Induced Mutations Unleash the Potentials of Plant Genetic Resources for Food and Agriculture
Abstract
:1. Addressing the Challenge of Feeding the 21st Century World
1.1. Enhanced Productivities on Farmers’ Fields Are Critical
1.2. The Unmasking of New Alleles Increases Chances for Success
2. Mutation
2.1. Common Types of Mutations Relevant to Crop Improvement
2.1.1. Single Base Substitutions or Point Mutations
- Samesense, silent, or wild-type mutation occurs when the substitution does not affect the amino acid that is encoded by a codon; this is because many amino acids are encoded by different codons. The mutation in this case can only be detected through sequencing (or some other appropriate molecular assays) as there is no detectable phenotypic change.
- Missense mutation refers to the situation where the new nucleotide modifies the codon such that an altered amino acid is encoded for in the protein. This results in detectable phenotypic changes.
- Nonsense mutation describes the substitution that results in a codon that instead of encoding an amino acid is modified into one of the stop codons. This results in the premature termination (or truncation) of the translation of codons to amino acid sequences in the proteins leading to a protein with shortened polypeptides that might be inactive. Severely truncated proteins, i.e., those with the truncation sites early in the gene sequence are inactive.
2.1.2. Insertions and Deletions (Indels)
2.2. Spontaneous Mutations
| Mutation that facilitated domestication | Examples of plant |
|---|---|
| Abolishment of bitterness and toxicity | almonds, lima beans, watermelons, potatoes, egg-plants, cabbages nuts |
| Abolishment of the need for sexual reproduction (seedlessness or parthenocarpy) | bananas, grapes, oranges, and pineapples |
| Loss of natural seed dispersal mechanism—shattering of pods and heads | peas, wheat, barley |
| Loss of the hard seed coat and other germination inhibitors (dormancy) | wheat, barley, peas |
| Facility for self-compatible hermaphroditism | grapes, papaya, etc. |
2.3. Induced Mutations
2.3.1. Physical Mutagens
| Mutagen | Typical Frequency (s−1) | Typical Energy (kJ/mol) | Typical Photon Energy(eV) |
|---|---|---|---|
| Particles | |||
| α-particles | 4.1 × 108 | ||
| β-particles | 1.5 × 107 | ||
| Electromagnetic radiation | |||
| cosmic rays | 6 × 1021 s−1 | 2.4 × 109 | |
| γ-rays | 3 × 102 s−1 | 1.2 × 108 | 1 MeV |
| x-rays | 3 × 1017 s−1 | 1.2 × 105 | 100 keV |
| ultraviolet | 3 × 1015 s−1 | 1200 | 4 eV |
2.3.2. Chemical Mutagens
| Chemical mutagenic agent | Mode of action |
|---|---|
| Base analogues e.g., 5-bromouracil (BU), 5-bromodeoxyuridine, 2-aminopurine (2AP) | Incorporates into DNA in place of the normal bases during DNA replication thereby causing transitions (purine to purine or pyrimidine to pyrimidine); and tautomerization (existing in two forms between which they interconvert e.g., guanine can exist in keto or enol forms). |
| Nitrous acid | Acts through deamination, the replacement of cytosine by uracil which can pair with adenine and thus from subsequent cycles of replication lead to transitions. |
| Alkylating agents such as:sulfonates e.g., ethylmethane sulfonate (EMS), diethyl sulfonate (DES); Sulphur mustards e.g., ethyl-2-chloroethyl sulphide;Nitrogen mustards e.g., 2-chloroethyl-dimethyl amine; and Epoxides e.g., ethylene oxideOthers are ethyleneimine, hydroxylamine (NH2OH), N-methyl-N’-nitro-N-nitrosoguanidine (MNNG), sodium azide and diazomethane. | They react with bases and add methyl or ethyl groups and, depending on the affected atom, the alkylated base may then degrade to yield a baseless site, which is mutagenic and recombinogenic, or mispair to result in mutations upon DNA replication. |
| Intercalating agents such as acridine orange, proflavin, ethidium bromide | They insert between bases of DNA thereby causing a “stretching” of the DNA duplex and the DNA polymerase in turn recognizes this stretch as an additional base and inserts an extra base opposite this stretched (intercalated) molecule. This results in frameshifts i.e., an alteration of the reading frame since codons are groups of three nucleotides. |
| Miscellaneous group of agents; Large molecules referred to as “bulky” lesions (e.g., N-acetoxy-N-2-acetyl-aminofluorine—NAAAF). | They bind to bases in DNA and cause them to be noncoding thereby preventing transcription and DNA replication; They cause intra- and inter-strand crosslinks (e.g., Psoralens); They also cause DNA strand breaks (e.g., peroxides). |
3. An Induced Mutagenesis Program
3.1. Choice of Mutagen
3.2. Practical Considerations in Induced Crop Mutagenesis
- ○
- The setting of realistic targets matched with the availability of the requisite appropriate human and material resources and the prior determination that induced crop mutagenesis offers a comparative advantage over other available technologies and strategies.
- ○
- A clear understanding of the genetics of the traits to be improved; for instance, polygenic traits (i.e., characters under the influence of many genes) have less chance of being modified with induced mutations than monogenic traits (i.e. characters under the influence of single genes).
- ○
- A knowledge of the reproductive biology of the crop, i.e., seed vs. vegetative propagation; if seed propagated, self- vs. cross-fertilization; and if vegetatively propagated, which propagation methods are possible (in vivo vs. in vitro).
- ○
- The determination of the plant propagule to be treated i.e. botanical seeds or gametes for seed propagated crops and nodal segments, stem cuttings, twigs, buds, or different types of in vitro cultures for vegetatively propagated crops and the assemblage of sufficient quantities.
- ○
- A knowledge of the ploidy levels of the crop especially when ploidy-related hybridization barriers could impact on the envisaged utility of the induced mutants.
- ○
- A determination of the genetic background of the target materials for induction of mutations. Usually, the best genetic background to be induced to mutate should in general be the best all-round genotype that is deficient in a single trait (the breeding objective); and is homozygous.
