1. Introduction
Brachiaria grass is one of the most important tropical grasses distributed throughout the tropics, especially in Africa [
1]. The genus
Brachiaria consists of about 100 documented species of which 7 perennial species of African origin have been used for pasture production in South America, Asia, South Pacific, and Australia [
2]. It has high biomass production potential and produces nutritious herbage resulting in increased livestock productivity [
3,
4].
Brachiaria is adapted to drought and low-fertility soils, sequesters carbon through its large root system, enhances nitrogen use efficiency, and subsequently minimizes eutrophication and greenhouse gas emissions [
5,
6,
7,
8].
Brachiaria plays important roles in soil erosion control and ecological restoration.
Brachiaria species have been an important component of sown pastures in humid lowlands and savannas of tropical America, with current estimated acreage of 99 million hectares in Brazil alone [
9].
In Africa, the evaluations of
Brachiaria species for pasture improvement started during the 1950s. These researches focused on
B. brizantha,
B. decumbens,
B. mutica, and
B. ruziziensis for forage production, agronomy (establishment, drought, cutting intervals, and fertilizers), compatibility with herbaceous and tree legumes, nutritive values, and their benefit to ruminant production. These studies concluded the suitability and broader adaptation of several
Brachiaria species to different agroecological zones in Africa [
10]. However, these practices were not widespread because of ample communal grazing lands, limited realization on roles of sown pasture in the livestock production, subsistence animal farming, and low government priority to pasture development. Recently, the mounting demand for livestock products in Africa has renewed interest of farmers, researchers, development agencies, and government organizations on forages, particularly in species with good adaptability to climate change such as Brachiaria grass. Therefore, there has been multiple repatriations of Brachiaria grass to Africa in the form of hybrids and improved landraces [
11,
12]. These materials have shown positive performance in terms of biomass production, improved forage availability and livestock productivity in Kenya and Rwanda. These results have revealed
Brachiaria as an ideal forage option for the livestock farmers in East Africa.
Despite high popularity, the Brachiaria acreage in Africa is low and relies on a few varieties that were developed for tropical Americas and Australia. Within a short period of introduction, some of these varieties have shown susceptibility to pests and diseases, raising question on the expansion of Brachiaria acreage in Africa with these varieties. There is therefore a need for an Africa-based
Brachiaria improvement program to develop varieties that are tolerant to biotic and abiotic stress for different environmental conditions. Germplasms of broad genetic base is the prerequisite for any crop improvement. The best approach to increase genetic variations in apomictic species such as
Brachiaria is tapping natural variations from the center of diversity. Since the 1950s, multiple missions were undertaken in Africa to collect
Brachiaria germplasms, with a current inventory of 987 accessions of 33 known
Brachiaria species [
13]. Considering distribution of
Brachiaria in Africa and size of the continent, the number of samples available in collection is definitely non-exhaustive and warrants further collection efforts. However, the existence of these genetic resources in Africa is continuously threatened by overgrazing and clearing of vegetation for crop production and nonagricultural uses and adverse effects of climate change.
Kenya is located within a region that represents a center of diversity for genus Brachiaria. Therefore, high natural variation is expected among Brachiaria populations in Kenya. This study aimed to create a collection of local Brachiaria ecotypes in Kenya, assess their genetic diversity using microsatellite markers, and examine their genetic relationships with eight commercial cultivars. The study will broaden geographical coverage and/or genetic base of the global Brachiaria collection and provide invaluable information for Brachiaria improvement and conservation programs.
3. Discussion
The genetic complexity, primarily apomictic mode of reproduction, and abundant natural variations in Africa urge for a two-pronged approach (selection and breeding) for improving Brachiaria grass in Africa. All-inclusive germplasm base with documented variations are prerequisite for the effective breeding programs. This study collected 79 Brachiaria ecotypes in Kenya and documented their genetic variations using microsatellite markers.
The PIC values for 22 SSR markers averaged to 0.5825, suggesting markers were capable of differentiating 79 Kenyan
Brachiaria ecotypes. The PIC value in this study is within the range reported by Silva et al. [
14], Jungmann et al. [
15], and Vigan et al. [
16], but was lower than that found by Jungmann et al. [
17] and Pessoa-Filho et al. [
18]. Similarly, the average numbers of allele detected per loci (5.45) was in the range reported by Silva et al. [
14], Jungmann et al. [
15], and Vigan et al. [
16], but was about half and one-third of that reported by Jungmann et al. [
17] and Pessoa-Filho et al. [
18], respectively. However, these comparisons between studies may not be conclusive due to differences in types and number of germplasms and markers used among studies.
