QTL Analysis for Drought Tolerance in Wheat: Present Status and Future Possibilities
Abstract
:1. Introduction
2. Methodology Used for Collecting Literature and Selecting QTLs
3. Morphological, Growth, and Agronomic Responses
4. Nature of Traits/Genes Involved in Drought Tolerance
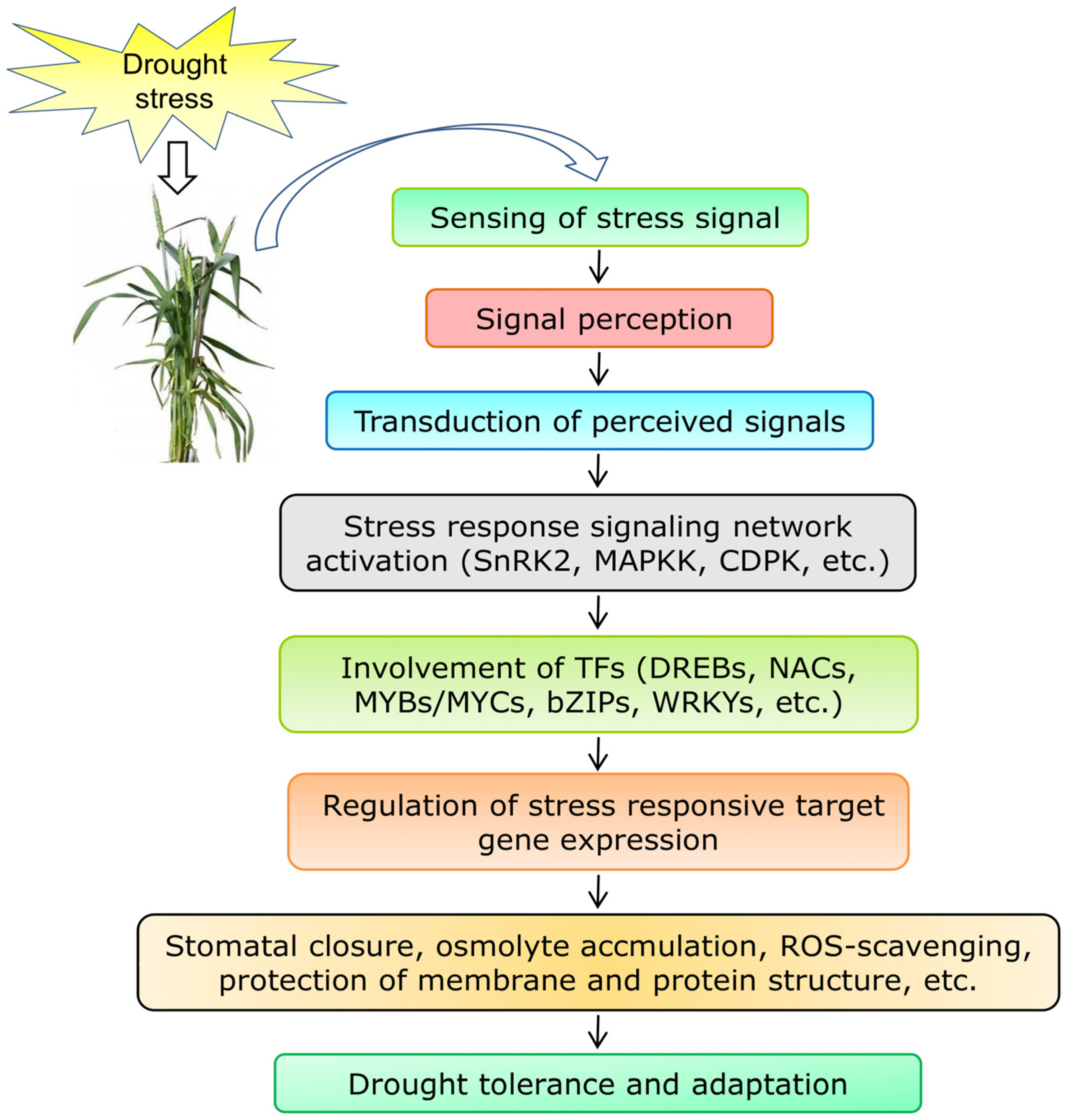
4.1. Genes Involved in Signal Perception, Transduction, and Regulation of Transcription
4.2. Genes Encoding Osmoprotectants/Antioxidants and Generating Reactive Oxygen Species (ROS)
5. Biparental Interval Mapping and Association Mapping
5.1. Major and Stable QTLs and Their Co-Localization with Meta-QTLs (MQTLs)
5.1.1. Biparental Interval Mapping for Agronomic Traits
5.1.2. Biparental Interval Mapping for Physiological Traits
5.2. Meta-QTLs and Their Associated Candidate Genes
5.3. Biparental Interval Mapping and Epistatic QTLs
5.4. MTAs Identified through GWAS
5.5. Genes Encoding Transcription Factors (TFs) and Involved in a Two-Component System (TCS)
6. Molecular Marker-Assisted Breeding
6.1. Marker-Assisted Backcrossing (MABC)
6.2. Marker-Assisted Recurrent Selection (MARS)
7. Future Perspectives
7.1. High Throughput Phenotyping
7.2. High Throughput Genotyping
7.3. Cloning of Genes Underlying QTLs for Drought Tolerance Related Traits
7.4. Genetical Genomics and eQTLs
7.5. EpiQTL for Drought Tolerance
7.6. Alien Genetic Variation for Drought Tolerance
7.7. Physiological Trait Based Breeding
8. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kang, Y.; Khan, S.; Ma, X. Climate change impacts on crop yield, crop water productivity and food security—A review. Prog. Nat. Sci. 2009, 19, 1665–1674. [Google Scholar] [CrossRef]
- Collins, N.C.; Tardieu, F.; Tuberosa, R. Quantitative trait loci and crop performance under abiotic stress: Where do we stand? Plant Physiol. 2008, 147, 469–486. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, M.P.; Ortiz, R. Adapting crops to climate change: A summary. In Climate Change and Crop Production; Reynolds, M.P., Ed.; CABI Series in Climate Change: Cambrigde, MA, USA, 2010; Volume 1, pp. 1–8. [Google Scholar]
- CIMMYT (Centro Internacional de Mejoramiento de Maíz y Trigo). CIMMYT Business Plan 2006–2010—Translating the Vision of Seeds of Innovation into a Vibrant Work Plan; CIMMYT: El Batan, Mexico, 2005. [Google Scholar]
- Rodell, M.; Velicogna, I.; Famiglietti, J.S. Satellite-based estimates of groundwater depletion in India. Nature 2009, 460, 999–1002. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, M.; Skovmand, B.; Trethowan, R.; Pfieffer, W. Evaluating a conceptual model for drought tolerance. In Molecular Approaches for Genetic Improvement of Cereals for Stable Production in Water-Limited Environments; Ribaut, J.M., Poland, D., Eds.; CIMMYT: El Batan, Mexico, 1999; pp. 49–53. [Google Scholar]
- Joshi, A.K.; Mishra, B.; Chatrath, R.; Ortiz Ferrara, G.; Singh, R.P. Wheat improvement in India: Present status, emerging challenges and future prospects. Euphytica 2007, 157, 431–446. [Google Scholar] [CrossRef]
- Gupta, P.K.; Balyan, H.S.; Gahlaut, V.; Kulwal, P.L. Phenotyping, genetic dissection, and breeding for drought and heat tolerance in common wheat: Status and prospects. Plant Breed. Rev. 2012, 36, 85–147. [Google Scholar]
- Farooq, M.; Hussain, M.; Siddique, K.H.M. Drought Stress in Wheat during Flowering and Grain-filling Periods. CRC Crit. Rev. Plant Sci. 2014, 33, 331–349. [Google Scholar] [CrossRef]
- Reynolds, M.P.; Balota, M.; Delgado, M.I.B.; Amani, I.; Fisher, R.A. Physiological and morphological traits associated with spring wheat yield under hot, irrigated conditions. Aust. J. Plant Physiol. 1994, 21, 717–730. [Google Scholar] [CrossRef]
- Reynolds, M.P.; Rebetzke, G. Application of plant physiology in wheat breeding. In The World Wheat Book: A History of Wheat Breeding; Bonjean, A.P., Angus, W.J., Van Ginkel, M., Eds.; TEC: Paris, France, 2011; Volume 2, pp. 877–906. [Google Scholar]
- Parry, M.A.J.; Reynolds, M.; Salvucci, M.E.; Raines, C.; Andralojc, P.J.; Zhu, X.-G.; Price, G.D.; Condon, A.G.; Furbank, R.T. Raising yield potential of wheat. II. Increasing photosynthetic capacity and efficiency. J. Exp. Bot. 2011, 62, 453–467. [Google Scholar] [CrossRef] [PubMed]
- Budak, H.; Hussain, B.; Khan, Z.; Ozturk, N.Z.; Ullah, N. From genetics to functional genomics: Improvement in drought signaling and tolerance in wheat. Front. Plant Sci. 2015, 6, 1012. [Google Scholar] [CrossRef] [PubMed]
- Sheoran, S.; Malik, R.; Narwal, S.; Tyagi, B.S.; Mittal, V.; Kharub, A.S.; Tiwari, V.; Sharma, I. Genetic and molecular dissection of drought tolerance in wheat and barley. J. Wheat Res. 2015, 7, 1–13. [Google Scholar]