- ○
- An informed decision on which induction method, i.e., physical vs. chemical mutagens, and the appropriate doses (including considerations of concentrations and durations for chemical mutagens). A pilot assay is usually necessary in order to determine the optimal dosage prior to the large-scale bulk treatment of the propagules.
- ○
- A clear definition of the plans, mechanisms and the identification of the requisite facilities for handling (including for planting and evaluating) the mutant population. This would also involve an understanding of, and planning for, the adequate population sizes.
- ○
- The scheduling, as the case may be, for the use of the identified infrastructure (irradiator, screen house, fields, laboratories, etc.) and other required inputs.
- ○
- As may be necessary, a definition of methodologies for the identification of the mutation events and subsequent selection of desired mutants. Options may include molecular biology assays with techniques that lend themselves to high throughput set-ups being preferred since they generally obviate the drudgery associated with handling large mutant populations, the majority of which will be jettisoned.
- ○
- Methodologies for the dissociation of chimeras and the production of stable mutants must also be clearly thought through. For putative mutants destined for plant breeding programs, a clear definition, along with relevant counterparts, of the means for incorporating the stable mutants into hybridization schemes is critical to success.
3.3. Optimal Doses for Mutagenic Treatment
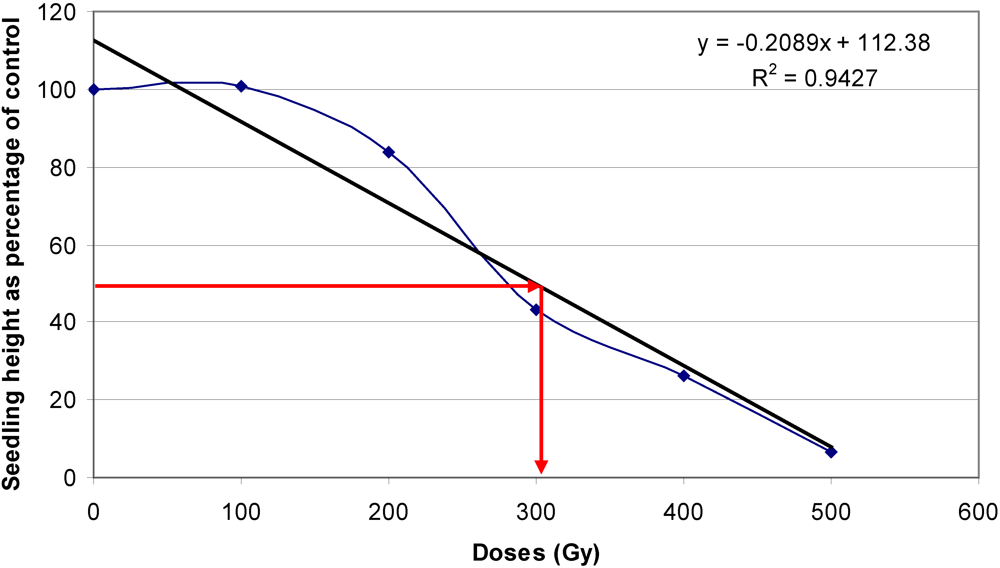
3.4. Incidence of Chimeras
3.5. Induced Mutants as Raw Materials for Crop Improvement

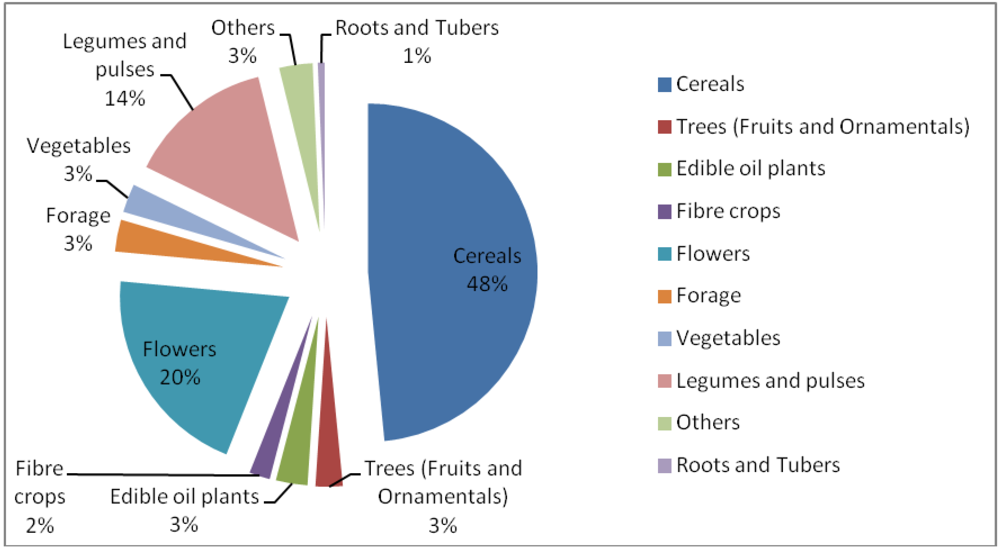
- ○
- Most varieties of durum wheat grown in Italy and used in making pasta that is marketed worldwide are induced mutants; their cultivation generates tens of millions of dollars in additional income to farmers and the seed industry per annum;
- ○
- The Rio Star variety of grape fruit in the USA is a mutant that accounts for 75% of the US grapefruit industry;
- ○
- The cultivation of the mutant Japanese pear variety, “Gold Nijesseiki”, contributes US$30m in additional income to farmers annually
- ○
- A mutant cotton variety, NIAB78, grown widely in Pakistan accounts for over US$20m in additional income to farmers annually
- ○
- Most of the rice grown in Asia (especially in China, Japan. Vietnam and India) and Australia are mutants. The additional income to farmers for growing and marketing these rice varieties is estimated in billions of US$ annually
- ○
- Mutant barley varieties with enhanced adaptation to extremely harsh environmental conditions in high altitudes are literally extending the frontiers of arable lands in Peru. Their cultivation provides much needed employment for resource-poor farmers.