The analysis of the distributions of alleles across populations is important for explaining genetic diversity and population relationships [
19]. Private alleles are important in plant breeding and conservation as they are present only in a single population among a broader collection of populations [
20]. Five ecotypes populations of Kenya were different for private alleles, with the highest number of private alleles in the ILRI population and the least in the Kiminini population. Such variations in the private alleles among populations most likely was the effect of the number of individuals per population, which ranged from 3 to 60 individuals. Although no information was available on species composition of each population, it is likely the presence of multiple species resulting in a high number of private alleles in some populations. Irrespective of populations,
HO was higher than
HE, indicating mixing of previously isolated populations. This is consistent with the human involvement in moving planting materials and outcrossing nature of some
Brachiaria species, for example,
B. ruziziensis.
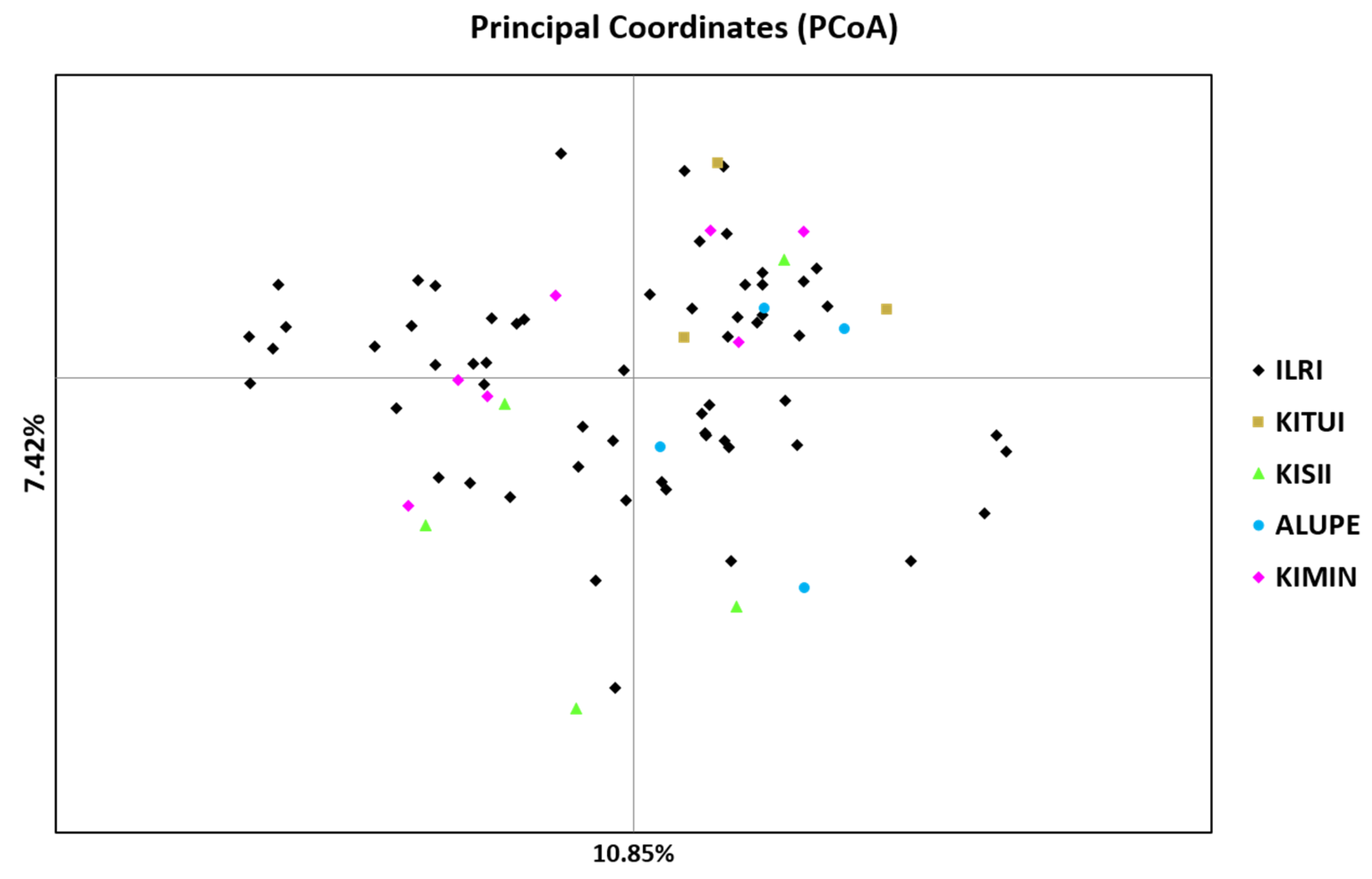
The study population varied in genetic distance and genetic identity coefficients. The highest genetic distance between Alupe and Kitui populations can be explained by the wider geographical distance between these two locations (675 km), but the genetic distance between other populations could not be associated to geographical proximity. Reports are available on forage research, including seed production of
B. ruziziensis in Kitale, Kenya [
21,
22], and involvement of Kenya Agricultural Research Institute and Kenya Seed Company in the past in production and trading of
B. ruziziensis seeds [
23]. It is likely that some of these
Brachiaria seeds might have reached farmers’ fields and other research stations in Kenya, including the ILRI, and afterwards naturalized in the wild. If this hold true, a low genetic distance (0.307) between the ILRI and Kiminini (20 km away from Kitale) populations could be because of shared genetic materials in early days.
The contribution of within-individual difference to total variation was 81%, whereas among-the-individual and among-populations differences contributed 17% and 2%, respectively (
Table 5). These observations were in agreement with Vigna et al. [
16] and Pessoa-Filho et al. [
18], who reported high contributions of within-the-accession/individual differences to total variation in