- Salvi, S.; Tuberosa, R. The crop QTLome comes of age. Curr. Opin. Biotechnol. 2015, 32, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Nezhadahmadi, A.; Prodhan, Z.H.; Faruq, G. Drought tolerance in wheat. Sci. World J. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Valliyodan, B.; Nguyen, H.T. Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Curr. Opin. Plant Biol. 2006, 9, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Gahlaut, V.; Jaiswal, V.; Kumar, A.; Gupta, P.K. Transcription factors involved in drought tolerance and their possible role in developing drought tolerant cultivars with emphasis on wheat (Triticum aestivum L.). Theor. Appl. Genet. 2016, 129, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Araus, J.L. Integrative physiological criteria associated with yield potential. In Proceedings of the Workshop on Increasing Yield Potential in Wheat: Breaking the Barriers, Obregon, Mexico, 28–30 March 1996.
- Tsunewaki, K.; Ebana, K. Production of near-isogenic lines of common wheat for glaucousness and genetic basis of this trait clarified by their use. Genes Genet. Syst. 1999, 74, 33–41. [Google Scholar] [CrossRef]
- Bennett, D.; Izanloo, A.; Edwards, J.; Kuchel, H.; Chalmers, K.; Tester, M.; Reynolds, M.P.; Schnurbusch, T.; Langridge, P. Identification of novel quantitative trait loci for days to ear emergence and flag leaf glaucousness in a bread wheat (Triticum aestivum L.) population adapted to southern Australian conditions. Theor. Appl. Genet. 2011, 124, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Richards, R.A. Defining selection criteria to improve yield under drought. Plant Growth Reg. 1996, 20, 157–166. [Google Scholar] [CrossRef]
- El-Hafid, R.; Smith, D.H.; Karrou, M.; Sami, K. Morphological attributes associated with early-season drought tolerance in spring durum wheat in Mediterranean environment. Euphytica 1998, 101, 273–282. [Google Scholar] [CrossRef]
- Pinto, R.S.; Reynolds, M.P.; Mathews, K.L.; McIntyre, C.L.; Olivares-Villegas, J.J.; Chapman, S.C. Heat and drought adaptive QTL in a wheat population designed to minimize confounding agronomic effects. Theor. Appl. Genet. 2010, 121, 1001–1021. [Google Scholar] [CrossRef] [PubMed]
- Tewolde, H.; Fernandez, C.J.; Erickson, C.A. Wheat cultivars adapted to post-heading high temperature stress. J. Agron. Crop Sci. 2006, 192, 111–120. [Google Scholar] [CrossRef]
- Ciuca, M.; Petcu, E. SSR markers associated with membrane stability in wheat (Triticum aestivum L.). Rom. Agric. Res. 2009, 26, 21–24. [Google Scholar]
- Kohli, M.M.; Mann, C.E.; Rajaram, S. Global status and recent progress in breeding wheat for the warmer areas. In Wheat for Non-Traditional, Warm Areas; Saunders, D.A., Ed.; CIMMYT: El Batan, Mexico, 1991; pp. 96–112. [Google Scholar]
- Hurd, E.A. Phenotype and drought tolerance in wheat. Agric. Meteorol. 1974, 14, 39–55. [Google Scholar] [CrossRef]
- Al-Khatib, K.; Paulsen, G.M. Photosynthesis and productivity during high temperature stress of wheat genotypes from major world regions. Crop Sci. 1990, 30, 1127–1132. [Google Scholar] [CrossRef]
- Al-Khatib, K.; Paulsen, G.M. High-temperature effects on photosynthetic processes in temperate and tropical cereals. Crop Sci. 1999, 39, 119–125. [Google Scholar] [CrossRef]
- Farooq, M.; Wahid, A.; Kobayashi, N.; Fujita, D.; Basra, S.M.A. Plant drought stress: Effects, mechanisms and management. Agron. Sustain. Dev. 2009, 29, 185–212. [Google Scholar] [CrossRef]
- Innes, P.; Blackwell, R.D.; Quarrie, S.A. Some effects of genetic variation in drought-induced abscisic acid accumulation on the yield and water use of spring wheat. J. Agric. Sci. 1984, 102, 341–351. [Google Scholar] [CrossRef]
- Abebe, T.; Guenzi, A.C.; Martin, B.; Cushman, C.J. Tolerance of mannitol-accumulating transgenic wheat to water stress and salinity. Plant Physiol. 2003, 131, 1748–1755. [Google Scholar] [CrossRef] [PubMed]
- Rosa, M.; Prado, C.; Podazza, G.; Interdonato, R.; González, J.A.; Hilal, M.; Prado, F.E. Soluble sugars. Plant Signal. Behav. 2009, 4, 388–393. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Rivero, R.M.; Kojima, M.; Gepstein, A.; Sakakibara, H.; Mittler, R.; Gepstein, S.; Blumwald, E. Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proc. Natl. Acad. Sci. USA 2007, 104, 19631–19636. [Google Scholar] [CrossRef] [PubMed]
- Rivero, R.M.; Shulaev, V.; Blumwald, E. Cytokinin-dependent photorespiration and the protection of photosynthesis during water deficit. Plant Physiol. 2009, 150, 1530–1540. [Google Scholar] [CrossRef] [PubMed]
- Calderini, D.; Savin, R.; Abeledo, L.; Reynolds, M.; Slafer, G. The importance of the period immediately preceding anthesis for grain weight determination in wheat. In Wheat in a Global Environment; Bedö, Z., Láng, L., Eds.; Springer: Dordrecht, The Netherlands, 2001; pp. 503–509. [Google Scholar]
- Araus, J.L.; Slafer, G.A.; Reynolds, M.P.; Royo, C. Plant breeding and drought in C3 cereals: What should we breed for? Ann. Bot. 2002, 89, 925–940. [Google Scholar] [CrossRef] [PubMed]
- Ribaut, J.-M.; Jiang, C.; Gonzalez-de-Leon, D.; Edmeades, G.; Hoisington, D. Identification of quantitative trait loci under drought conditions in tropical maize. 2. Yield components and marker-assisted selection strategies. Theor. Appl. Genet. 1997, 94, 887–896. [Google Scholar] [CrossRef]
- Painawadee, M.; Jogloy, S.; Kesmala, T.; Akkasaeng, C.; Patanothai, A. Heritability and correlation of drought resistance traits and agronomic traits in peanut (Arachis hypogaea L.). Asian J. Plant Sci. 2009, 8, 325–334. [Google Scholar] [CrossRef]