3.6. Induced Mutants as Tools for Understanding and Harnessing Heredity
| Crop | Host institution | Reference |
|---|---|---|
| Maize | The Maize Genetics Cooperation Stock Centre, University of Illinois, Urbana/Champaign, IL, USA | [51] |
| Arabidopsis | European Arabidopsis Stock Centre (or Nottingham Arabidopsis Stock Centre, NASC), University of Nottingham, Sutton Bonington Campus, UK | [52] |
| Arabidopsis Biological Resource Centre, (ABRC)Ohio State University, OH, USA | [53] | |
| Tomato | CM Rick Tomato Genetics Resource Centre, University of California at Davis, CA, USA | [54] |
| Cucurbits (cucumber, melon, Cucurbita, and watermelon) | Cucurbit Genetics Cooperative (CGC) North Carolina State University Raleigh, NC, USA | [55] |
| Rice | The Oryzabase of the National BioResource Project—Rice National Institute of Genetics, Japan | [56] |
| IR64 Rice Mutant Database of the International Rice Functional Genomics, International Rice Research Institute, Manila, Philippines | [57] | |
| Plant Functional Genomics Lab., POSTECH BIOTECH CENTER, San 31 Hyoja-dong, Nam-gu Pohang, Kyoungbuk, Korea | [58] | |
| Barley and wheat | Barley mutants, Scottish Crop Research Institute, Dundee, Scotland | [59] |
| Barley and Wheat Genetic Stock of the USDA-ARS | [60] | |
| USDA-ARS Cereal Crops Research Unit, Fargo, ND, USA | [61] | |
| Wheat Genetics Resource Center, Kansas State University, Manhattan, KS, USA | [62] | |
| Wheat Genetic Resources Database of the Japanese National BioResource Project | [63] | |
| Pea | Pea mutants, John Innes Centre, Norwich, UK | [64] |
3.7. Enhancing the Efficiency for Induced Mutagenesis
3.7.1. Reverse Genetics
| Plant | TILLING Facility | Reference |
|---|---|---|
| Rice, tomato, Arabidopsis | The UC Davis TILLING Core, University of California, Davis, CA, USA | [90] |
| Tomato | Metapontum Agrobios Centre, Metaponto di Bernalda, MT, Italy | [91] |
| Maize | The Maize TILLING Project, Purdue University, West Lafayette, IN, USA | [92] |
| Brassica napus | CAN-TILL, University of British Columbia, | [93] |
| Lotus, Medicago, Brassica rapa | RevGenUK, John Innes Centre, Norwich Research Park, Colney, Norwich, NR4 7UH, UK | [94] |
| Arabidopsis | Seattle TILLING Project, Fred Hutchinson Cancer Research Center, North Seattle, WA, USA | [95] |
| Tomato, Rapeseed, Pea, Medicago and Melon | Plant Genomic Research, INRA/CNRS—URGV, Evry cedex, France | [96] |
3.7.2. Cell and Tissue Biology
3.7.2.1. Cell suspension cultures
3.7.2.2. Doubled Haploidy
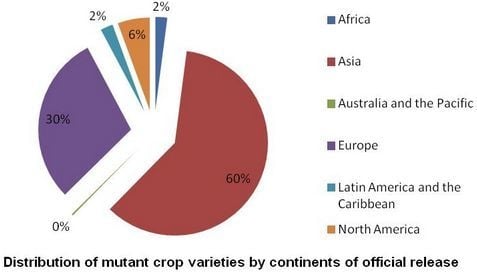
4. The FAO and IAEA Partnership for Nuclear Techniques in Agriculture
| Geographical Location | Number of Projects | ||
|---|---|---|---|
| National | Regional | Total | |
| Africa | 34 | 2 | 36 |
| Asia and the Pacific | 29 | 8 | 37 |
| Europe | 2 | 2 | 4 |
| Latin America and the Caribbean | 7 | 2 | 9 |
| Total | 72 | 14 | 86 |
5. Emerging Trends
5.1. Phenomics
5.2. Pre-Breeding
6. Conclusions and Future Perspectives
- Training of the new crop of plant breeders. Induced mutations, along with enabling biotechnologies, ought to be part of university curricula at both the honours and graduate levels. The annual training program of the Joint FAO/IAEA Division in induced crop mutations could provide the template for developing course curriculum that would be adaptable to other situations. In the same vein, students already undergoing formal plant breeding training programs in universities could also be deliberately targeted for inclusion in this training course. Equally, universities that have maintained strong programs in induced mutations may be supported to host trainees on ad-hoc basis.
- Research. Granted, biotechnologies, such as reverse genetics and cell biology, facilitate the induction, detection and transfer of desirable mutation events. But, the protocols, most of which are genotype-dependent, have been developed for the well-researched major crops. More investments are clearly required for adapting the protocols or developing new ones for the “orphan” crops that are the mainstay of the cropping systems of many food insecure countries. Easily adaptable protocols for vegetatively propagated crops are particularly in short supply. Also, protocols that are less resource-intensive and are adaptable to the resource-poor research environments in developing countries are needed. For instance, for TILLING, neither the endonuclease cleavage of heteroduplexes nor the newer sequencing methods used in the identification of point mutations can be routinely carried out in the laboratories of developing countries that are technology challenged, resource-poor, lack skilled personnel and have weak infrastructures and utilities.
- Community of practice. FAO and the IAEA, through their ca. half a century partnership, is investing considerable resources in training, technology transfer, strengthening of infrastructure, provision of services and in research. To achieve the greatest impacts through engagements at scale, a critical mass of partners that are engaged in similar endeavors and whose activities complement those of this partnership need to be assembled. Such a community could arise around a theme, such as climate change, for instance. Induced mutations can unmask alleles of genes that can be introgressed into breeding materials. This would clearly complement several ongoing activities by a multitude of potential partners, even on global scales, but efforts need to be invested in coalescing these efforts. The TCP and CRP models of the Joint FAO/IAEA Division of Nuclear Techniques for Food and Agriculture can be further scaled up to achieve this purpose.
Conflict of Interest
References
- Food and Agriculture Organization of the United Nations, How to Feed the World in 2050; Food and Agriculture Organization of the United Nations: Rome, Italy, 2009.
