Drought Stress-Related Physiological Changes and Histone Modifications in Barley Primary Leaves at HSP17 Gene
Abstract
:1. Introduction
2. Results
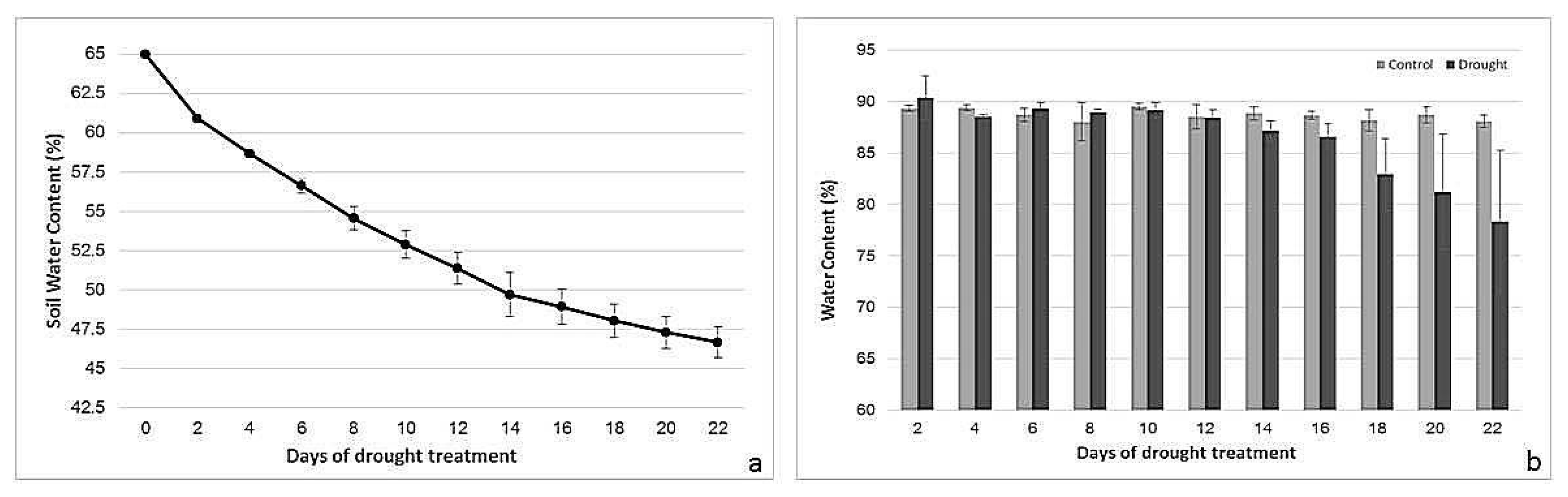
2.1. Drought Treatment: Soil Water Content (SWC) and Leaf Water Content (LWC)
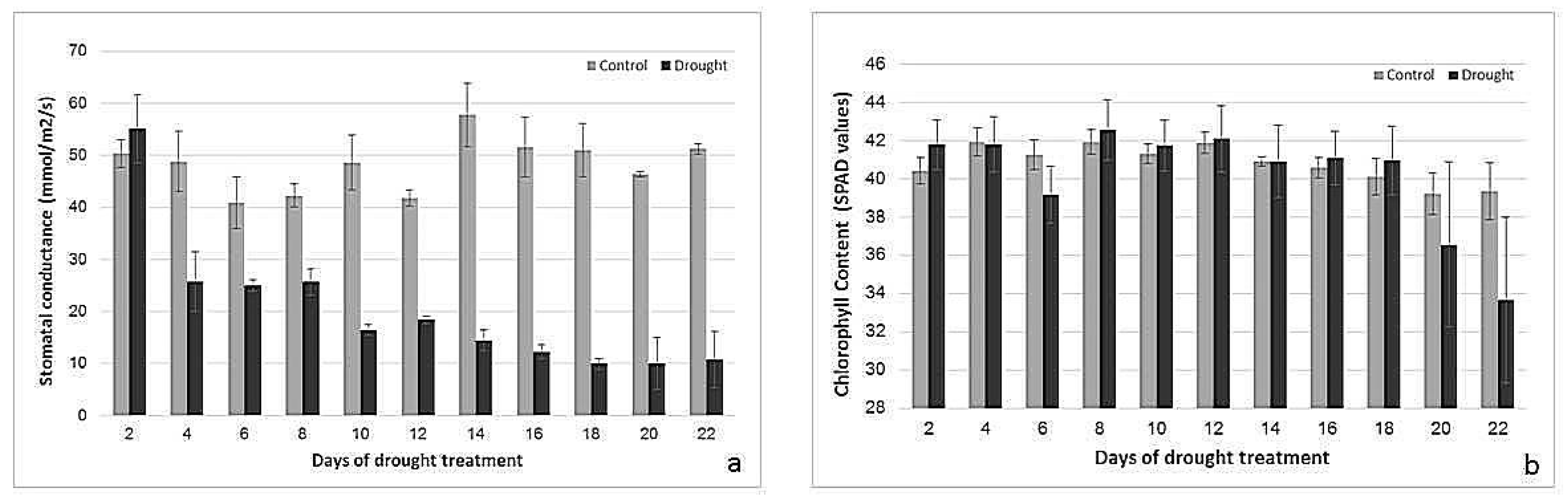
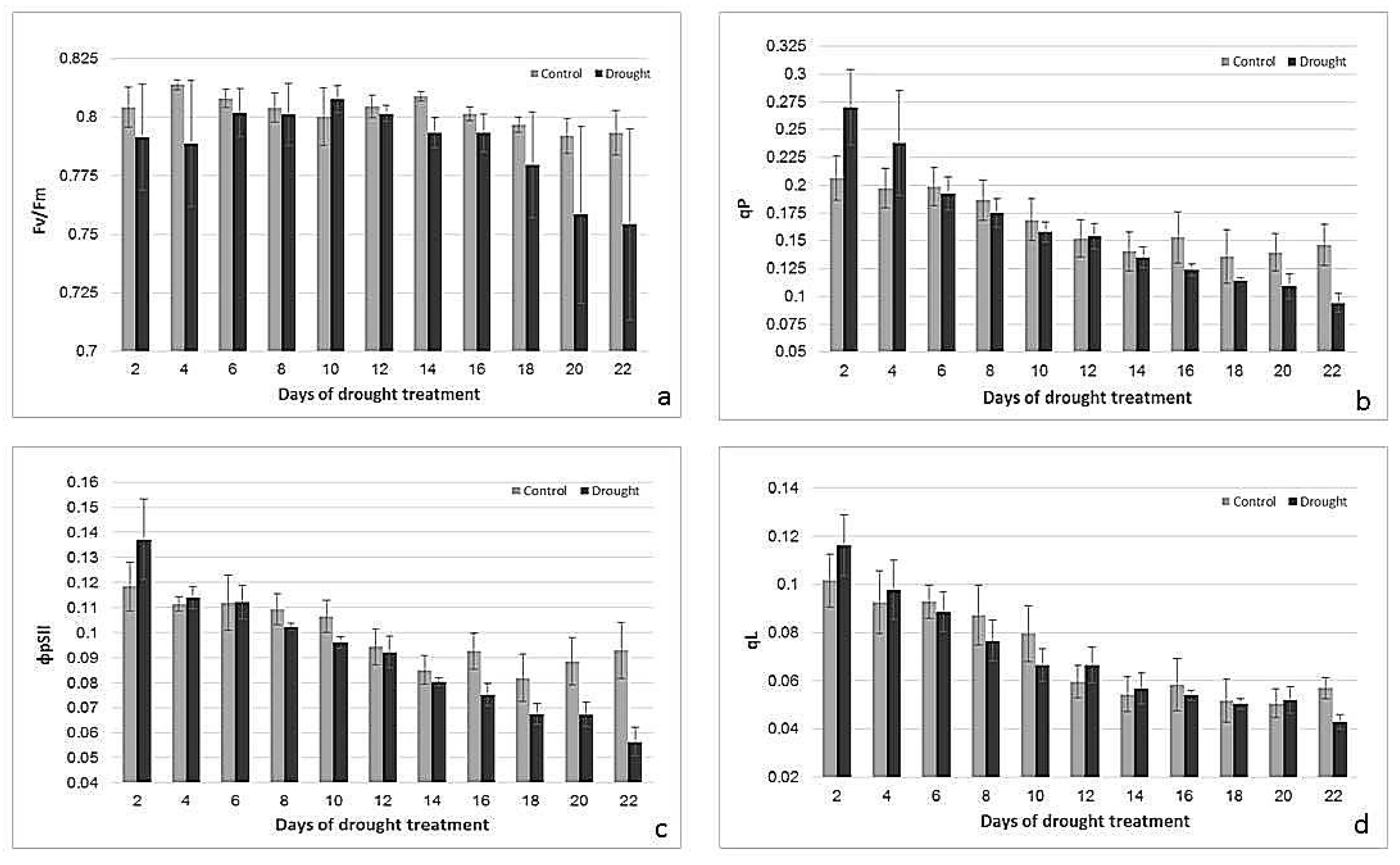
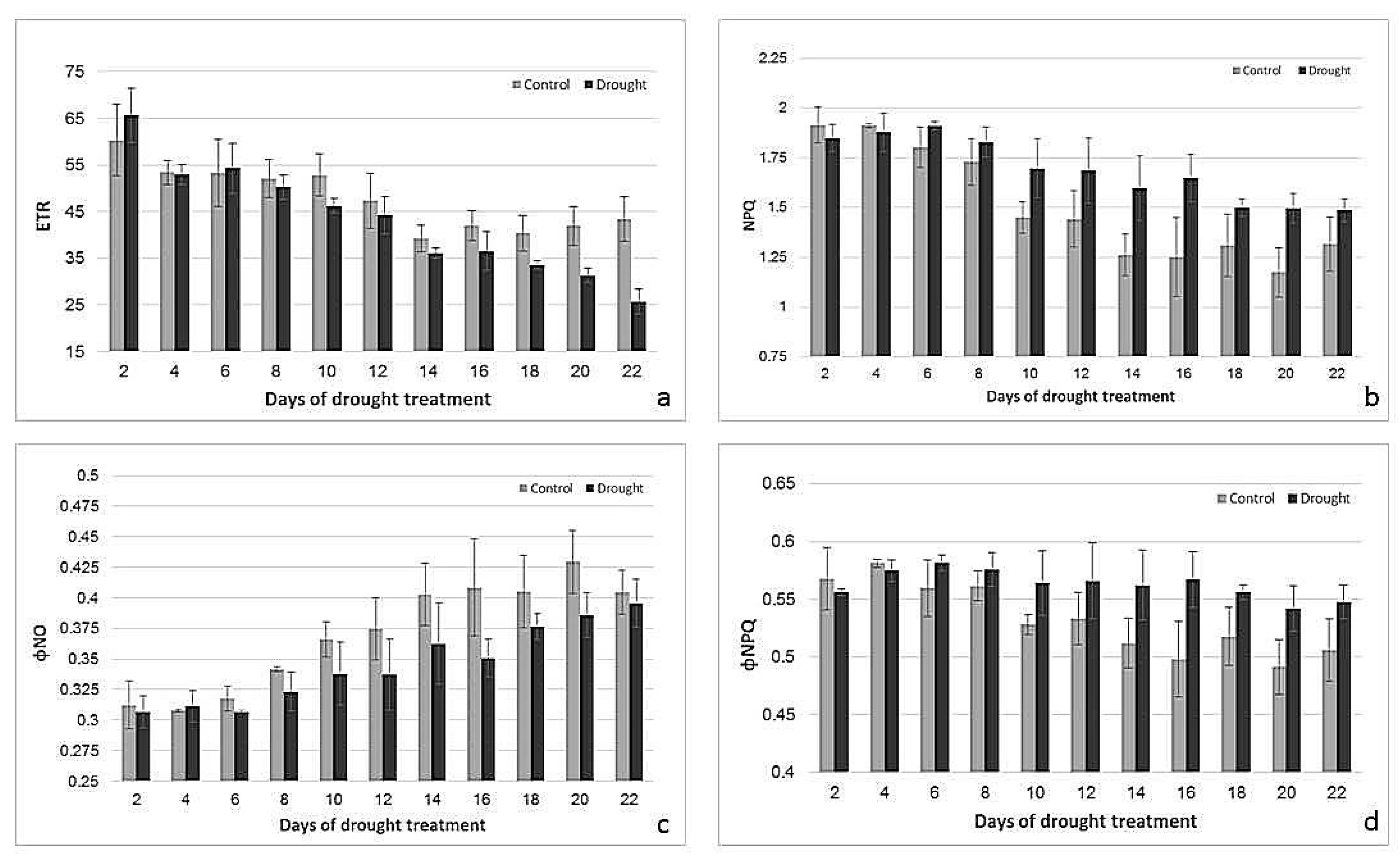
2.2. Effects of Drought Treatment on Stomatal Conductance, Chlorophyll Content, and Photosynthetic Parameters
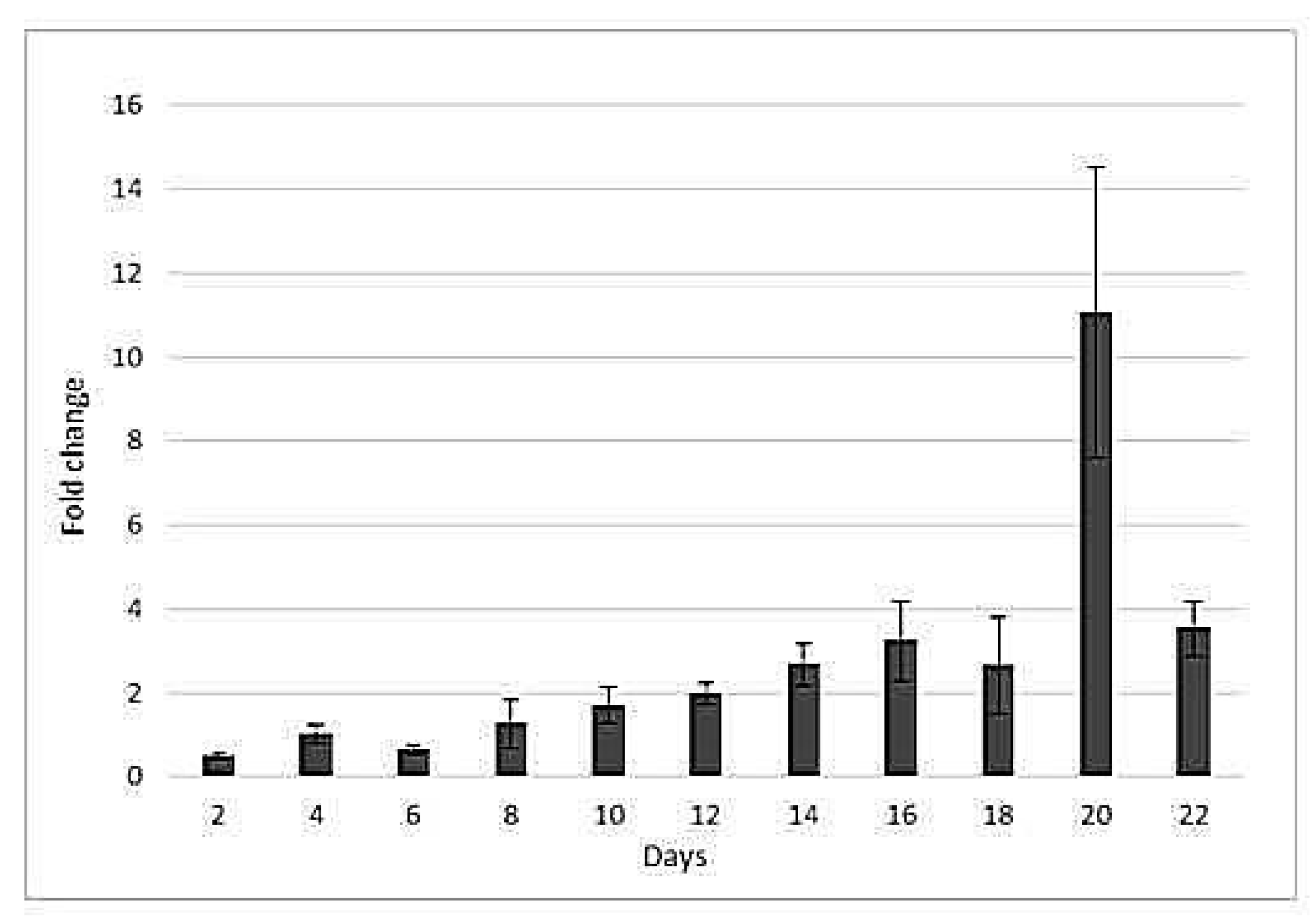
2.3. Drought-Responsive Expression of HSP17Gene
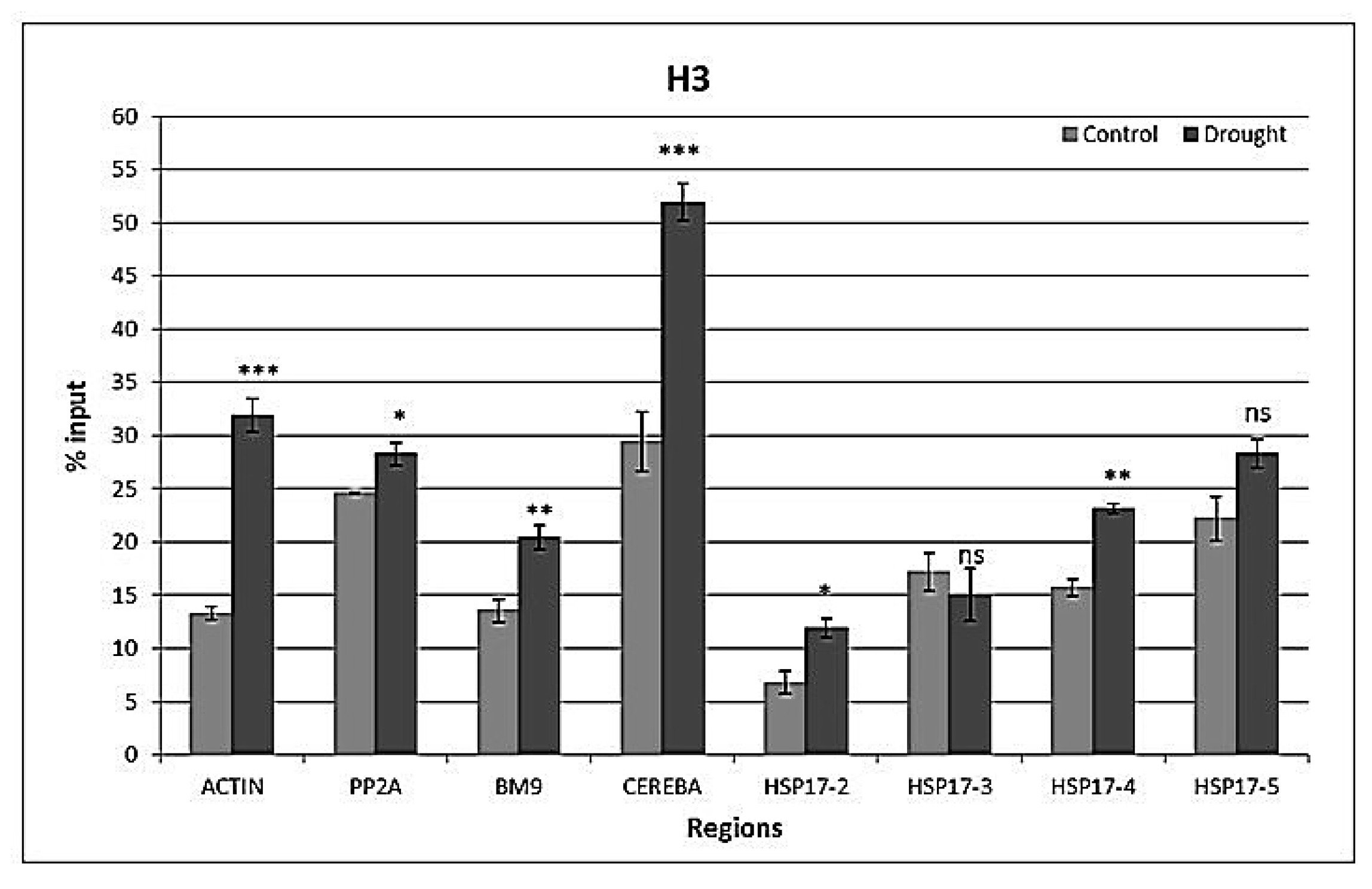
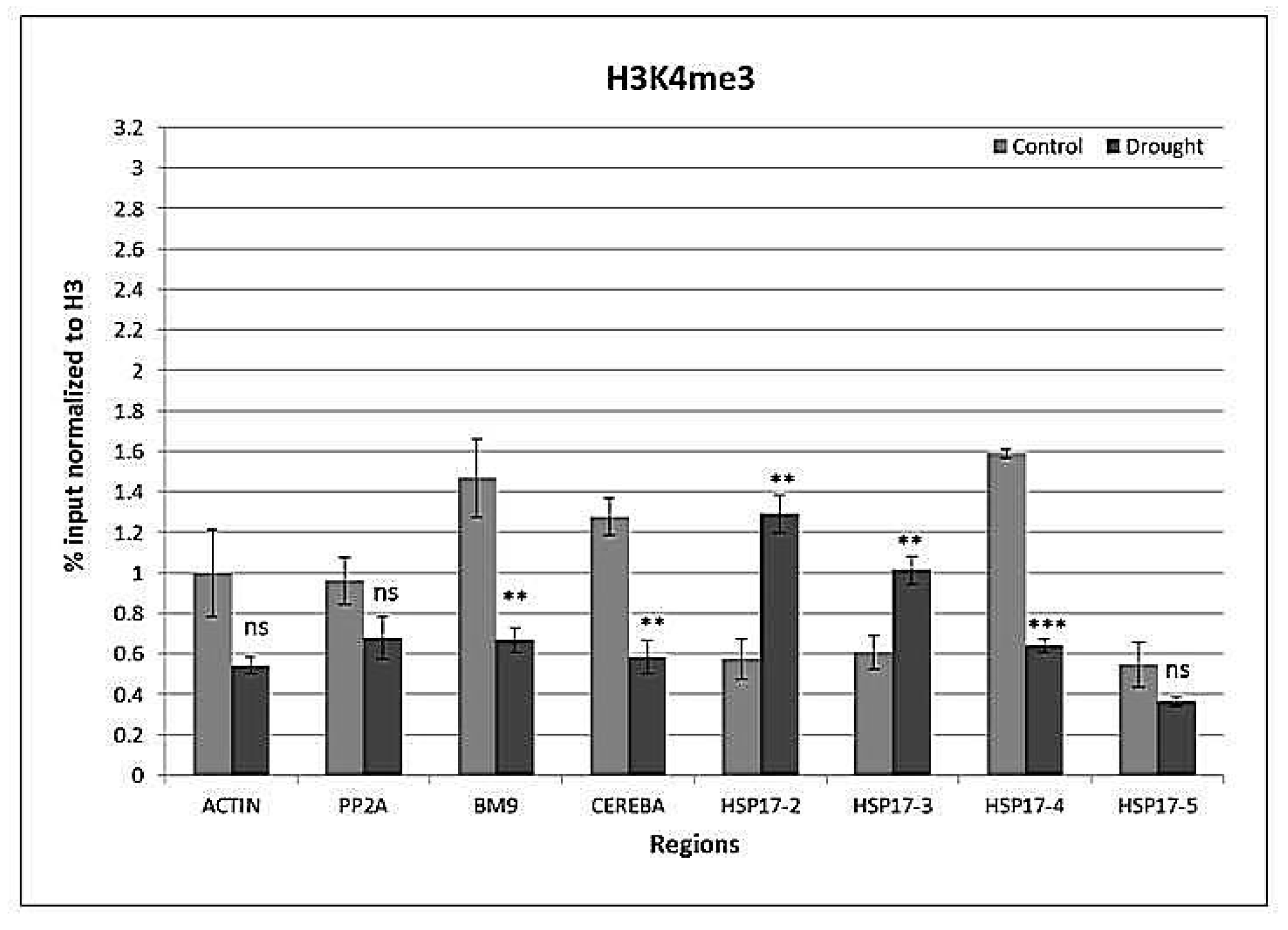
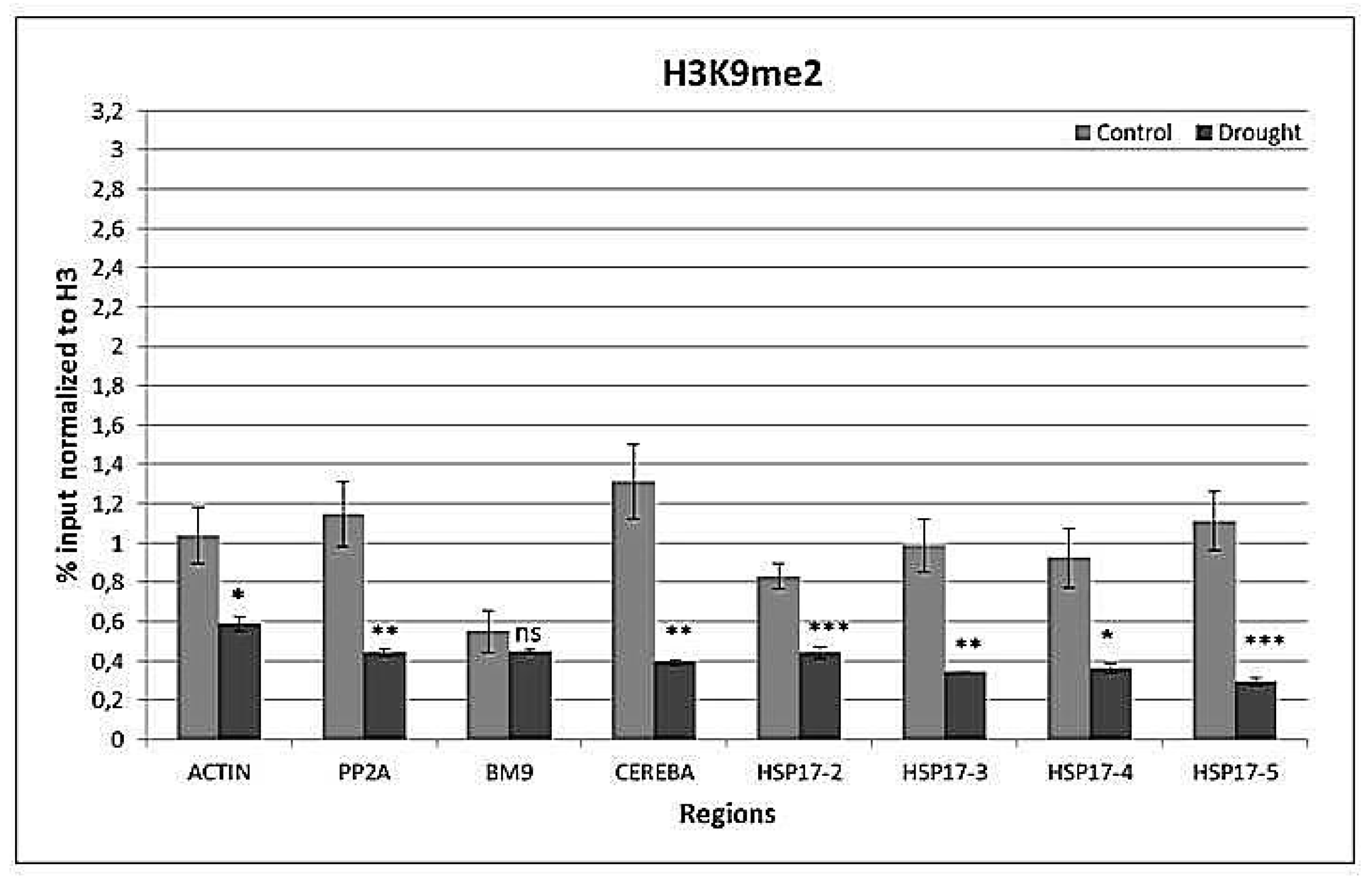
2.4. Drought-Responsive Histone Modifications at HSP17 and Control Genes
3. Discussion
3.1. Physiological Responses to Drought
3.2. hsp17 Gene Expression
3.3. Drought-Responsive Alterations of Histone Modification Patterns
4. Material and Methods
4.1. Plant Material and Growth Conditions
4.2. Physiological Measurements
4.3. RNA Isolation and cDNA Synthesis
4.4. Expression Analysis by Real-Time PCR
4.5. ChIP