- Sellammal, R.; Robin, S.; Raveendran, M. Association and heritability studies for drought resistance under varied moisture stress regimes in backcross inbred population of rice. Rice Sci. 2014, 21, 150–161. [Google Scholar] [CrossRef]
- Khanna-Chopra, R.; Singh, K. Drought resistance in crops: Physiological and genetic basis of traits for crop productivity. In Stress Responses in Plants; Tripathi, B.N., Muller, M., Eds.; Springer: Cham, Switzerland, 2015; pp. 267–292. [Google Scholar]
- Acuna-Galindo, M.A.; Mason, R.E.; Subramanian, N.K.; Hays, D.B. Meta-analysis of wheat QTL regions associated with adaptation to drought and heat stress. Crop Sci. 2015, 55, 477–492. [Google Scholar] [CrossRef]
- Shukla, S.; Singh, K.; Patil, R.V.; Kadam, S.; Bharti, S.; Prasad, P.; Singh, N.K.; Khanna-Chopra, R. Genomic regions associated with grain yield under drought stress in wheat (Triticum aestivum L.). Euphytica 2015, 203, 449–467. [Google Scholar] [CrossRef]
- Kirigwi, F.M.; Van Ginkel, M.; Brown-Guedira, G.; Gill, B.S.; Paulsen, G.M.; Fritz, A.K. Markers associated with a QTL for grain yield in wheat under drought. Mol. Breed. 2007, 20, 401–413. [Google Scholar] [CrossRef]
- Quarrie, S.A.; Pekic Quarrie, S.; Radosevic, R.; Rancic, D.; Kaminska, A.; Barnes, J.D.; Leverington, M.; Ceoloni, C.; Dodig, D. Dissecting a wheat QTL for yield present in a range of environments: From the QTL to candidate genes. J. Exp. Bot. 2006, 57, 2627–2637. [Google Scholar] [CrossRef] [PubMed]
- Golabadi, M.; Arzani, A.; Mirmohammadi Maibody, S.A.M.; Tabatabaei, B.E.S.; Mohammadi, S.A. Identification of microsatellite markers linked with yield components under drought stress at terminal growth stages in durum wheat. Euphytica 2011, 177, 207–221. [Google Scholar] [CrossRef]
- Lopes, M.S.; Reynolds, M.P.; McIntyre, C.L.; Mathews, K.L.; Jalal Kamali, M.R.; Mossad, M.; Feltaous, Y.; Tahir, I.S.A.; Chatrath, R.; Ogbonnaya, F.; et al. QTL for yield and associated traits in the Seri/Babax population grown across several environments in Mexico, in the West Asia, North Africa, and South Asia regions. Theor. Appl. Genet. 2013, 126, 971–984. [Google Scholar] [CrossRef] [PubMed]
- Maccaferri, M.; Sanguineti, M.C.; Corneti, S.; Ortega, J.L.A.; Ben Salem, M.; Bort, J.; DeAmbrogio, E.; Del Moral, L.F.G.; Demontis, A.; El-Ahmed, A.; et al. Quantitative trait loci for grain yield and adaptation of durum wheat (Triticum durum Desf.) across a wide range of water availability. Genetics 2008, 178, 489–511. [Google Scholar] [CrossRef] [PubMed]
- Salem, K.F.M.; Roder, M.S.; Borner, A. Identification and mapping quantitative trait loci for stem reserve mobilisation in wheat (Triticum aestivum L.). Cereal Res. Commun. 2007, 35, 1367–1374. [Google Scholar] [CrossRef]
- Bennett, D.; Izanloo, A.; Reynolds, M.; Kuchel, H.; Langridge, P.; Schnurbusch, T. Genetic dissection of grain yield and physical grain quality in bread wheat (Triticum aestivum L.) under water-limited environments. Theor. Appl. Genet. 2012, 125, 255–271. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Sehgal, S.K.; Kumar, U.; Prasad, P.V.V.; Joshi, A.K.; Gill, B.S. Genomic characterization of drought tolerance-related traits in spring wheat. Euphytica 2012, 186, 265–276. [Google Scholar] [CrossRef]
- Quarrie, S.A.; Steed, A.; Calestani, C.; Semikhodskii, A.; Lebreton, C.; Chinoy, C.; Steele, N.; Pljevljakusić, D.; Waterman, E.; Weyen, J.; et al. A high-density genetic map of hexaploid wheat (Triticum aestivum L.) from the cross Chinese Spring × SQ1 and its use to compare QTLs for grain yield across a range of environments. Theor. Appl. Genet. 2005, 110, 865–880. [Google Scholar] [CrossRef] [PubMed]
- Thomas, H.; Smart, C.M. Crops that stay green. Ann. Appl. Biol. 1993, 123, 193–219. [Google Scholar] [CrossRef]
- Christopher, J.T.; Christopher, M.J.; Borrell, A.K.; Fletcher, S.; Chenu, K. Stay-green traits to improve wheat adaptation in well-watered and water-limited environments. J. Exp. Bot. 2016, 67, 5159–5172. [Google Scholar] [CrossRef] [PubMed]
- Rebetzke, G.J.; Jimenez-Berni, J.A.; Bovill, W.D.; Deery, D.M.; James, R.A. High-throughput phenotyping technologies allow accurate selection of stay-green. J. Exp. Bot. 2016, 67, 4919–4924. [Google Scholar] [CrossRef] [PubMed]
- Quarrie, S.A.; Gulli, M.; Calestani, C.; Steed, A.; Marmiroli, N. Localization of drought-induced abscisic acid production on the long arm of chromosome 5 A of wheat. Theor. Appl. Genet. 1994, 89, 794–800. [Google Scholar] [CrossRef] [PubMed]
- Barakat, M.N.; Saleh, M.S.; Al-Doss, A.A.; Moustafa, K.A.; Elshafei, A.A.; Zakri, A.M.; Al-Qurainy, F.H. Mapping of QTLs associated with abscisic acid and water stress in wheat. Biol. Plant 2015, 59, 291–297. [Google Scholar] [CrossRef]
- Ibrahim, S.E.; Schubert, A.; Pillen, K.; Léon, J. Comparison of QTLs for drought tolerance traits between two advanced backcross populations of spring wheat. Int. J. Agric. Sci. 2012, 2, 216–227. [Google Scholar]