- Tester, M.; Langridge, P. Breeding technologies to increase crop production in a changing world. Science 2010, 327, 818–822. [Google Scholar]
- Ejeta, G. Revitalizing agricultural research for global food security. Food Sec. 2009, 1, 391–401. [Google Scholar]
- Nelson, G.C.; Rosegrant, M.W.; Koo, J.; Robertson, R.; Sulser, T.; Zhu, T.; Ringler, C.; Msangi, S.; Palazzo, A.; Batka, M.; Magalhaes, M.; Valmonte-Santos, R.; Ewing, M.; Lee, D. Climate Change: Impact on Agriculture and Costs of Adaptation; International Food Policy Research Institute: Washington, DC, USA, 2009; p. 19. [Google Scholar]
- Hertel, T.W.; Burke, M.B.; Lobell, D.B. The poverty implications of climate-induced crop yield changes by 2030. Glob. Environ. Change 2010, 20, 577–585. [Google Scholar]
- Rosegrant, M.W. Impacts of climate change on food security and livelihoods. In Food Security and Climate Change in Dry Areas, Proceedings of an International Conference, Amman, Jordan, 1–4 February 2010; Solh, M., Saxena, M.C., Eds.; International Center for Agricultural Research in the Dry Areas: Aleppo, Syria, 2011; pp. 24–26. [Google Scholar]
- Food and Agriculture Organization of the United Nations, Global Survey of Agricultural Mitigation Projects; Food and Agriculture Organization of the United Nations: Rome, Italy, 2010.
- Beddington, J.; Asaduzzaman, M.; Fernandez, A.; Clark, M.; Guillou, M.; Jahn, M.; Erda, L.; Mamo, T.; van Bo, N.; Nobre, C.A.; Scholes, R.; Sharma, R.; Wakhungu, J. Achieving Food Security in the Face of Climate Change: Summary for Policy Makers from the Commission on Sustainable Agriculture and Climate Change; CGIAR Research Program on Climate Change, Agriculture and Food Security (CCAFS): Copenhagen, Denmark, 2011. [Google Scholar]
- Intergovernmental Panel on Climate Change, Summary for Policymakers. In Managing the Risks of Extreme Events and Disasters to Advance Climate Change Adaptation, A Special Report of Working Groups I and II of the Intergovernmental Panel on Climate Change; Field, C.B.; Barros, V.; Stocker, T.F.; Qin, D.; Dokken, D.J.; Ebi, K.L.; Mastrandrea, M.D.; Mach, K.J.; Plattner, G.K.; Allen, S.K.; Tignor, M.; Midgley, P.M. (Eds.) Cambridge University Press: Cambridge, UK, 2012.
- Chatham House, The Feeding of the Nine Billion: Global Food Security for the 21st Century; Chatham House: London, UK, 2009.
- Food and Agriculture Organization of the United Nations, Save and Grow—A Policy Maker’s Guide to the Sustainable Intensification of Smallholder Crop Production; Food and Agriculture Organization of the United Nations: Rome, Italy, 2011.
- McCouch, S. Diversifying selection in plant breeding. PLoS Biol. 2004, 2, 1507–1512. [Google Scholar] [CrossRef] [Green Version]
- Waines, G.J.; Ehdaie, B. Domestication and crop physiology: roots of green-revolution wheat. Ann. Bot. 2007, 100, 991–998. [Google Scholar] [CrossRef]
- Mba, C.; Guimaraes, P.; Ghosh, K. Re-orienting crop improvement for the changing climatic conditions of the 21st century. Agric. Food Secur. 2012, 1, 7. [Google Scholar] [CrossRef]
- All Nobel Prizes in Physics. Available online: http://nobelprize.org/nobel_prizes/physics/laureates (accessed on 2 November 2012).
- Muller, H.J. Artificial transmutation of the gene. Science 1927, 66, 84–87. [Google Scholar]
- Stadler, L.J. Genetic Effects of X-rays in maize. Proc. Nat. Acad. Sci. 1928, 14, 69–72. [Google Scholar] [CrossRef]
- Stadler, L.J. Mutations in barley induced by X-rays and radium. Science 1928, 68, 186–187. [Google Scholar]
- Stadler, L.J. Some genetic effects of X-rays in plants. J. Heredity 1930, 21, 2–19. [Google Scholar]
- Stadler, L.J. The experimental modification of heredity in crop plants 1. Induced chromosomal irregularities. 2. Induced mutations. Sci. Agr. 1931, 11, 557–572, 645–661. [Google Scholar]
- Stadler, L.J. On the genetic nature of induced mutations in plants. Proc. 6th Int. Congr. Gen. 1932, 1, 274. [Google Scholar]
- Mba, C.; Afza, R.; Shu, Q.Y. Mutagenic radiations: X-rays, ionizing particles and ultraviolet. In Plant Mutation Breeding and Biotechnology; Shu, Q., Forster, B.P., Nakagawa, H., Eds.; CABI: Oxfordshire, UK, 2012; pp. 83–90. [Google Scholar]
- Mba, C.; Shu, Q.Y. Gamma Irradiation. In Plant Mutation Breeding and Biotechnology; Shu, Q., Forster, B.P., Nakagawa, H., Eds.; CABI: Oxfordshire, UK, 2012; pp. 91–98. [Google Scholar]
- Mei, M.; Deng, H.; Lu, Y.; Zhuang, C.; Liu, Z.; Qiu, Q.; Qiu, Y.; Yang, T.C. Mutagenic effects of heavy ion radiation in plants. Adv. Space Res. 1994, 14, 363–372. [Google Scholar] [CrossRef]
- Wu, L.; Yu, Z. Radiobiological effects of a low-energy ion beam on wheat. Radiat. Environ. Biophys. 2001, 40, 53–57. [Google Scholar] [CrossRef]
- Mei, M.; Qiu, Y.; He, Y.; Bucker, H.; Yang, C.H. Mutational effects of space flight on Zea mays seeds. Adv. Space Res. 1994, 14, 33–39. [Google Scholar]
- Mei, M.; Qiu, Y.; Sun, Y.; Huang, R.; Yao, J.; Zhang, Q.; Hong, M.; Ye, J. Morphological and molecular changes of maize plants after seeds been flown on recoverable satellite. Adv. Space Res. 1998, 22, 1691–1697. [Google Scholar] [CrossRef]
- Ren, W.; Zhang, Y.; Deng, B.; Guo, H.; Cheng, L.; Liu, Y. Effect of space flight factors on alfalfa seeds. Afr. J. Biotechnol. 2010, 9, 7273–7279. [Google Scholar]
- Liu, L.; Zhao, L.; Guo, H. Current status and perspective outlook of space induced mutation breeding in crop plants. Rev. China Agric. Sci. Technol. 2007, 9, 26–29. [Google Scholar]
- Auerbach, C. Tests of carcinogenic substances in relation to the production of mutationins Drosophila melanogaster. Proc. R. Soc. Edinb. 1940, B60, 164–173. [Google Scholar]
- Auerbach, C.; Robson, J.M. Production of mutations by allyl isothiocyanate. Nature 1944, 154, 81. [Google Scholar] [CrossRef]
- Auerbach, C.; Robson, J.M. Chemical production of mutations. Nature 1946, 157, 302. [Google Scholar] [CrossRef]
- Auerbach, C. The induction by mustard gas of chromosomal instabilities in Drosophila melanogaster. Proc. R. Soc. Edinb. 1947, B62, 307–320. [Google Scholar]
- Auerbach, C.; Robson, J.M. The production of mutations by chemical substances. Proc. R. Soc. Edinb. 1947, B62, 27. [Google Scholar]
- Auerbach, C.; Robson, J.M. Tests for chemical substances for mutagenic action. Proc. R. Soc. Edinb. 1947, B62, 284–291. [Google Scholar]
- Auerbach, C. Chemical induction of mutations. In Hereditas 1949, Suppl. Vol. for Proc. 8th Intern. Congr. Genet. pp. 128–147.
