, S.T.D. NMR assignments of a hypothetical pseudo-knotted protein HP0242 from Helicobacter pylori. Biomol. NMR Assign. 2014, 8, 287–289. [Google Scholar] [CrossRef] [PubMed]
- Wang, I.; Chen, S.Y.; Hsu, S.T.D. Unraveling the folding mechanism of the smallest knotted protein, MJ0366. J. Phys. Chem. B 2015, 119, 4359–4370. [Google Scholar] [CrossRef] [PubMed]
- Bustamante, A.; Rivera, M.; Molina, J.; Baez, M. The Folding Mechanism of an Artificial Knotted Protein Characterized by Optical Tweezers. FASEB J. 2017, 31, lb87. [Google Scholar]
- Hsu, S.T.D. Protein knotting through concatenation significantly reduces folding stability. Sci. Rep. 2016, 6, 39357. [Google Scholar] [CrossRef] [PubMed]
- Mallam, A.L.; Onuoha, S.C.; Grossmann, J.G.; Jackson, S.E. Knotted fusion proteins reveal unexpected possibilities in protein folding. Mol. Cell 2008, 30, 642–648. [Google Scholar] [CrossRef] [PubMed]
- Mallam, A.L.; Morris, E.R.; Jackson, S.E. Exploring knotting mechanisms in protein folding. Proc. Natl. Acad. Sci. USA 2008, 105, 18740–18745. [Google Scholar] [CrossRef] [PubMed]
- Mallam, A.L.; Rogers, J.M.; Jackson, S.E. Experimental detection of knotted conformations in denatured proteins. Proc. Natl. Acad. Sci. USA 2010, 107, 8189–8194. [Google Scholar] [CrossRef] [PubMed]
- Andrews, B.T.; Capraro, D.T.; Sulkowska, J.I.; Onuchic, J.N.; Jennings, P.A. Hysteresis as a marker for complex, overlapping landscapes in proteins. J. Phys. Chem. Lett. 2012, 4, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, S.J.M.; Mallam, A.L.; Jackson, S.E.; Hsu, S.T.D. Backbone NMR assignments of a topologically knotted protein in urea-denatured state. Biomol. NMR Assign. 2014, 8, 439–442. [Google Scholar] [CrossRef] [PubMed]
- Andersson, F.I.; Pina, D.G.; Mallam, A.L.; Blaser, G.; Jackson, S.E. Untangling the folding mechanism of the 52-knotted protein UCH-L3. FEBS J. 2009, 276, 2625–2635. [Google Scholar] [CrossRef] [PubMed]
- Andersson, F.I.; Werrell, E.F.; McMorran, L.; Crone, W.J.; Das, C.; Hsu, S.T.D.; Jackson, S.E. The effect of Parkinson’s-disease-associated mutations on the deubiquitinating enzyme UCH-L1. J. Mol. Biol. 2011, 407, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Lou, S.C.; Wetzel, S.; Zhang, H.; Crone, E.W.; Lee, Y.T.; Jackson, S.E.; Hsu, S.T.D. The knotted protein UCH-L1 exhibits partially unfolded forms under native conditions that share common structural features with its kinetic folding intermediates. J. Mol. Biol. 2016, 428, 2507–2520. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.T.C.; Chang, C.Y.; Chen, S.Y.; Pan, Y.R.; Ho, M.R.; Hsu, S.T.D. Entropic stabilization of a deubiquitinase provides conformational plasticity and slow unfolding kinetics beneficial for functioning on the proteasome. Sci. Rep. 2017, 7, 45174. [Google Scholar] [CrossRef] [PubMed]