- Quarrie, S.; Kaminska, A.; Barnes, J.; Dodig, D.; Gennaro, A. A QTL for grain yield on 7AL of wheat is activated by ABA and low nutrient treatments during flag leaf ontogeny. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2007, 146, S253. [Google Scholar] [CrossRef] [Green Version]
- Mao, D.; Liu, T.; Xu, C.; Li, X.; Xing, Y. Epistasis and complementary gene action adequately account for the genetic bases of transgressive segregation of kilo-grain weight in rice. Euphytica 2011, 180, 261–271. [Google Scholar] [CrossRef]
- Upadhyaya, H.D.; Sharma, S.; Singh, S.; Singh, M. Inheritance of drought resistance related traits in two crosses of groundnut (Arachis hypogaea L.). Euphytica 2011, 177, 55–66. [Google Scholar] [CrossRef]
- Singh, A.; Knox, R.E.; DePauw, R.M.; Singh, A.K.; Cuthbert, R.D.; Campbell, H.L.; Shorter, S.; Bhavani, S. Stripe rust and leaf rust resistance QTL mapping, epistatic interactions, and co-localization with stem rust resistance loci in spring wheat evaluated over three continents. Theor. Appl. Genet. 2014, 127, 2465–2477. [Google Scholar] [CrossRef] [PubMed]
- Govindaraj, P.; Vinod, K.K.; Arumugachamy, S.; Maheswaran, M. Analysing genetic control of cooked grain traits and gelatinization temperature in a double haploid population of rice by quantitative trait loci mapping. Euphytica 2009, 166, 165–176. [Google Scholar] [CrossRef]
- Wu, X.; Chang, X.; Jing, R. Genetic Analysis of Carbon Isotope Discrimination and its Relation to Yield in a Wheat Doubled Haploid Population. J. Integr. Plant Biol. 2011, 53, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Gahlaut, V. Genetic Dissection of Water Stress Tolerance in Bread Wheat. Ph.D. Thesis, Chaudhary Charan Singh University, Meerut, India, 2016. [Google Scholar]
- Yang, D.L.; Jing, R.L.; Chang, X.P.; Li, W. Identification of quantitative trait loci and environmental interactions for accumulation and remobilization of water-soluble carbohydrates in wheat (Triticum aestivum L.) stems. Genetics 2007, 176, 571–584. [Google Scholar] [CrossRef] [PubMed]
- Kadam, S.; Singh, K.; Shukla, S.; Goel, S.; Vikram, P.; Pawar, V. Genomic associations for drought tolerance on the short arm of wheat chromosome 4B. Funct. Integr. Genom. 2012, 12, 447–464. [Google Scholar] [CrossRef] [PubMed]
- Rebetzke, G.J.; Ellis, M.H.; Bonnett, D.G.; Richards, R.A. Molecular mapping of genes for coleoptile growth in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 2007, 114, 1173–1183. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Li, M.; Liu, Y.; Chang, L.; Cheng, H.; Chen, J.; Chai, S. Identification of quantitative trait loci and water environmental interactions for developmental behaviors of leaf greenness in Wheat. Front. Plant Sci. 2016, 7, 273. [Google Scholar] [CrossRef] [PubMed]
- Edae, E.A.; Byrne, P.F.; Haley, S.D.; Lopes, M.S.; Reynolds, M.P. Genome-wide association mapping of yield and yield components of spring wheat under contrasting moisture regimes. Theor. Appl. Genet. 2014, 127, 791–807. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.Q.; Khan, S.H.; Khan, A.S.; Kazi, A.M.; Basra, S.M.A. Identification of QTLs for drought tolerance traits on wheat chromosome 2A using association mapping. Int. J. Agric. Biol. 2014, 16, 862–870. [Google Scholar]
- Zhang, K.; Wang, J.; Zhang, L.; Rong, C.; Zhao, F.; Peng, T.; Li, H.; Cheng, D.; Liu, X.; Qin, H.; et al. Association analysis of genomic loci important for grain weight control in elite common wheat varieties cultivated with variable water and fertiliser supply. PLoS ONE 2013, 8, e57853. [Google Scholar] [CrossRef] [PubMed]
- Ain, Q.; Rasheed, A.; Anwar, A.; Mahmood, T.; Imtiaz, M.; Mahmood, T.; Xia, X.; He, Z.; Quraishi, U.M. Genome-wide association for grain yield under rainfed conditions in historical wheat cultivars from Pakistan. Front. Plant Sci. 2015, 6, 743. [Google Scholar] [CrossRef] [PubMed]
- Edae, E.A.; Byrne, P.F.; Manmathan, H.; Haley, S.D.; Moragues, M.; Lopes, M.S.; Reynolds, M.P. Association mapping and nucleotide sequence variation in five drought tolerance candidate genes in spring wheat. Plant Gen. 2013, 6, 547–562. [Google Scholar] [CrossRef]
- Gahlaut, V.; Mathur, S.; Dhariwal, R.; Khurana, J.P.; Tyagi, A.K.; Balyan, H.S.; Gupta, P.K. A multi-step phosphorelay two-component system impacts on tolerance against dehydration stress in common wheat. Funct. Integr. Genom. 2014, 14, 707–716. [Google Scholar] [CrossRef] [PubMed]
- Jain, N.; Singh, G.P.; Singh, P.K.; Ramya, P.; Krishna, H.; Ramya, K.T.; Todkar, L.; Amasiddha, B.; Prashant, K.C.; Vijay, P.; et al. Molecular approaches for wheat improvement under drought and heat stress. Indian J. Genet. 2014, 74, 578–583. [Google Scholar] [CrossRef]
- Merchuk-Ovnat, L.; Barak, V.; Fahima, T.; Odron, F.; Lidzbarsky, G.A.; Krugman, T.; Saranga, Y. Ancestral QTL alleles from wild emmer wheat improve drought resistance and productivity in modern wheat cultivars. Front. Plant Sci. 2016, 7, 452. [Google Scholar] [CrossRef] [PubMed]
- CGIAR Challenge Programme. Available online: http://www.generationcp.org/communications/media/feature-stories/breaking-new-ground-in-mars-gcp-launches-challenge-initiative-on-wheat-in-asia (accessed on 20 October 2016).