B. brizantha (84%) and
B. ruziziensis (88%) populations. Similarly, Garcia et al. [
24] and Azevedo et al. [
25] reported 73% and 65% of total variation attributed to within species or cluster, respectively. However, Jungmann et al. [
26] reported 44% of the variation in
B. humidicola accessions as being due to the subdivision of the germplasms into five groups. The
FST and effective number of migrants per generation (
Nm) values of 0.021 and 11.580 indicated a relatively low genetic differentiation among populations [
27] and relatively high level of gene flow among the Kenyan ecotypes populations [
28], respectively. A low genetic differentiation among the study populations could be associated with apomictic mode of reproduction, variable ploidy causing meiotic anomalies leading to reduced pollen fertility, and dispersal of seeds by migratory herbivorous and human activities such as hay transportation for feeding animals [
16,
26,
29,
30,
31,
32]. Polyploid plants are effective colonizers that can occupy pioneer habitats and generate individuals that are able to exploit new niches or outcompete progenitor species, whereas apomictic polyploid plants can fix heterosis [
16,
26,
30].
This study is an effort to build a collection of
Brachiaria ecotypes in Kenya and identify the potential values of these genetic resources in the
Brachiaria breeding program. It is important to acknowledge some methodological limitations of this study while inferring population genetic parameters such as unequal and/or small sample sizes (3–60 individuals per population), unknown species and ploidy status of ecotypes, and dominant scoring scheme used in recording SSR fragments. The current
Brachiaria taxonomy is far from satisfactory and the problem of generic identity, and species composition across entire taxa needs to be undertaken [
1,
22]. Application of robust genetic markers and bioinformatics procedures in genetic analysis of
Brachiaria spp. have been constrained by a limited understanding of
Brachiaria genetics, cytogenetic and reproductive biology, and unavailability of reference genome. The agricultural and environmental importance of
Brachiaria has recently spurred several studies on
Brachiaria, including sequencing of
B. ruziziensis genome. Therefore, Kenyan ecotypes collected in this study should be conserved and characterized further with the advent of new genomics and bioinformatics tools developed for species with complex genome.