- Sułkowska, J.I.; Sułkowski, P.; Onuchic, J. Dodging the crisis of folding proteins with knots. Proc. Natl. Acad. Sci. USA 2009, 106, 3119–3124. [Google Scholar] [CrossRef] [PubMed]
- Sułkowska, J.I.; Noel, J.K.; Onuchic, J.N. Energy landscape of knotted protein folding. Proc. Natl. Acad. Sci. USA 2012, 109, 17783–17788. [Google Scholar] [CrossRef] [PubMed]
- Covino, R.; Škrbić, T.; Faccioli, P.; Micheletti, C.; Beccara, S. The role of non-native interactions in the folding of knotted proteins: Insights from molecular dynamics simulations. Biomolecules 2013, 4, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Beccara, S.; Škrbić, T.; Covino, R.; Micheletti, C.; Faccioli, P. Folding pathways of a knotted protein with a realistic atomistic force field. PLoS Comput. Biol. 2013, 9, e1003002. [Google Scholar] [CrossRef] [PubMed]
- Prentiss, M.C.; Wales, D.J.; Wolynes, P.G. The energy landscape, folding pathways and the kinetics of a knotted protein. PLoS Comput. Biol. 2010, 6, e1000835. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Terakawa, T.; Wang, W.; Takada, S. Energy landscape and multiroute folding of topologically complex proteins adenylate kinase and 2ouf-knot. Proc. Natl. Acad. Sci. USA 2012, 109, 17789–17794. [Google Scholar] [CrossRef] [PubMed]
- Noel, J.K.; Onuchic, J.N.; Sulkowska, J.I. Knotting a protein in explicit solvent. J. Phys. Chem. Lett. 2013, 4, 3570–3573. [Google Scholar] [CrossRef]
- Dabrowski-Tumanski, P.; Jarmolinska, A.; Sulkowska, J. Prediction of the optimal set of contacts to fold the smallest knotted protein. J. Phys. Condens. Matter 2015, 27, 354109. [Google Scholar] [CrossRef] [PubMed]
- Noel, J.K.; Sułkowska, J.I.; Onuchic, J.N. Slipknotting upon native-like loop formation in a trefoil knot protein. Proc. Natl. Acad. Sci. USA 2010, 107, 15403–15408. [Google Scholar] [CrossRef] [PubMed]
- Wallin, S.; Zeldovich, K.B.; Shakhnovich, E.I. The folding mechanics of a knotted protein. J. Mol. Biol. 2007, 368, 884–893. [Google Scholar] [CrossRef] [PubMed]
- Sułkowska, J.I.; Noel, J.K.; Ramírez-Sarmiento, C.A.; Rawdon, E.J.; Millett, K.C.; Onuchic, J.N. Knotting pathways in proteins. Biochem. Soc. Trans. 2013, 41, 523–527. [Google Scholar] [CrossRef] [PubMed]