- Granier, C.; Aguirrezabal, L.; Chen, K.; Cookson, S.J.; Duazat, M.; Hamard, P.; Thioux, J.J.; Rolland, G.; Bouchier-Combaud, S.; Lebaudy, A. Phenopsis, an automated platform for reproducible phenotyping of plant response to soil water deficit in Arabidopsis thaliana permitted the identification of an accession with low sensitivity to soil water deficit. New Phytol. 2006, 169, 623–635. [Google Scholar] [CrossRef] [PubMed]
- Walter, A.; Scharr, H.; Gilmer, F.; Zierer, R.; Nagel, K.A.; Ernst, M.; Wiese, A.; Virnich, O.; Christ, M.M.; Uhlig, B.; et al. Dynamics of seedling growth acclimation towards altered light conditions can be quantified via GROWSCREEN: A setup and procedure designed for rapid optical phenotyping of different plant species. New Phytol. 2007, 174, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Biskup, B.; Scharr, H.; Fischbach, A.; Wiese-Klinkenberg, A.; Schurr, U.; Walter, A. Diel growth cycle of isolated leaf discs analyzed with a novel, high-throughput three-dimensional imaging method is identical to that of intact leaves. Plant Physiol. 2009, 149, 1452–1461. [Google Scholar] [CrossRef] [PubMed]
- Eberius, M.; Lima-Guerra, J. High-throughput plant phenotyping: Data acquisition, transformation, and analysis. In Bioinformatics; Edwards, D., Stajich, J., Hansen, D., Eds.; Springer: New York, NY, USA, 2009; pp. 259–278. [Google Scholar]
- Jansen, M.; Gilmer, F.; Biskup, B.; Nagel, K.A.; Rascher, U.; Fischbach, A.; Briem, S.; Dreissen, G.; Tittmann, S.; Braun, S.; et al. Simultaneous phenotyping of leaf growth and chlorophyll fluorescence via GROWSCREEN FLUORO allows detection of stress tolerance in Arabidopsis thaliana and other rosette plants. Funct. Plant Biol. 2009, 36, 902–914. [Google Scholar] [CrossRef]
- Arvidsson, S.; Pérez-Rodríguez, P.; Mueller-Roeber, B. A growth phenotyping pipeline for Arabidopsis thaliana integrating image analysis and rosette area modeling for robust quantification of genotype effects. New Phytol. 2011, 191, 895–907. [Google Scholar] [CrossRef] [PubMed]
- Golzarian, M.R.; Frick, R.A.; Rajendran, K.; Berger, B.; Roy, S.; Tester, M.; Lun, D.S. Accurate inference of shoot biomass from high-throughput images of cereal plants. Plant Methods 2011, 7, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagel, K.A.; Putz, A.; Gilmer, F.; Heinz, K.; Fischbach, A.; Pfeifer, J.; Faget, M.; Blossfeld, S.; Ernst, M.; Dimaki, C.; et al. GROWSCREEN-Rhizo is a novel phenotyping robot enabling simultaneous measurements of root and shoot growth for plants grown in soil-filled rhizotrons. Funct. Plant Biol. 2012, 39, 891–904. [Google Scholar] [CrossRef]
- Li, L.; Zhang, Q.; Huang, D. A review of imaging techniques for plant phenotyping. Sensors 2014, 14, 20078–20111. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Neumann, K.; Friedel, S.; Kilian, B.; Chen, M.; Altmann, T.; Klukas, C. Dissecting the phenotypic components of crop plant growth and drought responses based on high-throughput image Analysis. Plant Cell 2014, 26, 4636–4655. [Google Scholar] [CrossRef] [PubMed]
- Neumann, K.; Klukas, C.; Friedel, S.; Rischbeck, P.; Chen, D.; Entzian, A.; Stein, N.; Graner, A.; Kilian, B. Dissecting spatio-temporal biomass accumulation in barley under different water regimes using high-throughput image analysis. Plant Cell Environ. 2015, 38, 1980–1996. [Google Scholar] [CrossRef] [PubMed]
- Ahamed, T.; Tian, L.; Jiang, Y.; Zhao, B.; Liu, H.; Ting, K.C. Tower remote-sensing system for monitoring energy crops; image acquisition and geometric corrections. Biosyst. Eng. 2012, 112, 93–107. [Google Scholar] [CrossRef]
- Deery, D.; Jimenez-Berni, J.; Jones, H.; Sirault, X.; Furbank, R. Proximal remote sensing buggies and potential applications for field-based phenotyping. Agronomy 2014, 5, 349–379. [Google Scholar] [CrossRef]
- Sankaran, S.; Khot, L.R.; Espinoza, C.Z.; Jarolmasjed, S.; Santhuvalli, V.R.; Vandemark, G.J.; Miklas, P.N.; Carter, A.H.; Pumphrey, M.O.; Knowles, N.R.; et al. Low-altitude, high-resolution aerial imaging systems for row and field crop phenotyping: A review. Eur. J. Agron. 2015, 70, 112–123. [Google Scholar] [CrossRef]
- Bai, G.; Ge, Y.; Hussain, W.; Baenziger, P.S.; Graef, G. A multi-sensor system for high throughput field phenotyping in soybean and wheat breeding. Comput. Electron. Agric. 2016, 128, 181–192. [Google Scholar] [CrossRef]
- White, J.W.; Andrade-Sanchez, P.; Gore, M.A.; Bronson, K.F.; Coffelt, T.A.; Conley, M.M.; Feldmann, K.A.; French, A.N.; Heun, J.T.; Hunsaker, D.J.; et al. Field-based phenomics for plant genetics research. Field Crop. Res. 2012, 133, 101–112. [Google Scholar] [CrossRef]
- Crain, J.L.; Wei, Y.; Barker, J.; Thompson, S.M.; Alderman, P.D.; Reynolds, M.; Zhang, N.; Poland, J. Development and deployment of a portable field phenotyping platform. Crop Sci. 2016, 56, 965–975. [Google Scholar] [CrossRef]
- Selvaraj, M.G.; Ogawa, S.; Ishitani, M. Root Phenomics-New Windows to understand plant performance and increase crop productivity. J. Plant Biochem. Physiol. 2013, 1, 116. [Google Scholar] [CrossRef]
- Mooney, S.J.; Pridmore, T.P.; Helliwell, J.; Bennett, M.J. Developing X-ray computed tomography to non-invasively image 3-D root systems architecturein soil. Plant Soil. 2012, 352, 1–22. [Google Scholar] [CrossRef]
- Schadt, E.E.; Linderman, M.D.; Sorenson, J.; Lawrence Lee, L.; Garry, P.; Nolan, G.P. Computational solutions to large-scale data management and analysis. Nat. Rev. Genet. 2010, 11, 647–657. [Google Scholar] [CrossRef] [PubMed]
- Knecht, A.C.; Campbell, M.T.; Caprez, A.; Swanson, D.R.; Walia, H. Image Harvest: An open-source platform for high-throughput plant image processing and analysis. J. Exp. Bot. 2016, 67, 3587–3599. [Google Scholar] [CrossRef] [PubMed]
- Wheat Genomic Resources in a Post-Reference Sequence Era. Available online: http://www.wheatinitiative.org/events/wheat-genomic-resources-post-reference-sequence-era (accessed on 20 October 2016).