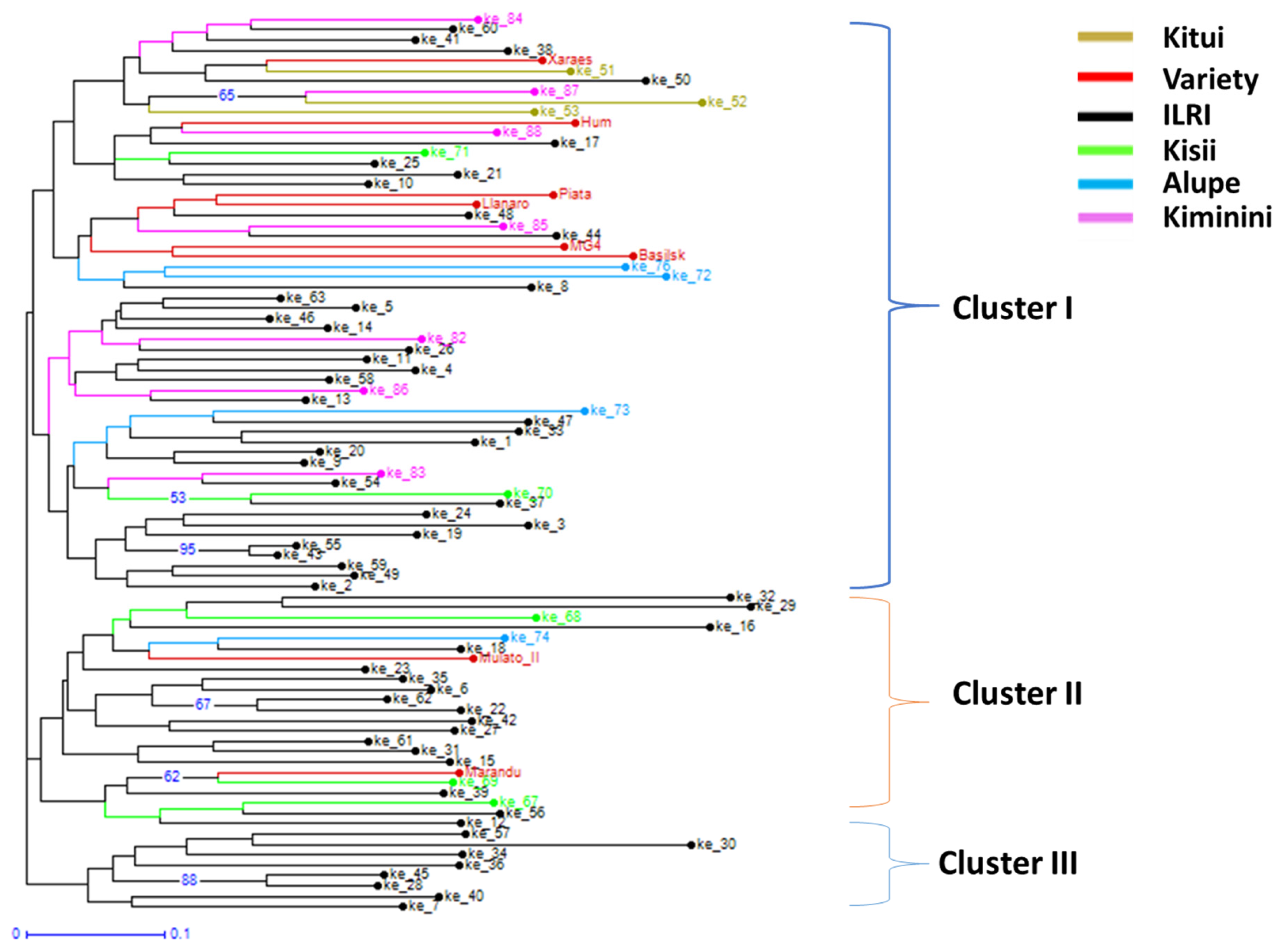
This is among the very first studies of this century in sub-Saharan Africa that involved collection of local Brachiaria ecotypes from different parts of Kenya and examination of their genetic differences using microsatellite markers. The genetic diversity data revealed that ecotypes, though representing a few locations of Kenya, contained much more diversity than currently available 8 improved Brachiaria varieties, which represent three species (B. brizantha, B. decumbens, and B. humidicola) and three-way cross-hybrid Mulato II (B. brizantha × B. decumbens × B. ruziziensis). These results clearly indicate a need for (I) further collection of local ecotypes in Kenya and other east and central African countries that represent center of diversity of Brachiaria species to enrich the Brachiaria genepool in the gene bank collections; (II) genetic characterization of local ecotypes and currently available gene bank materials to understand diversity and ascertain the need for further collection; and (III) morphological characterization of available genetic resource to identify/develop varieties suitable for different production environments.