- Wang, I.; Chen, S.Y.; Hsu, S.T.D. Folding analysis of the most complex Stevedore’s protein knot. Sci. Rep. 2016, 6, 31514. [Google Scholar] [CrossRef] [PubMed]
- Faísca, P.F.; Travasso, R.D.; Charters, T.; Nunes, A.; Cieplak, M. The folding of knotted proteins: Insights from lattice simulations. Phys. Biol. 2010, 7, 016009. [Google Scholar] [CrossRef] [PubMed]
- Soler, M.A.; Nunes, A.; Faísca, P.F. Effects of knot type in the folding of topologically complex lattice proteins. J. Chem. Phys. 2014, 141, 025101. [Google Scholar] [CrossRef] [PubMed]
- Soler, M.A.; Faísca, P.F. Effects of knots on protein folding properties. PLoS ONE 2013, 8, e74755. [Google Scholar] [CrossRef] [PubMed]
- Niewieczerzal, S.; Sulkowska, J.I. Knotting and unknotting proteins in the chaperonin cage: Effects of the excluded volume. PLoS ONE 2017, 12, e0176744. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Dabrowski-Tumanski, P.; Niewieczerzal, S.; Sulkowska, J.I. The exclusive effects of chaperonin on the behavior of the 52 knotted proteins. PLoS Comput. Biol. 2017. under review. [Google Scholar]
- Soler, M.A.; Rey, A.; Faísca, P.F. Steric confinement and enhanced local flexibility assist knotting in simple models of protein folding. Phys. Chem. Chem. Phys. 2016, 18, 26391–26403. [Google Scholar] [CrossRef] [PubMed]
- Chwastyk, M.; Cieplak, M. Cotranslational folding of deeply knotted proteins. J. Phys. Condens. Matter 2015, 27, 354105. [Google Scholar] [CrossRef] [PubMed]
- Chwastyk, M.; Cieplak, M. Multiple folding pathways of proteins with shallow knots and co-translational folding. J. Chem. Phys. 2015, 143. [Google Scholar] [CrossRef] [PubMed]
- Suma, A.; Rosa, A.; Micheletti, C. Pore translocation of knotted polymer chains: How friction depends on knot complexity. ACS Macro Lett. 2015, 4, 1420–1424. [Google Scholar] [CrossRef]
- Szymczak, P. Translocation of knotted proteins through a pore. Eur. Phys. J. Spec. Top. 2014, 223, 1805–1812. [Google Scholar] [CrossRef]
- Szymczak, P. Tight knots in proteins: Can they block the mitochondrial pores? Biochem. Soc. Trans. 2013, 41, 620–624. [Google Scholar] [CrossRef] [PubMed]
- Plesa, C.; Verschueren, D.; Pud, S.; van der Torre, J.; Ruitenberg, J.W.; Witteveen, M.J.; Jonsson, M.P.; Grosberg, A.Y.; Rabin, Y.; Dekker, C. Direct observation of DNA knots using a solid-state nanopore. Nat. Nanotechnol. 2016, 11, 1093–1097. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chwastyk, M.; Cieplak, M. Topological transformations in proteins: Effects of heating and proximity of an interface. Sci. Rep. 2017, 7, 39851. [Google Scholar] [CrossRef] [PubMed]