- Pallotta, M.; Schnurbusch, T.; Hayes, J.; Hay, A.; Baumann, U.; Paull, J.; Langridge, P.; Sutton, T. Molecular basis of adaptation to high soil boron in wheat landraces and elite cultivars. Nature 2014, 514, 88–91. [Google Scholar] [CrossRef] [PubMed]
- Rawat, N.; Pumphrey, M.O.; Liu, S.; Zhang, X.; Tiwari, V.K.; Ando, K.; Trick, H.N.; Bockus, W.W.; Akhunov, E.; Anderson, J.A.; et al. Wheat Fhb1 encodes a chimeric lectin with agglutinin domains and a pore-forming toxin-like domain conferring resistance to Fusarium head blight. Nat. Genet. 2016, 48, 1576–1580. [Google Scholar] [CrossRef] [PubMed]
- Lowry, D.B.; Logan, T.L.; Santuari, L.; Hardtke, C.S.; Richards, J.H.; DeRose-Wilson, L.J.; McKay, J.K.; Sen, S.; Juenger, T.E. Expression quantitative trait locus mapping across water availability environments reveals contrasting associations with genomic features in Arabidopsis. Plant Cell 2013, 25, 3266–3279. [Google Scholar] [CrossRef] [PubMed]
- Aprile, A.; Havlickova, L.; Panna, R.; Marè, C.; Borrelli, G.M.; Marone, D.; Perrotta, C.; Rampino, P.; De Bellis, L.; Curn, V.; et al. Different stress responsive strategies to drought and heat in two durum wheat cultivars with contrasting water use efficiency. BMC Genom. 2013, 14, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Dhar, M.K.; Vishal, P.; Sharma, R.; Kaul, S. Epigenetic dynamics: Role of epimarks and underlying machinery in plants exposed to abiotic stress. Int. J. Genom. 2014, 2014, 187146. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, T.; Seki, M. Epigenetic memory for stress response and adaptation in plants. Plant Cell Physiol. 2014, 55, 1859–1863. [Google Scholar] [CrossRef] [PubMed]
- Bilichak, A.; Kovalchuk, I. Transgenerational response to stress in plants and its application for breeding. J. Exp. Bot. 2016, 67, 2081–2092. [Google Scholar] [CrossRef] [PubMed]
- Cortijo, S.; Wardenaar, R.; Colomé-Tatché, M.; Gilly, A.; Etcheverry, M.; Labadie, K.; Caillieux, E.; Hospital, F.; Aury, J-M.; Wincker, P.; et al. Mapping the epigenetic basis of complex traits. Science 2014, 343, 1145. [Google Scholar] [CrossRef] [PubMed]
- Trethowan, R.M.; Mujeeb-Kazi, A. Novel germplasm resources for improving environmental stress tolerance of hexaploid wheat. Crop Sci. 2008, 48, 1255–1265. [Google Scholar] [CrossRef]
- Becker, S.R.; Byrne, P.F.; Reid, S.D.; Bauerle, W.L.; McKay, J.K.; Haley, S.D. Root traits contributing to drought tolerance of synthetic hexaploid wheat in a greenhouse study. Euphytica 2016, 207, 213–224. [Google Scholar] [CrossRef]
- Sharma, S.; Xu, S.; Ehdaie, B.; Hoops, A.; Close, T.J.; Lukaszewski, A.J.; Waines, J.G. Dissection of QTL effects for root traits using a chromosome arm-specific mapping population in bread wheat. Theor. Appl. Genet. 2011, 122, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Farshadfar, E.; Rahmani, S.; Jowkar, M.M. Evaluation of genetic diversity and QTLs controlling drought tolerance indicators in Agropyron using wheat-Agropyron disomic addition lines. J. Biodivers. Environ. Sci. 2015, 6, 290–299. [Google Scholar]
- Reynolds, M.; Langridge, P. Physiological breeding. Curr. Opin. Plant Biol. 2016, 31, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, M.; Manes, Y.; Izanloo, A.; Langridge, P. Phenotyping approaches for physiological breeding and gene discovery in wheat. Ann. Appl. Biol. 2009, 155, 309–320. [Google Scholar] [CrossRef]
- Reynolds, M.; Foulkes, J.; Furbank, R.; Griffiths, S.; King, J.; Murchie, E.; Parry, M.; Slafer, G. Achieving yield gains in wheat. Plant Cell Environ. 2012, 35, 1799–1823. [Google Scholar] [CrossRef] [PubMed]
- Richards, R.A.; Rebetzke, G.J.; Watt, M.; Condon, A.G.; Spielmeyer, W.; Dolferus, R. Breeding for improved water productivity in temperate cereals: Phenotyping, quantitative trait loci, markers and the selection environment. Funct. Plant Biol. 2010, 37, 85–97. [Google Scholar] [CrossRef]
- Internantional wheat yield partnership. Available online: http://iwyp.org/ (accessed on 18 October 2016).