- Auerbach, C. Induction of changes in genes and chromosomes. Problems in chemical mutagenesis. Cold Spring Harbor Symp. Quant. Biol. 1951, 16, 199–213. [Google Scholar] [CrossRef]
- Auerbach, C. Genetical effects of radiations and chemicals. Experientia 1957, 13, 217–224. [Google Scholar] [CrossRef]
- Mba, C.; Afza, R.; Bado, S.; Jain, S.M. Induced mutagenesis in plants using physical and chemical agents. In Plant Cell Culture: Essential Methods; Davey, M.R., Anthony, P., Eds.; John Wiley & Sons, Ltd.: Chichester, UK, 2010. [Google Scholar]
- Mba, C.; Afza, R.; Jankowicz-Cieslak, J.; Bado, S.; Matijevic, M.; Huynh, O.; Till, B.T. Enhancing Genetic Diversity Through Induced Mutagenesis in Vegetatively Propagated Plants. In Induced Plant Mutations in the Genomics Era; Shu, Q.Y., Ed.; Food and Agriculture Organization of the United Nations: Rome, Italy, 2009; pp. 293–296. [Google Scholar]
- Van Harten, A.M. Mutation Breeding. Theory and practical Applications; Cambridge University Press: Cambridge, UK, 1998; p. 353. [Google Scholar]
- Mutant Varieties Databse of the Joint FAO/IAEA Division of Nuclear Techniques in Food and Agriculture. Available online: http://mvgs.iaea.org/AboutMutantVarities.aspx (accessed on 2 November 2012).
- Plant Breeding and Genetics Section of the Joint FAO/IAEA Division of Nuclear Techniques in Food and Agriculture. Available online: http://www-naweb.iaea.org/nafa/pbg/index.html (accessed on 2 November 2012).
- Maluszynski, M.; Nichterlein, K.; van Zanten, L.; Ahloowalia, B.S. Officially released mutant varieties—The FAO/IAEA database. Mutat. Breed. Rev. 2000, 12, 1–88. [Google Scholar]
- Maluszynski, M.; Szarejko, I. Induced mutations in the Green and Gene Revolutions. In the Wake of the Double Helix: From the Green Revolution to the Gene Revolution, Proceedings of the International Congress, Bologna, Italy, 27–31 May 2003; Tuberosa, R., Phillips, R.L., Gale, M., Eds.; 2005; pp. 403–425. [Google Scholar]
- Ahloowalia, B.S.; Maluszynski, M.; Nichterlein, K. Global impact of mutation-derived varieties. Euphytica 2004, 135, 187–204. [Google Scholar]
- Kharkwal, M.C.; Shu, Q.Y. The Role of Induced Mutations in World Food Security. In Induced Plant Mutations in the Genomics Era; Shu, Q.Y., Ed.; Food and Agriculture Organization of the United Nations: Rome, Italy, 2009; pp. 33–38. [Google Scholar]
- Askew, M.F. Novel oil, fibre and protein crops in UK, A Future Perspective. Brighton Crop Protect. Conf. 1993, 2, 658. [Google Scholar]
- Larkin, P.J. Introduction. In Somaclonal Variation and Induced Mutations in Crop Improvement; Jain, S.M., Brar, D.S., Ahloowalia, B.S., Eds.; Kluwer Academic Publishers: Dordrecht, Holland, 1998; pp. 3–13. [Google Scholar]
- National Plant Germplasm System of the United States Department of Agriculture. Available online: http://www.ars-grin.gov/npgs/holdings.html (accessed on 2 November 2012).
- The Maize Genetics Cooperation Stock Centre, University of Illinois, Urbana/Champaign, IL, USA. Available online: http://maizecoop.cropsci.uiuc.edu/mgc-info.php (accessed on 2 November 2012).
- European Arabidopsis Stock Centre (also known as Nottingham Arabidopsis Stock Centre, NASC), University of Nottingham, Sutton Bonington Campus, UK. Available online: http://arabidopsis.info/ (accessed on 2 November 2012).
- Arabidopsis Biological Resource Centre, (ABRC), Ohio State University, OH, USA. Available online: http://abrc.osu.edu/ (accessed on 2 November 2012).
- CM Rick Tomato Genetics Resource Centre, University of California at Davis, CA, USA. Available online: http://tgrc.ucdavis.edu/ (accessed on 2 November 2012).
- Cucurbit Genetics Cooperative (CGC), North Carolina State UniversityRaleigh, NC, USA. Available online: http://cuke.hort.ncsu.edu/cgc/index.html (accessed on 2 November 2012).