4.6. Data Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Mahajan, S.; Tuteja, N. Cold, salinity and drought stresses: An overview. Arch. Biochem. Biophys. 2005, 444, 139–158. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, S.; Sawant, K. Drought stress adaptation: Metabolic adjustment and regulation of gene expression. Plant Breed. 2013, 132, 21–32. [Google Scholar] [CrossRef]
- Maxwell, K.; Johnson, G.N. Chlorophyll fluorescence—A practical guide. J. Exp. Bot. 2000, 51, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Murchie, E.H.; Lawson, T. Chlorophyll fluorescence analysis: A guide to good practice and understanding some new applications. J. Exp. Bot. 2013, 64, 3983–3998. [Google Scholar] [CrossRef] [PubMed]
- Athar, H.; Ashraf, M. Photosynthesis under drought stress. In Photosynthesis, 2nd ed.; Pessarakli, M., Ed.; CRC Press: New York, NY, USA, 2005; pp. 795–810. [Google Scholar]
- De Carvalho, M.H.C. Drought stress and reactive oxygen species. Plant Signal Behav. 2008, 3, 156–165. [Google Scholar] [CrossRef]
- Chaves, M.M.; Flexas, J.; Pinheiro, C. Photosynthesis under drought and salt stress: Regulation mechanisms from whole plant to cell. Ann. Bot. 2009, 103, 551–560. [Google Scholar] [CrossRef] [PubMed]
- Guo, P.; Baum, M.; Grando, S.; Ceccarelli, S.; Bai, G.; Li, R.; von Korff, M.; Varshney, R.K.; Graner, A.; Valkoun, J. Differentially expressed genes between drought-tolerant and drought-sensitive barley genotypes in response to drought stress during the reproductive stage. J. Exp. Bot. 2009, 60, 3531–3544. [Google Scholar] [CrossRef] [PubMed]
- Behringer, D.; Zimmermann, H.; Ziegenhagen, B.; Liepelt, S. Differential gene expression reveals candidate genes for drought stress response in Abies alba (Pinaceae). PLoS ONE 2015, 10, e0124564. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Fan, C.; Li, Y.; Zhang, J.; Sun, J.; Chen, Y.; Tian, C.; Su, X.; Lu, M.; Liang, C.; et al. Effects of drought and salt-stresses on gene expression in Caragana korshinskii seedlings revealed by RNA-seq. BMC Genomics 2016, 17, 200. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Zhu, J.K. Epigenetic regulation of stress responses in plants. Curr. Opin. Plant Biol. 2009, 12, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Han, S.K.; Wagner, D. Role of chromatin in water stress responses in plants. J. Exp. Bot. 2014, 65, 2785–2799. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, T.; Seki, M. Epigenetic memory for stress response and adaptation in plants. Plant Cell Physiol. 2014, 55, 1859–1863. [Google Scholar] [CrossRef] [PubMed]
- Filek, M.; Łabanowska, M.; Koscielniak, J.; Biesaga-Koscielniak, J.; Kurdziel, M.; Szarejko, I.; Hartikainen, H. Characterization of barley leaf tolerance to drought stress by chlorophyll fluorescence and electron paramagnetic resonance studies. J. Agron. Crop Sci. 2015, 201, 228–240. [Google Scholar] [CrossRef]
- Ferdous, J.; Sanchez-Ferrerov, J.C.; Langridge, P.; Milne, L.; Chowdhury, J.; Brien, C.; Tricker, P.J. Differential expression of microRNAs and potential targets under drought stress in barley. Plant Cell Environ. 2017, 40, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Rollins, J.A.; Habte, E.; Templer, S.E.; Colby, T.; Schmidt, J.; von Korff, M. Leaf proteome alterations in the context of physiological and morphological responses to drought and heat stress in barley (Hordeum vulgare L.). J. Exp. Bot. 2013, 64, 3201–3212. [Google Scholar] [CrossRef] [PubMed]
- Templer, S.E.; Ammon, A.; Pscheidt, D.; Ciobotea, O.; Schuy, C.; McCollum, C.; Sonnewald, U.; Hanemann, A.; Förster, J.; Ordon, F.; et al. Metabolite profiling of barley flag leaves under drought and combined heat and drought stress reveals metabolic QTLs for metabolites associated with antioxidant defense. J. Exp. Bot. 2017, 68, 1697–1713. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.M.; To, T.K.; Ishida, J.; Morosawa, T.; Kawashima, M.; Matsui, A.; Toyoda, T.; Kimura, H.; Shinozaki, K.; Seki, M. Alterations of lysine modifications on the histone H3 N-tail under drought stress conditions in Arabidopsis thaliana. Plant Cell Physiol. 2008, 49, 1580–1588. [Google Scholar] [CrossRef] [PubMed]
- Zong, W.; Zhong, X.; You, J.; Xiong, L. Genome-wide profiling of histone H3K4-tri-methylation and gene expression in rice under drought stress. Plant Mol. Biol. 2013, 81, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, R.M.; Ricardi, M.M.; Iusem, N.D. Epigenetic marks in an adaptive water stress-responsive gene in tomato roots under normal and drought conditions. Epigenetics 2013, 8, 864–872. [Google Scholar] [CrossRef] [PubMed]