- Rutkoski, J.E.; Heffner, E.L.; Sorrells, M.E. Genomic selection for durable stem rust resistance in wheat. Euphytica 2010, 179, 161–173. [Google Scholar] [CrossRef]
- Poland, J.; Endelman, J.; Dawson, J.; Rutkoski, J.; Wu, S.; Manes, Y.; Dreisigacker, S.; Crossa, J.; Sánchez-Villeda, H.; Mark Sorrells, M.; et al. Genomic selection in wheat breeding using genotyping-by-sequencing. Plant Genome 2012, 5, 103–113. [Google Scholar] [CrossRef]
- Longin, C.F.H.; Mi, X.; Würschum, T. Genomic selection in wheat: Optimum allocation of test resources and comparison of breeding strategies for line and hybrid breeding. Theor. Appl. Genet. 2015, 128, 1297–1306. [Google Scholar] [CrossRef] [PubMed]
| S. No. | Adaptation Mechanism/Trait | Ease of Use (+/++/+++) | Reference |
|---|---|---|---|
| I. Avoidance | |||
| 1. | Leaf rolling | +++ | [19] |
| 2. | Leaf glaucousness | +++ | [20,21] |
| 3. | Shoot vigor | +++ | [22,23] |
| 4. | Transpirational cooling (cooler canopy) | ++ | [11,24] |
| 5. | Stomatal conductance | + | [10] |
| 6. | Early maturation | +++ | [25] |
| 7. | Membrane stability | + | [10,26] |
| 8. | Green flag leaf area (stay green) | +++ | [27] |
| 9. | Root vigor and architecture | + | [28] |
| II. Tolerance | |||
| 1. | Photosynthetic rate | + | [29,30] |
| 2. | Chlorophyll content | ++ | [31] |
| 3. | ABA accumulation | + | [32] |
| 4. | Osmoprotectant accumulation | + | [33] |
| 5. | Soluble sugar content | + | [34] |
| 6. | Generation of reactive oxygen species (ROS) | + | [35] |
| S. No. | Trait Class/Trait | Number of QTLs/MTAs | ||||
|---|---|---|---|---|---|---|
| IM | GWAS | CGAM | Total | Range of PVE (%) | ||
| I. Agronomic Trait | ||||||
| 1. | Grain yield | 84 | 7 | - | 91 | 02.6–39.9 |
| 2. | Thousand grain weight | 60 | 15 | - | 75 | 00.9–45.2 |
| 3. | Test weight | 13 | - | - | 13 | 03.0–10.0 |
| 4. | Grains m−2 | 36 | - | - | 36 | 03.0–21.4 |
| 5. | Grain width | 02 | - | - | 02 | N/A |
| 6. | Days to heading | 20 | 10 | 01 | 31 | 02.4–42.4 |
| 7. | Days to flowering | 03 | - | - | 03 | 07.2–11.4 |
| 8. | Days to maturity | 12 | 04 | 01 | 17 | 01.7–30.1 |
| 9. | Grain-filling duration | - | 02 | 01 | 03 | 07.1 |
| 10. | Spike density | - | 03 | - | 03 | N/A |
| 11. | Final biomass | - | - | 01 | 01 | 07.9 |
| 12. | Spikes m−2 | 01 | - | 01 | 02 | 06.1–09.1 |
| 13. | Grain weight per spike | 01 | - | 01 | 02 | 06.4–06.7 |
| 14. | Grain number per spike | 02 | 03 | 01 | 06 | 04.5–12.7 |
| 15. | Flag leaf width | - | - | 02 | 02 | 03.6–08.6 |
| 16. | Culm length | 07 | - | - | 07 | 04.1–17.5 |
| 17. | Harvest index | 14 | - | 01 | 15 | 01.7–22.4 |
| 18. | Spike harvest index | 01 | - | - | 01 | 10.1 |
| 19. | Spike dry matter | 05 | - | - | 05 | 06.6–19.1 |
| 20. | Total dry matter | 03 | - | - | 03 | 09.0–11.0 |
| II. Physiological Traits | ||||||
| 1. | Stem reserve mobilization | 03 | - | - | 03 | 21.0–42.2 |
| 2. | Coleoptile length | 68 | 02 | - | 70 | 00.3–65.0 |
| 3. | Canopy temperature | 25 | - | - | 25 | 02.0–13.2 |
| 4. | Normalized difference vegetative index | 06 | 02 | 01 | 09 | 02.0–09.0 |
| 5. | Glaucousness | 04 | - | - | 04 | 04.1–13.1 |
| 6. | Water soluble carbohydrates | 76 | 06 | - | 82 | 01.1–30.0 |
| 7. | Early vigor | 10 | - | - | 10 | 03.0–18.0 |
| 8. | SPAD/chlorophyll content | 82 | - | - | 82 | 02.7–59.1 |
| 9. | Cell membrane stability | 08 | - | - | 08 | 25.0–44.0 |
| 10. | Carbon isotope discrimination | 54 | - | - | 54 | 00.8–27.4 |
| 11. | ABA content | 17 | - | - | 17 | 05.1–30.0 |
| 12. | Leaf green area | - | 03 | 02 | 05 | 04.0–04.2 |
| 13. | Leaf senescence | - | - | 01 | 01 | 6.3 |
| 14. | Relative water content | 01 | 03 | - | 04 | 06.5–17.8 |
| 15. | Osmotic adjustment | 02 | - | - | 02 | N/A |
| 16. | Osmotic potential | 10 | - | - | 10 | 02.7–08.9 |
| 17. | Photosynthetic active radiation | 14 | - | - | 14 | N/A |
| 18. | Transpiration | 14 | - | - | 14 | N/A |
| 19. | Leaf rolling | 10 | - | - | 10 | 01.6–07.8 |
| III. Root and Related Traits | ||||||
| 1. | Root length | 11 | - | - | 11 | 05.0–15.6 |
| 2. | Total root biomass | 02 | - | - | 02 | 09.4–10.8 |
| 3. | Root number | 03 | - | - | 03 | 07.3–07.8 |
| 4. | Root dry weight | 05 | - | - | 05 | 03.5–07.5 |
| 5. | Root to shoot ratio | 02 | - | - | 02 | 08.0–11.0 |
| Total | 691 | 60 | 14 | 763 | ||
| S. No. | Trait/QTL | Linked Marker | Position (cM) | Env. a | PVE (R2) b | Reference |
|---|---|---|---|---|---|---|
| I. Agronomic Traits | ||||||
| 1. Grain Yield | ||||||
| (a) | qGYWD.3B.2 | Xgpw7774 | 97.6 | 4/7 | 19.6 | [45] |
| (b) | 4A | Xwmc420 | 90.4 | Mean/2 | 20.0 | [46] |
| (c) | 4A-a | Xgwm397 | 06.0 | 5/7 | 23.9 | [24] |
| (d) | Qyld.csdh.7AL | Xgwm322 | 155.9 | 11/21 | 20.0 * | [47] |
| 2. 1000-Grain Weight | ||||||