- Soler, M.A.; Faísca, P.F. How difficult is it to fold a knotted protein? In silico insights from surface-tethered folding experiments. PLoS ONE 2012, 7, e52343. [Google Scholar] [CrossRef] [PubMed]
- Haglund, E.; Pilko, A.; Wollman, R.; Jennings, P.A.; Onuchic, J.N. Pierced Lasso Topology Controls Function in Leptin. J. Phys. Chem. B 2017, 121, 706–718. [Google Scholar] [CrossRef] [PubMed]
- Leroy, E.; Boyer, R.; Auburger, G.; Leube, B.; Ulm, G.; Mezey, E.; Harta, G.; Brownstein, M.J.; Jonnalagada, S.; Chernova, T.; et al. The ubiquitin pathway in Parkinson’s disease. Nature 1998, 395, 451–452. [Google Scholar] [CrossRef] [PubMed]
- Haglund, E.; Sulkowska, J.I.; Noel, J.K.; Lammert, H.; Onuchic, J.N.; Jennings, P.A. Pierced lasso bundles are a new class of knot-like motifs. PLoS Comput. Biol. 2014, 10, e1003613. [Google Scholar] [CrossRef] [PubMed]
- Bronsoms, S.; Pantoja-Uceda, D.; Gabrijelcic-Geiger, D.; Sanglas, L.; Aviles, F.X.; Santoro, J.; Sommerhoff, C.P.; Arolas, J.L. Oxidative folding and structural analyses of a kunitz-related inhibitor and its disulfide intermediates: Functional implications. J. Mol. Biol. 2011, 414, 427–441. [Google Scholar] [CrossRef] [PubMed]
- Knappe, T.A.; Linne, U.; Robbel, L.; Marahiel, M.A. Insights into the biosynthesis and stability of the lasso peptide capistruin. Chem. Biol. 2009, 16, 1290–1298. [Google Scholar] [CrossRef] [PubMed]
- Blond, A.; Cheminant, M.; Destoumieux-Garzón, D.; Ségalas-Milazzo, I.; Peduzzi, J.; Goulard, C.; Rebuffat, S. Thermolysin-linearized microcin J25 retains the structured core of the native macrocyclic peptide and displays antimicrobial activity. FEBS J. 2002, 269, 6212–6222. [Google Scholar] [CrossRef]
- Rosengren, K.J.; Blond, A.; Afonso, C.; Tabet, J.C.; Rebuffat, S.; Craik, D.J. Structure of thermolysin cleaved microcin J25: Extreme stability of a two-chain antimicrobial peptide devoid of covalent links. Biochemistry 2004, 43, 4696–4702. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chwastyk, M.; Cieplak, M. Structural entanglements in protein complexes. J. Chem. Phys. 2017, 146, 225102. [Google Scholar] [CrossRef]
- Baccelli, I.; Luti, S.; Bernardi, R.; Scala, A.; Pazzagli, L. Cerato-platanin shows expansin-like activity on cellulosic materials. Appl. Microbiol. Biotechnol. 2014, 98, 175. [Google Scholar] [CrossRef] [PubMed]
- Sułkowska, J.I.; Sułkowski, P.; Szymczak, P.; Cieplak, M. Stabilizing effect of knots on proteins. Proc. Natl. Acad. Sci. USA 2008, 105, 19714–19719. [Google Scholar] [CrossRef] [PubMed]