- Kwon, C.S.; Lee, D.; Choi, G.; Chung, W.I. Histone occupancy-dependent and -independent removal of H3K27 trimethylation at cold-responsive genes in Arabidopsis. Plant J. 2009, 60, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Chwialkowska, K.; Nowakowska, U.; Mroziewicz, A.; Szarejko, I.; Kwasniewski, M. Water-deficiency conditions differently modulate the methylome of roots and leaves in barley (Hordeum vulgare L.). J. Exp. Bot. 2016, 67, 1109–1121. [Google Scholar] [CrossRef] [PubMed]
- Papaefthimiou, D.; Tsaftaris, A.S. Characterization of a drought inducible trithorax-like H3K4 methyltransferase from barley. Biol. Plant. 2012, 56, 683–692. [Google Scholar] [CrossRef]
- Papaefthimiou, D.; Tsaftaris, A.S. Significant induction by drought of HvPKDM7-1, a gene encoding a jumonji-like histone demethylase homologue in barley (H. vulgare). Acta Physiol. Plant. 2012, 34, 1187–1198. [Google Scholar] [CrossRef]
- Ahmed, I.M.; Nadira, U.A.; Bibi, N.; Zhang, G.; Wu, F. Tolerance to combined stress of drought and salinity in barley. In Combined Stresses in Plants Physiological, Molecular, and Biochemical Aspects; Mahalingam, R., Ed.; Springer: Basel, Switzerland, 2015; pp. 93–121. ISBN 978-3-319-07899-1. [Google Scholar]
- Munne-Bosch, S.; Alegre, L. Die and let live: Leaf senescence contributes to plant survival under drought stress. Funct. Plant Biol. 2004, 31, 203–216. [Google Scholar] [CrossRef]
- Lim, P.O.; Kim, H.J.; Nam, H.G. Leaf senescence. Annu. Rev. Plant Biol. 2007, 58, 115–136. [Google Scholar] [CrossRef] [PubMed]
- Prochazkova, D.; Sairam, R.K.; Srivastava, G.C.; Singh, D.V. Oxidative stress and antioxidant activity as the basis of senescence in maize leaves. Plant Sci. 2001, 161, 765–771. [Google Scholar] [CrossRef]
- Rivero, R.M.; Kojima, M.; Gepstein, A.; Sakakibara, H.; Mittler, R.; Gepstein, S.; Blumwald, E. Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proc. Natl. Acad. Sci. USA 2007, 104, 19631–19636. [Google Scholar] [CrossRef] [PubMed]