- The Oryzabase of the National BioResource Project—Rice National Institute of Genetics, Japan. Available online: http://www.shigen.nig.ac.jp/rice/oryzabaseV4/ (accessed on 2 November 2012).
- IR64 Rice Mutant Database of the International Rice Functional Genomics, International Rice Research Institute, Manila, Philippines. Available online: http://irfgc.irri.org/index.php?option=com_content&task=view&id=22&Itemid=44 (accessed on 2 November 2012).
- Plant Functional Genomics Lab., POSTECH BIOTECH CENTER, San 31 Hyoja-dong, Nam-gu Pohang, Kyoungbuk, Korea. Available online: http://www.postech.ac.kr/life/pfg/ (accessed on 2 November 2012).
- Barley mutants, Scottish Crop Research Institute, Dundee, Scotland. Available online: http://bioinf.scri.ac.uk/barley/?page_id=14 (accessed on 2 November 2012).
- Barley and Wheat Genetic Stock of the USDA-ARS. Available online: http://www.ars.usda.gov/main/docs.htm?docid=2922 (accessed on 2 November 2012).
- USDA-ARS Cereal Crops Research Unit, Fargo, ND, USA. Available online: http://wheat.pw.usda.gov/ggpages/GeneticStocks/Fargo_ARS_genetic_stocks.html (accessed on 2 November 2012).
- Wheat Genetics Resource Center, Kansas State University, Manhattan, KS, USA. Available online: http://www.k-state.edu/wgrc/ (accessed on 2 November 2012).
- Wheat Genetic Resources Database of the Japanese National BioResource Project. Available online: http://www.shigen.nig.ac.jp/wheat/komugi/strains/aboutNbrp.jsp (accessed on 2 November 2012).
- Pea mutants, John Innes Centre, Norwich, UK. Available online: http://www.jic.ac.uk/GERMPLAS/pisum/index.htm (accessed on 02 November 2012).
- McCallum, C.M.; Comai, L.; Greene, E.A.; Henikoff, S. Targeted screening for induced mutations. Nat. Biotechnol. 2000, 18, 455–457. [Google Scholar] [CrossRef]
- Henikoff, S.; Till, B.J.; Comai, L. TILLING. Traditional Mutagenesis Meets Functional Genomics. Plant Physiol. 2004, 135, 630–636. [Google Scholar] [CrossRef]
- McCallum, C.M.; Comai, L.; Greene, E.A.; Henikoff, S. Targeting induced local lesions IN genomes (TILLING) for plant functional genomics. Plant Physiol. 2000, 123, 439–442. [Google Scholar] [CrossRef]
- Colbert, T.; Till, B.J.; Tompa, R.; Reynolds, S.; Steine, M.N.; Yeung, A.T.; McCallum, C.M.; Comai, L.; Henikoff, S. High-throughput screening for induced point mutations. Plant Physiol. 2001, 126, 480–484. [Google Scholar] [CrossRef]
- Greene, E.A.; Codomo, C.A.; Taylor, N.E.; Henikoff, J.G.; Till, B.J.; Reynolds, S.H.; Enns, L.C.; Burtner, C.; Johnson, J.E.; Odden, A.R.; et al. Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in Arabidopsis. Genetics 2003, 164, 731–740. [Google Scholar]
- Till, B.J.; Reynolds, S.H.; Greene, E.A.; Codomo, C.A.; Enns, L.C.; Johnson, J.E.; Burtner, C.; Odden, A.R.; Young, K.; Taylor, N.E.; Henikoff, J.G.; Comai, L.; Henikoff, S. Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Res. 2003, 13, 524–530. [Google Scholar] [CrossRef]
- Wu, J.L.; Wu, C.; Lei, C.; Baraoidan, M.; Bordeos, A.; Madamba, M.R.S.; Ramos-Pamplona, M.; Mauleon, R.; Portugal, A.; Ulat, V.J.; Bruskiewich, R.; Wang, G.; Leach, J.; Khush, G.; Leung, H. Chemical and irradiation-induced mutants of indica rice IR64 for forward and reverse genetics. Plant Mol. Biol. 2005, 59, 85–97. [Google Scholar] [CrossRef]
- Sato, Y.; Shirasawa, K.; Takahashi, Y.; Nishimura, M.; Nishio, T. Mutant Selection from Progeny of Gamma-ray-irradiated Rice by DNA Heteroduplex Cleavage using Brassica Petiole Extract. Breed. Sci. 2006, 56, 179–183. [Google Scholar] [CrossRef]
- Till, B.J.; Cooper, J.; Tai, T.H.; Colowit, P.; Greene, E.A.; Henikoff, S.; Comai, L. Discovery of chemically induced mutations in rice by TILLING. BMC Plant Biol. 2007, 7, 19. [Google Scholar] [CrossRef]
- Suzuki, T.; Eiguchi, M.; Kumamaru, T.; Satoh, H.; Matsusaka, H.; Moriguchi, K.; Nagato, Y. MNU-induced mutant pools and high performance TILLING enable finding of any gene mutation in rice. Mol. Genet. Genomics 2008, 279, 213–223. [Google Scholar] [CrossRef]
- Till, B.J.; Reynolds, S.H.; Weil, C.; Springer, N.; Burtner, C.; Young, K.; Bowers, E.; Codomo, C.A.; Enns, L.C.; Odden, A.R.; Greene, E.A.; Comai, L.; Henikoff, S. Discovery of induced point mutations in maize genes by TILLING. BMC Plant Biol. 2004, 4, 12. [Google Scholar] [CrossRef] [Green Version]
- Slade, A.J.; Fuerstenberg, S.I.; Loeffler, D.; Steine, M.N.; Facciotti, D. A reverse genetic, nontransgenic approach to wheat crop improvement by TILLING. Nat. Biotechnol. 2005, 23, 75–81. [Google Scholar]
- Dong, C.; Dalton-Morgan, J.; Vincent, K.; Sharp, P. A Modified TILLING Method for Wheat Breeding. Plant Genome 2009, 2, 39–47. [Google Scholar] [CrossRef]