- Sayre, T.C.; Lee, T.M.; King, N.P.; Yeates, T.O. Protein stabilization in a highly knotted protein polymer. Protein Eng. Des. Sel. 2011, 24, 627–630. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Yang, L.; Liu, P.; Gao, Y.Q.; Zhao, X.S. Single-Molecule Detection Reveals Knot Sliding in TrmD Denaturation. Chem. Eur. J. 2013, 19, 5909–5916. [Google Scholar] [CrossRef] [PubMed]
- Sułkowska, J.I.; Sułkowski, P.; Szymczak, P.; Cieplak, M. Tightening of knots in proteins. Phys. Rev. Lett. 2008, 100, 058106. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.T.; Yamada, T.; Carlsson, U.; Ikai, A. The importance of being knotted: Effects of the C-terminal knot structure on enzymatic and mechanical properties of bovine carbonic anhydrase II. FEBS Lett. 2002, 519, 35–40. [Google Scholar] [CrossRef]
- Sułkowska, J.I.; Cieplak, M. Mechanical stretching of proteins? A theoretical survey of the Protein Data Bank. J. Phys. Condens. Matter 2007, 19, 283201. [Google Scholar] [CrossRef]
- Dzubiella, J. Sequence-specific size, structure, and stability of tight protein knots. Biophys. J. 2009, 96, 831–839. [Google Scholar] [CrossRef] [PubMed]
- Bornschlögl, T.; Anstrom, D.M.; Mey, E.; Dzubiella, J.; Rief, M.; Forest, K.T. Tightening the knot in phytochrome by single-molecule atomic force microscopy. Biophys. J. 2009, 96, 1508–1514. [Google Scholar] [CrossRef] [PubMed]
- Sułkowska, J.I.; Sułkowski, P.; Onuchic, J.N. Jamming proteins with slipknots and their free energy landscape. Phys. Rev. Lett. 2009, 103, 268103. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Genchev, G.Z.; Lu, H.; Li, H. Mechanically untying a protein slipknot: Multiple pathways revealed by force spectroscopy and steered molecular dynamics simulations. J. Am. Chem. Soc. 2012, 134, 10428–10435. [Google Scholar] [CrossRef] [PubMed]
- Sikora, M.; Sułkowska, J.I.; Cieplak, M. Mechanical strength of 17 134 model proteins and cysteine slipknots. PLoS Comput. Biol. 2009, 5, e1000547. [Google Scholar] [CrossRef] [PubMed]
- Sułkowska, J.I.; Sułkowski, P.; Szymczak, P.; Cieplak, M. Untying knots in proteins. J. Am. Chem. Soc. 2010, 132, 13954–13956. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Larrea, D.; Bayley, H. Protein co-translocational unfolding depends on the direction of pulling. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Berko, D.; Tabachnick-Cherny, S.; Shental-Bechor, D.; Cascio, P.; Mioletti, S.; Levy, Y.; Admon, A.; Ziv, T.; Tirosh, B.; Goldberg, A.L.; et al. The direction of protein entry into the proteasome determines the variety of products and depends on the force needed to unfold its two termini. Mol. Cell 2012, 48, 601–611. [Google Scholar] [CrossRef] [PubMed]