| (a) | 3B | Xbarc101 | 86.1 | Mean/2 | 45.2 | [48] |
| (b) | QTgw-7D-b | XC29-P13 | 12.5 | 10/11 | 21.9 | [49] |
| 3. Days to Heading | ||||||
| (a) | QDh-7D.b | XC29-P13 | 12.5 | 11/11 | 22.7 | [49] |
| (b) | QHd.idw-2A.2 | Xwmc177 | 46.1 | 13/16 | 32.2 | [50] |
| 4. Days to Maturity | ||||||
| (a) | QDm-7D.b | X7D-acc/cat-10 | 2.7 | 10/11 | 22.7 | [49] |
| II. Physiological Traits | ||||||
| 1. Stem Reserve Mobilization | ||||||
| (a) | QSrm.ipk-2D | Xgwm249a | 142.0 | 2/2 | 42.2 | [51] |
| (b) | QSrm.ipk-5D | Xfbb238b | 19.0 | 2/2 | 37.5 | [51] |
| (c) | QSrm.ipk-7D | Xfbb189b | 338.0 | 2/2 | 21.0 | [51] |
| 2. Water Soluble Carbohydrate | ||||||
| (a) | QWsc-c.aww-3A | Xwmc0388A | 64.9 | 2/2 | 19.0 | [52] |
| 3. SPAD/Chlorophyll Content | ||||||
| (a) | Qchl.ksu-3B | Xbarc68 | 67.2 | 2/3 | 59.1 | [53] |
| MQTL | Chr. | Linked Marker | Traits for Individual QTL Representing MQTL | Co-Localized Candidate Gene ID a | Predicted Function |
|---|---|---|---|---|---|
| MQTL2 | 1A | Xwmc11 | CID, CL, KN, SG, WSC, YLD | 1. Ta.11441.3 | 1. ADP-ribosylation factor1 |
| 2. Ta.24298.1 | 2. Prolamin, 2, 26 kDa globulin, Alpha globulin | ||||
| 3. Ta.1257.2 | 3. Prolamin subfamily 2 | ||||
| MQTL3 | 1A | Xwmc51 | PS, WSC | - | - |
| MQTL11 | 2A | Xwmc296 | Bio, CID, CL, GF, HI, WSC, WS | - | - |
| MQTL14 | 2B | Xwmc489 | HI, PH, KN, SG | - | - |
| MQTL16 | 2B | Xbarc7 | BIO, CL, HI, WS | - | - |
| MQTL18 | 2B | Xgwm47 | PH, SG, WSC, YLD | 1. Ta.8144.1 | 1. Gamma-SNAP |
| 2. Ta.9253.1 | 2. SIT4 phosphatase | ||||
| MQTL21 | 2D | Xwmc601 | CID, CL, WSC | - | - |
| MQTL22 | 2D | Xgwm539 | CID, SG, TKW, YLD | - | - |
| MQTL23 | 3A | Xwmc11 | TKW, WS | - | - |
| MQTL29 | 3D | Xgwm314 | CL, PH, PS, SD, TKW, YLD | - | - |
| MQTL42 | 5B | Xwmc73 | PH, YLD | 1. Ta.9194.1 | 1. L-ascorbate:Na symporter |
| MQTL46 | 5D | Xgwm358 | CL, PS, WSC | - | - |
| MQTL50 | 6A | Xgwm427 | CID, TKW | - | - |
| MQTL51 | 6B | Xgwm508 | HI, KN, WS, YLD | 1. Ta.13551.1 | 1. SurE |
| 2. Ta.5227.2 | 2. S-adenosylmethionine synthetase 1 | ||||
| MQTL53 | 6B | Xbrac198 | CL, WSC | - | - |
| MQTL56 | 6D | Xwmc773 | CID, YLD | - | - |
| MQTL61 | 7B | Xgwm400 | HD, BIO, CID, HI, MD, WS, YLD | - | - |
| MQTL64 | 7D | Xcfd66 | PS, WSC | - | - |
| MQTL66 | 7D | Xwmc659 | PS | 1. Ta.1055.1 | 1. Catalase isozyme A |
| S. No. | Trait Class/Trait | QTL × QTL Pairs | PVE (%) Range | Reference |
|---|---|---|---|---|
| I. Agronomic Trait | ||||
| 1. | Grain yield | 04 | 0.51 | [45,50,66] |
| 2. | Thousand grain weight | 24 | 0.59–8.26 | [45,66,67,68] |
| 3. | Days to flowering | 12 | 0.30–1.40 | [45,67,69] |
| II. Physiological Traits | ||||
| 1. | Coleoptile length | 04 | 0.50–2.70 | [70] |
| 2. | Water soluble carbohydrates | 24 | 0.84–5.61 | [68] |
| 3. | Carbon isotope discrimination | 02 | N/A | [66] |
| 4. | SPAD/chlorophyll content | 38 | 1.08–3.29 | [71] |
| Total | 108 | 0.30–8.26 | ||
| Trait | QTL_i QTL/Chromosome | Associated Marker; Postion (cM) | QTL-j QTL/Chromosome | Associated Marker; Position (cM) | PVE |
|---|---|---|---|---|---|
| TGW | 1. QTgwg.cgb-1B | P3622-280; 0 | QTgwg.cgb-5A | Xwmc524; 0 | 5.16 |
| 2. QTgwg.cgb-4A.2 | CWM145; 9 | QTgwg.cgb-4A.3 | XP4232-260; 3 | 8.26 | |
| 3. QTgwg.cgb-6A.2 | Xgwm334; 0 | QTgwg.cgb-6A.3 | XP3474-260; 2 | 5.79 | |
| 4. QTgwm.cgb-2B.1 | P6411-216; 0 | QTgwm.cgb-7B.4 | Xwmc276; 1 | 6.61 | |
| WSC | 1. QSwscg.cgb-2B | WMC441; 5 | QSwscg.cgb-6B | Xwmc182; 0 | 5.61 |
| S. No. | Marker Name | Chr. | Trait | Candidate Gene |
|---|---|---|---|---|
| 1. | Tdurum_ contig80278_ 250 | 1AL | GY | Galactosylgalactos ylxylosylprotein 3-beta-Glucuronosyl transferase 1 |
| 2. | Excalibur_ c8052_541 | 1BS | DTH | e3 ubiquitin-protein ligase herc2 |
| 3. | RAC875_rep_ c77617_1454 | 2AL | TGW | Serine threonine-protein phosphatase 6 Regulatory subunit 3-like isoform x1 |
| 4. | BS00022025_ 51 | 3BL | TGW | Glycosyltransferase- like protein |
| 5. | RAC875_ c23144_1560 | 4BL | GY | Upf0202 protein at1g10490-like |
| 6. | tplb0024a09_ 742 | 7DS | GY | Rna polymerase ii transcription partial |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gupta, P.K.; Balyan, H.S.; Gahlaut, V. QTL Analysis for Drought Tolerance in Wheat: Present Status and Future Possibilities. Agronomy 2017, 7, 5. https://doi.org/10.3390/agronomy7010005
Gupta PK, Balyan HS, Gahlaut V. QTL Analysis for Drought Tolerance in Wheat: Present Status and Future Possibilities. Agronomy. 2017; 7(1):5. https://doi.org/10.3390/agronomy7010005
Chicago/Turabian StyleGupta, Pushpendra Kumar, Harindra Singh Balyan, and Vijay Gahlaut. 2017. "QTL Analysis for Drought Tolerance in Wheat: Present Status and Future Possibilities" Agronomy 7, no. 1: 5. https://doi.org/10.3390/agronomy7010005