- Hohmann, U.; Jacobs, G.; Jung, C. An EMS mutagenesis protocol for sugar beet and isolation of non-bolting mutants. Plant Breed. 2005, 124, 317–321. [Google Scholar] [CrossRef]
- Caldwell, D.G.; McCallum, N.; Shaw, P.; Muehlbauer, G.J.; Marshall, D.F.; Waugh, R. A structured mutant population for forward and reverse genetics in barley (Hordeum vulgare L.). Plant J. 2004, 40, 143–150. [Google Scholar] [CrossRef]
- Cooper, J.L.; Till, B.J.; Laport, R.G.; Darlow, M.C.; Kleffner, J.M.; Jamai, A.; El-Mellouki, T.; Liu, S.; Ritchie, R.; Nielsen, N.; Bilyeu, K.D.; Meksem, K.; Comai, L.; Henikoff, S. TILLING to detect induced mutations in soybean. BMC Plant Biol. 2008, 8, 9. [Google Scholar] [CrossRef]
- Triques, K.; Sturbois, B.; Gallais, S.; Dalmais, M.; Chauvin, S.; Clepet, C.; Aubourg, S.; Rameau, C.; Caboche, M.; Bendahmane, A. Characterization of Arabidopsis thaliana mismatch specific endonucleases: Application to mutation discovery by TILLING in pea. Plant J. 2007, 51, 1116–1125. [Google Scholar] [CrossRef]
- Galeano, C.H.; Gomez, M.; Rodriguez, L.M.; Blair, M.W. CEL I Nuclease digestion for SNP discovery and marker development in common bean (Phaseolus vulgaris L.). Crop Sci. 2009, 49, 381–394. [Google Scholar] [CrossRef]
- Porch, T.G.; Blair, M.W.; Lariguet, P.; Galeano, C.; Pankhurst, C.E.; Broughton, W.J. Generation of a mutant population for TILLING common bean genotype BAT 93. J. Am. Soc. Hort. Sci. 2009, 134, 348–355. [Google Scholar]
- Minoia, S.; Petrozza, A.; D’Onofrio, O.; Piron, F.; Mosca, G.; Sozio, G.; Cellini, F.; Bendahmane, A.; Carriero, F. A new mutant genetic resource for tomato crop improvement by TILLING technology. BMC Res. Notes 2010, 3, 69. [Google Scholar] [CrossRef]
- Jankowicz-Cieslak, J.; Huynh, O.A.; Brozynska, M.; Nakitandwe, J.; Till, B.J. Induction, rapid fixation and retention of mutations in vegetatively propagated banana. Plant Biotechnol. J. 2012. [Google Scholar] [CrossRef]
- Till, B.J.; Jankowicz-Cieslak, J.; Sagi, L.; Huynh, O.A.; Utsushi, H.; Swennen, R.; Terauchi, R.; Mba, C. Discovery of nucleotide polymorphisms in the Musa gene pool by Ecotilling. Theor. Appl. Genet. 2010, 121, 1381–1389. [Google Scholar] [CrossRef]
- Tadele, Z.; Mba, C.; Till, B.J. TILLING for mutations in model plants and crops. In Molecular Techniques in Crop Improvement, 2nd; Jain, S.M., Brar, D.S., Eds.; Springer Publishing Inc.: Dordrecht, Holland, 2009; pp. 307–322. [Google Scholar]
- Tsai, H.; Howell, T.; Nitcher, R.; Missirian, V.; Watson, B.; Ngo, K.J.; Lieberman, M.; Fass, J.; Uauy, C.; Tran, R.K.; Khan, A.I.; Filkov, V.; Tai, T.H.; Dubcovsky, J.; Comai, L. Discovery of Rare Mutations in Populations: TILLING by Sequencing. Plant Physiol. 2011, 156, 1257–1268. [Google Scholar] [CrossRef]
- Till, B.J.; Afza, R.; Bado, S.; Huynh, O.A.; Jankowicz-Cieslak, J.; Matijevic, M.; Mba, C. Global TILLING Projects. In Induced Plant Mutations in the Genomics Era, Proceedings of International Symposium on Induced Mutations in Plants, Vienna, Austria, 11–15 August 2008; Shu, Q.Y., Ed.; Food and Agriculture Organization of the United Nations: Rome, Italy, 2009; pp. 237–239. [Google Scholar]
- The UC Davis TILLING Core, University of California, Davis, CA, USA. Available online: http://tilling.ucdavis.edu/index.php/Main_Page (accessed on 2 November 2012).
- LycoTILL—Tomato Mutant Database, Metapontum Agrobios Centre, Metaponto di Bernalda, MT, Italy. Available online: http://www.agrobios.it/tilling/index.html (accessed on 14 January 2013).
- The Maize TILLING Project, Purdue University, West Lafayette, IN, USA. Available online: http://genome.purdue.edu/maizetilling/ (accessed on 2 November 2012).
- CAN-TILL, University of British Columbia. Available online: http://www3.botany.ubc.ca/can-till/Projects.html#napus (accessed on 2 November 2012).
- RevGenUK, John Innes Centre, Norwich Research Park, Colney, Norwich, NR4 7UH, UK. Available online: http://revgenuk.jic.ac.uk/about.htm (accessed on 2 November 2012).
- Seattle TILLING Project, Fred Hutchinson Cancer Research Center, North Seattle, WA, USA. Available online: http://tilling.fhcrc.org/ (accessed on 2 November 2012).
- Plant Genomic Research, INRA/CNRS—URGV, Evry cedex, France. Available online: http://www.versailles.inra.fr/urgv/contact.htm (accessed on 2 November 2012).
- Nickell, L.G. The continuous submerged cultivation of plant tissue as single cells. Proc. Nat. Acad. Sci. 1956, 42, 848–850. [Google Scholar] [CrossRef]
- Lopez-Torres, J. Personal Communication. Instituto de Investigaciones de Viandas Tropicales (INIVIT).