- Wojciechowski, M.; Gómez-Sicilia, À.; Carrión-Vázquez, M.; Cieplak, M. Unfolding knots by proteasome-like systems: Simulations of the behaviour of folded and neurotoxic proteins. Mol. Biosyst. 2016, 12, 2700–2712. [Google Scholar] [CrossRef] [PubMed]
- San Martín, Á.; Rodriguez-Aliaga, P.; Molina, J.A.; Martin, A.; Bustamante, C.; Baez, M. Knots can impair protein degradation by ATP-dependent proteases. Proc. Natl. Acad. Sci. USA 2017, 114, 9864–9869. [Google Scholar] [CrossRef] [PubMed]
- Christian, T.; Sakaguchi, R.; Perlinska, A.P.; Lahoud, G.; Ito, T.; Taylor, E.A.; Yokoyama, S.; Sulkowska, J.I.; Hou, Y.M. Methyl transfer by substrate signaling from a knotted protein fold. Nat. Struct. Mol. Biol. 2016, 23, 941–948. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, S.A.; Harp, J.M.; Devarakonda, S.; Kim, Y.; Rastinejad, F.; Khorasanizadeh, S. The active site of the SET domain is constructed on a knot. Nat. Struct. Mol. Biol. 2002, 9, 833–838. [Google Scholar] [CrossRef] [PubMed]
- Dabrowski-Tumanski, P.; Stasiak, A.; Sulkowska, J.I. In Search of Functional Advantages of Knots in Proteins. PLoS ONE 2016, 11, e0165986. [Google Scholar] [CrossRef] [PubMed]
- Yuzenkova, J.; Delgado, M.; Nechaev, S.; Savalia, D.; Epshtein, V.; Artsimovitch, I.; Mooney, R.A.; Landick, R.; Farias, R.N.; Salomon, R.; et al. Mutations of bacterial RNA polymerase leading to resistance to microcin J25. J. Biol. Chem. 2002, 277, 50867–50875. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, J.; Sineva, E.; Knight, J.; Levy, R.M.; Ebright, R.H. Antibacterial peptide microcin J25 inhibits transcription by binding within and obstructing the RNA polymerase secondary channel. Mol. Cell 2004, 14, 739–751. [Google Scholar] [CrossRef] [PubMed]
- Adelman, K.; Yuzenkova, J.; La Porta, A.; Zenkin, N.; Lee, J.; Lis, J.T.; Borukhov, S.; Wang, M.D.; Severinov, K. Molecular mechanism of transcription inhibition by peptide antibiotic Microcin J25. Mol. Cell 2004, 14, 753–762. [Google Scholar] [CrossRef] [PubMed]
- Haglund, E. Engineering covalent loops in proteins can serve as an on/off switch to regulate threaded topologies. J. Phys. Condens. Matter 2015, 27, 354107. [Google Scholar] [CrossRef] [PubMed]
- Tkaczuk, K.L.; Dunin-Horkawicz, S.; Purta, E.; Bujnicki, J.M. Structural and evolutionary bioinformatics of the SPOUT superfamily of methyltransferases. BMC Bioinform. 2007, 8, 73. [Google Scholar] [CrossRef] [PubMed]
- Potestio, R.; Micheletti, C.; Orland, H. Knotted vs. unknotted proteins: Evidence of knot-promoting loops. PLoS Comput. Biol. 2010, 6, e1000864. [Google Scholar] [CrossRef] [PubMed]
- Lua, R.C.; Grosberg, A.Y. Statistics of knots, geometry of conformations, and evolution of proteins. PLoS Comput. Biol. 2006, 2, e45. [Google Scholar] [CrossRef] [PubMed]
- Wüst, T.; Reith, D.; Virnau, P. Sequence determines degree of knottedness in a coarse-grained protein model. Phys. Rev. Lett. 2015, 114, 028102. [Google Scholar] [CrossRef] [PubMed]
- Coronel, L.; Orlandini, E.; Micheletti, C. Non-monotonic knotting probability and knot length of semiflexible rings: The competing roles of entropy and bending energy. Soft Matter 2017. [Google Scholar] [CrossRef] [PubMed]
- Flory, P.; Volkenstein, M. Statistical Mechanics of Chain Molecules. Biopolymers 1969. [Google Scholar] [CrossRef]
- Creighton, T.E. Proteins: Structures and Molecular Properties; Macmillan: New York, NY, USA, 1993. [Google Scholar]
- Chothia, C. One thousand families for the molecular biologist. Nature 1992, 357, 543–544. [Google Scholar] [CrossRef] [PubMed]
- White, T.A.; Kell, D.B. Comparative genomic assessment of novel broad-spectrum targets for antibacterial drugs. Comp. Funct. Genom. 2004, 5, 304–327. [Google Scholar] [CrossRef] [PubMed]
- Vogel, G.V. Meet WHO’s dirty dozen: The 12 bacteria for which new drugs are most urgently needed. Science 2017. [Google Scholar] [CrossRef]
- Zhou, H.X. Loops, linkages, rings, catenanes, cages, and crowders: Entropy-based strategies for stabilizing proteins. Acc. Chem. Res. 2004, 37, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Gierut, A.M.; Niemyska, W.; Dabrowski-Tumanski, P.; Sułkowski, P.; Sulkowska, J.I. PyLasso: A PyMOL plugin to identify lassos. Bioinformatics 2017. [Google Scholar] [CrossRef]
- Jarmolinska, A.I.; Kadlof, M.; Dabrowski-Tumanski, P.; Sulkowska, J.I. GapRepairer—Repair protein structures and their topology. Bioinformatics 2017. under review. [Google Scholar]
- Abe, H.; Ida, D. Mean-square radius of gyration and scattering function of semiflexible ring polymers of the trefoil knot. Polymers 2016, 8, 271. [Google Scholar] [CrossRef]
- Rawdon, E.; Dobay, A.; Kern, J.C.; Millett, K.C.; Piatek, M.; Plunkett, P.; Stasiak, A. Scaling behavior and equilibrium lengths of knotted polymers. Macromolecules 2008, 41, 4444–4451. [Google Scholar] [CrossRef]
- Mansfield, M.L.; Douglas, J.F. Properties of knotted ring polymers. I. Equilibrium dimensions. J. Chem. Phys. 2010, 133, 044903. [Google Scholar] [CrossRef] [PubMed]