- Forster, B.P.; Heberle-Bors, E.; Kasha, K.J.; Touraev, A. The resurgence of haploids in higher plants. Trends Plant Sci. 2007, 12, 368–375. [Google Scholar] [CrossRef]
- Jauhar, P.P.; Xu, S.S.; Baenziger, P.S. Haploidy in cultivated wheats: Induction and utility in basic and applied research. Crop Sci. 2009, 49, 737–755. [Google Scholar] [CrossRef]
- Castillo, A.M.; Cistué, L.; Vallés, M.P.; Sanz, J.M.; Romagosa, I.; Molina-Cano, J.L. Efficient production of androgenic doubled-haploid mutants in barley by the application of sodium azide to anther and microspore cultures. Plant Cell Rep. 2001, 20, 105–111. [Google Scholar] [CrossRef]
- Forster, B.P.; Thomas, W.T.B. Doubled haploids in genetics and plant breeding. Plant Breed. Rev. 2005, 25, 57–88. [Google Scholar]
- Mkuya, M.S.; Hua-min, S.; Wen-zhen, L.; Zong-xiu, S. Effect of 137Cs gamma rays to panicles on rice anther culture. Rice Science 2005, 12, 299–302. [Google Scholar]
- Kasha, K.J. Chromosome Doubling and Recovery of Doubled Haploid Plants. In Biotechnology in Agriculture and Forestry; Palmer, C.E., Keller, W.A., Kasha, K.J., Eds.; Springer-Verlag: Berlin, Germany, 2005; Volume 56, pp. 123–152. [Google Scholar]
- Takahata, Y.; Fukuoka, H.; Wakui, K. Utilization of Microspore-Derived Embryos. In Biotechnology in Agriculture and Forestry; Palmer, C.E., Keller, W.A., Kasha, K.J., Eds.; Springer-Verlag: Berlin, Germany, 2005; Volume 56, pp. 153–169. [Google Scholar]
- Szarejko, I.; Forster, B.P. Doubled haploidy and induced mutation. Euphytica 2007, 158, 359–370. [Google Scholar] [CrossRef]
- Maluszynski, M.; Kasha, K.J.; Szarejko, I. Published doubled haploid protocols in plant species. In Doubled Hapolid Production in Crop Plants: A Manual; Maluszynski, M., Kasha, K.J., Forster, B.P., Szarejko, I., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2003; pp. 309–335. [Google Scholar]
- Joint FAO/IAEA Division of Nuclear Techniques in Food and Agriculture. Available online: http://www-naweb.iaea.org/nafa/about-nafa/index.html (accessed on 2 November 2012).
- Coordinated Research Activities of the International Atomic Energy Agency. Available online: http://www-crp.iaea.org/default.asp (accessed on 2 November 2012).
- Active crop improvement related Coordinated Research Projects managed by the Plant Breeding and Genetics Section of the Joint FAO/IAEA Division of Nuclear Techniques in Food and Agriculture. Available online: http://www-naweb.iaea.orp/rg/nafa/pbg/cactive-crps-pbg.html (accessed on 2 November 2012).
- Plant Breeding and Genetics Laboratory of the Joint FAO/IAEA Division of Nuclear Techniques in Food and Agriculture. Available online: http://www-naweb.iaea.org/nafa/pbg/pbg-laboratory.html (accessed on 2 November 2012).
- Houle, D.; Govindaraju, D.R.; Omholt, S. Phenomics: The next challenge. Nat. Rev. Genet. 2010, 11, 855–866. [Google Scholar] [CrossRef]
- Finkel, E. With ‘Phenomics’, plant scientists hope to shift breeding into overdrive. Science 2009, 325, 380–381. [Google Scholar] [CrossRef]
- Berger, B.; Parent, B.; Tester, M. High-throughput shoot imaging to study drought responses. J. Exp. Bot. 2010. [Google Scholar] [CrossRef]
- The Australian Plant Phenomics Facility. Available online: http://www.plantphenomics.org.au/ (accessed on 2 November 2012).
- The Biotron Experimental Climate Change Research Centre. Available online: http://www.thebiotron.ca/ (accessed on 2 November 2012).
- Ecotron. Available online: http://www.ecotron.cnrs.fr/ (accessed on 2 November 2012).
- The Ecophysiology Laboratory of Plant under Environmental Stress. Available online: http://www1.montpellier.inra.fr/ibip/lepse/english/ (accessed on 2 November 2012).
- Lemna Tec. Available online: http://www.lemnatec.com/ (accessed on 2 November 2012).
- Jülich Plant Phenotyping Centre. Available online: http://www2.fz-juelich.de/icg/icg-3/jppc (accessed on 2 November 2012).
- Martynov, S.P.; Dobrotvorskaya, T.V. Genealogical analysis of diversity of Russian winter wheat cultivars (Triticum aestivum L.). Genet. Resour. Crop Evol. 2006, 53, 379–386. [Google Scholar] [CrossRef]
- Nass, L.L.; Paterniani, E. Pre-breeding: A link between genetic resources and maize breeding. Sci. agric. 2000, 57, 581–587. [Google Scholar] [CrossRef]
- Pre-breeding theme by the Global Crop Diversity Trust. Available online: http://www.croptrust.org/content/pre-breeding (accessed on 2 November 2012).
- The e-learning course on pre-breeding by the Global Partnership Initiative on Plant Breeding Capacity Building. Available online: http://km.fao.org/gipb/e-learning/gipb-pre-breeding-course/en/ (accessed on 2 November 2012).
- Mba, C.; Guimaraes, E.P.; Guei, G.R.; Hershey, C.; Paganini, M.; Pick, B.; Ghosh, K. Mainstreaming the continuum approach to the management of plant genetic resources for food and agriculture through national strategy. Plant Genet. Resour. 2011. [Google Scholar] [CrossRef]
© 2013 by Food and Agriculture Organization of the United Nations; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Share and Cite
Mba, C. Induced Mutations Unleash the Potentials of Plant Genetic Resources for Food and Agriculture. Agronomy 2013, 3, 200-231. https://doi.org/10.3390/agronomy3010200
Mba C. Induced Mutations Unleash the Potentials of Plant Genetic Resources for Food and Agriculture. Agronomy. 2013; 3(1):200-231. https://doi.org/10.3390/agronomy3010200
Chicago/Turabian StyleMba, Chikelu. 2013. "Induced Mutations Unleash the Potentials of Plant Genetic Resources for Food and Agriculture" Agronomy 3, no. 1: 200-231. https://doi.org/10.3390/agronomy3010200