- Plunkett, P.; Piatek, M.; Dobay, A.; Kern, J.C.; Millett, K.C.; Stasiak, A.; Rawdon, E.J. Total curvature and total torsion of knotted polymers. Macromolecules 2007, 40, 3860–3867. [Google Scholar] [CrossRef]
- Uehara, E.; Deguchi, T. Statistical and hydrodynamic properties of topological polymers for various graphs showing enhanced short-range correlation. J. Chem. Phys. 2016, 145, 164905. [Google Scholar] [CrossRef] [PubMed]
- Mansfield, M.L.; Douglas, J.F. Properties of knotted ring polymers. II. Transport properties. J. Chem. Phys. 2010, 133, 044904. [Google Scholar] [CrossRef] [PubMed]
- Klotz, A.R.; Narsimhan, V.; Soh, B.W.; Doyle, P.S. Dynamics of DNA Knots during Chain Relaxation. Macromolecules 2017, 50, 4074–4082. [Google Scholar] [CrossRef]
- Dai, L.; Doyle, P.S. Trapping a Knot into Tight Conformations by Intra-Chain Repulsions. Polymers 2017, 9, 57. [Google Scholar] [CrossRef]
- Micheletti, C.; Marenduzzo, D.; Orlandini, E.; Summers, D. Knotting of random ring polymers in confined spaces. J. Chem. Phys. 2006, 124, 064903. [Google Scholar] [CrossRef] [PubMed]
- Richard, D.; Stalter, S.; Siebert, J.T.; Rieger, F.; Trefz, B.; Virnau, P. Entropic Interactions between Two Knots on a Semiflexible Polymer. Polymers 2017, 9, 55. [Google Scholar] [CrossRef]
- Andersson, F.I.; Jackson, S.E.; Hsu, S.T.D. Backbone assignments of the 26 kDa neuron-specific ubiquitin carboxyl-terminal hydrolase L1 (UCH-L1). Biomol. NMR Assign. 2010, 4, 41–43. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Jackson, S.E. Characterization of the Folding of a 5 2-Knotted Protein Using Engineered Single-Tryptophan Variants. Biophys. J. 2016, 111, 2587–2599. [Google Scholar] [CrossRef] [PubMed]
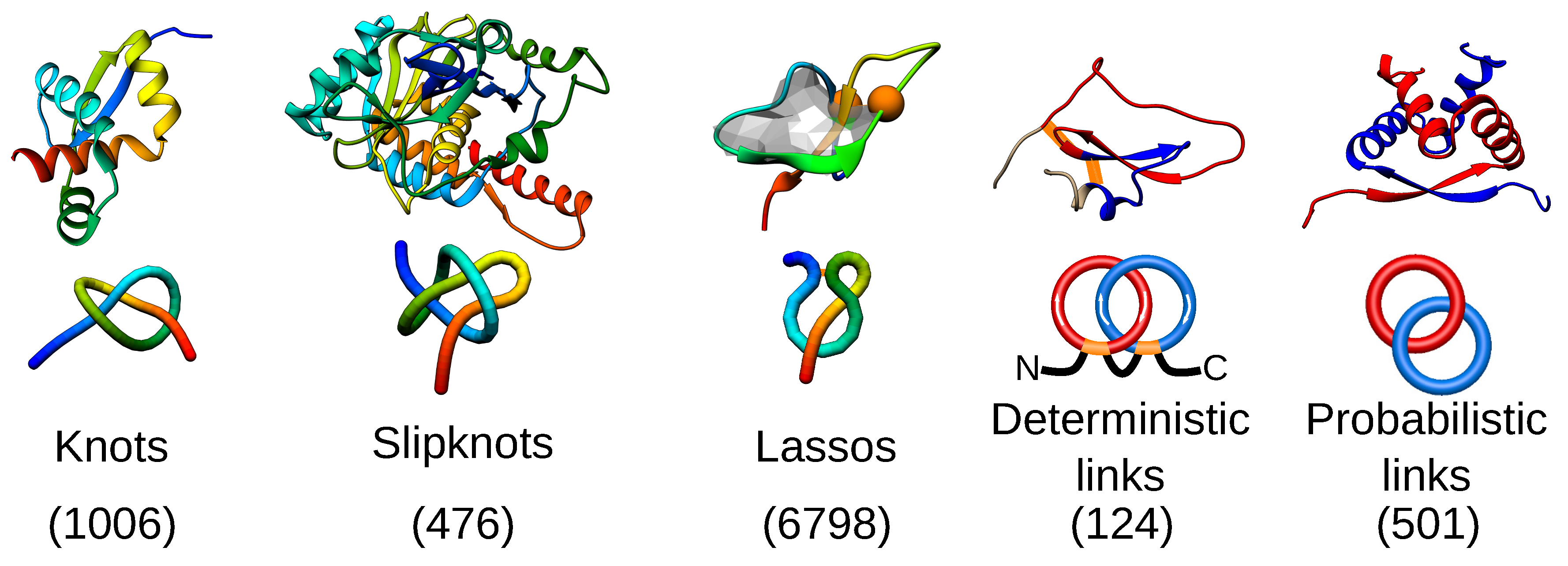
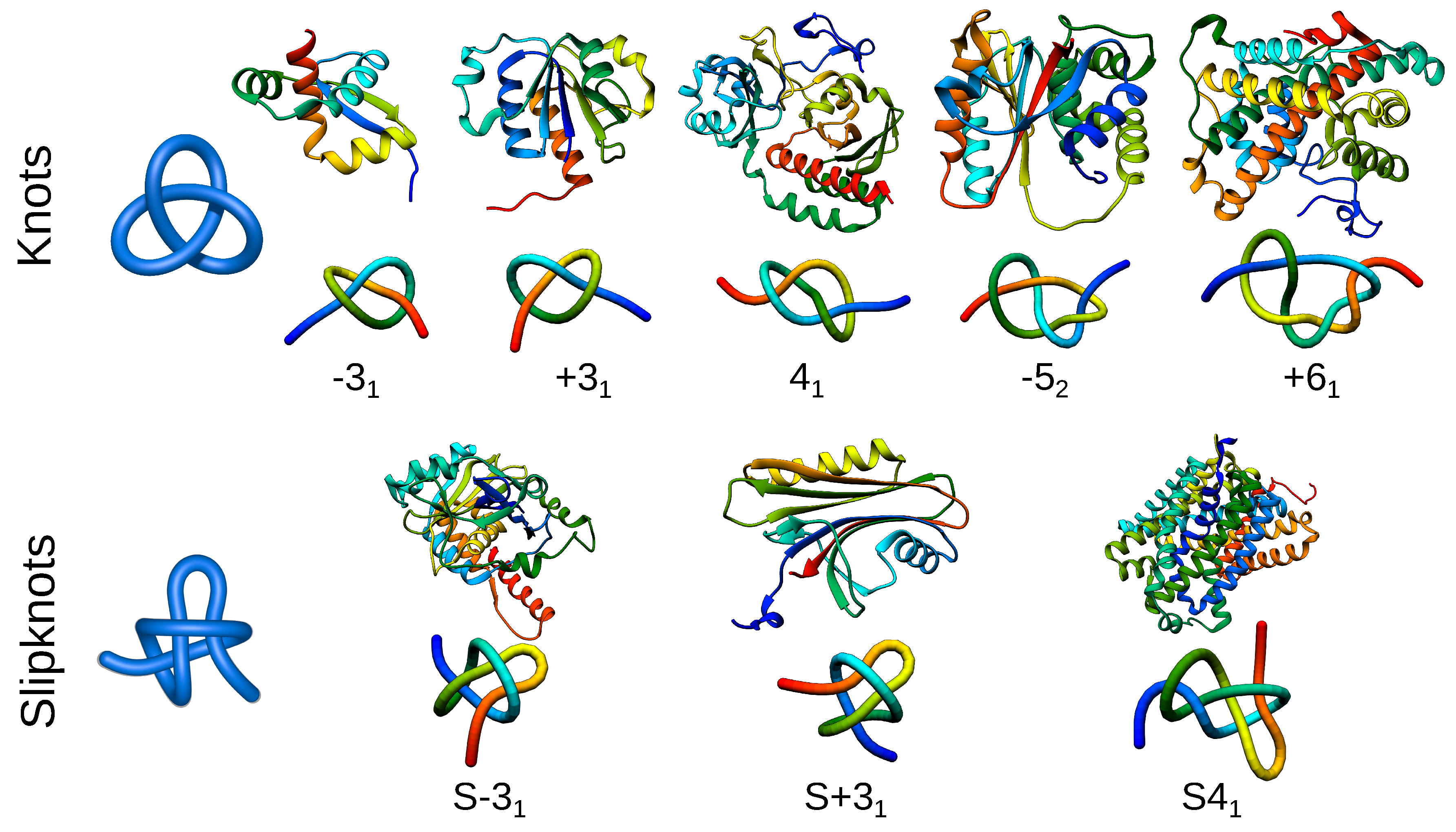


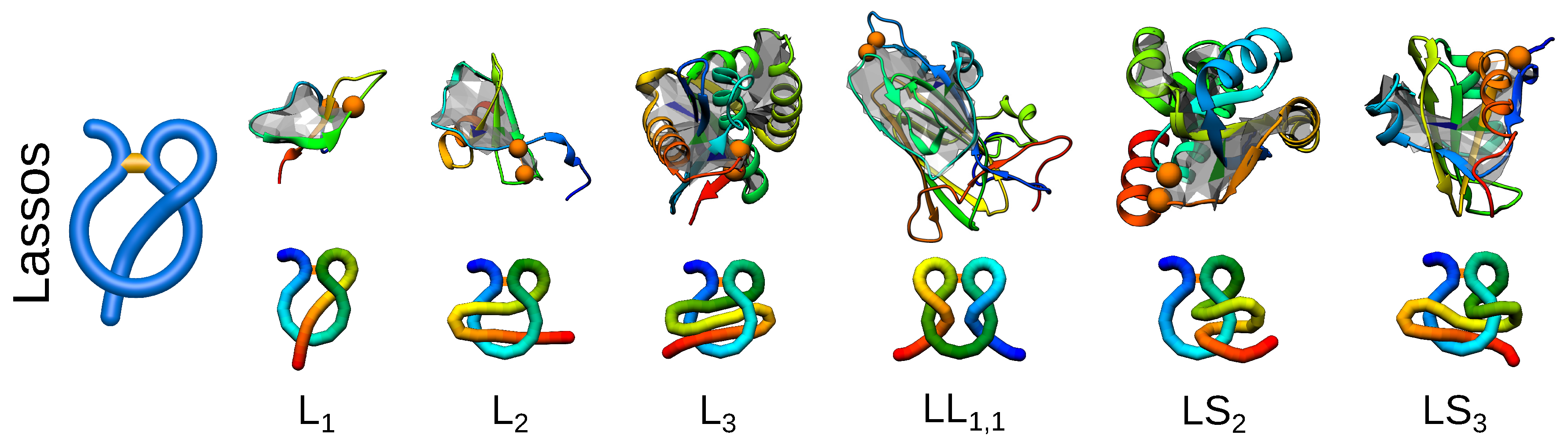

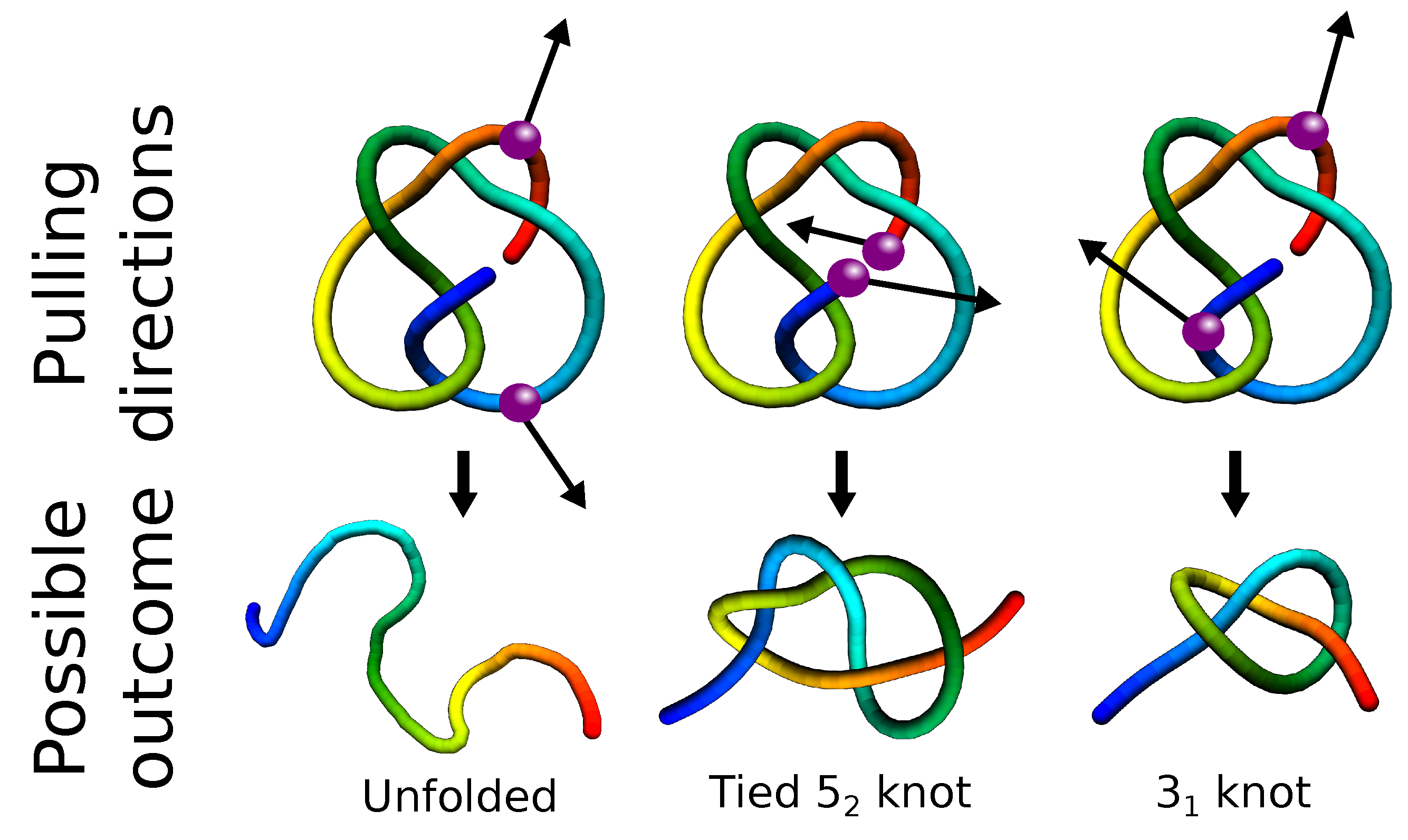

| Property | Advantages | Disadvantages |
|---|---|---|
| Folding |
|
|
| Stability |
|
|
| Function |
| |
| Conservation |
|
|
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dabrowski-Tumanski, P.; Sulkowska, J.I. To Tie or Not to Tie? That Is the Question. Polymers 2017, 9, 454. https://doi.org/10.3390/polym9090454
Dabrowski-Tumanski P, Sulkowska JI. To Tie or Not to Tie? That Is the Question. Polymers. 2017; 9(9):454. https://doi.org/10.3390/polym9090454
Chicago/Turabian StyleDabrowski-Tumanski, Pawel, and Joanna I. Sulkowska. 2017. "To Tie or Not to Tie? That Is the Question" Polymers 9, no. 9: 454. https://doi.org/10.3390/polym9090454