- Chaves, M.M.; Pereira, J.S.; Maroco, J.; Rodrigues, M.L.; Ricardo, C.P.; Osorio, M.L.; Carvalho, I.; Faria, T.; Pinheiro, C. How plants cope with water stress in the field. Photosynthesis and growth. Ann. Bot. 2002, 89, 907–916. [Google Scholar] [CrossRef] [PubMed]
- Blum, A. Drought resistance, water-use efficiency, and yield potential-are they compatible, dissonant, or mutually exclusive? Aust. J. Agric. Res. 2005, 56, 1159–1168. [Google Scholar] [CrossRef]
- Arunyanark, A.; Jogloy, S.; Akkasaeng, C.; Vorasoot, N.; Kesmala, T.; Nageswara Rao, R.C.; Wright, G.C.; Patanothai, A. Chlorophyll stability is an indicator of drought tolerance in peanut. J. Agron. Crop Sci. 2008, 194, 113–125. [Google Scholar] [CrossRef]
- Li, R.H.; Guo, P.G.; Michael, B.; Stefania, G.; Cecceralli, S. Evaluation of chlorophyll content and fluorescence parameters as indicators of drought tolerance in barley. Agric. Sci. China 2006, 5, 751–757. [Google Scholar] [CrossRef]
- Kramer, D.M.; Johnson, G.; Kiirats, O.; Edwards, G.E. New fluorescence parameters for the determination of q(a) redox state and excitation energy fluxes. Photosynth. Res. 2004, 79, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Gorbe, E.; Calatayud, A. Applications of chlorophyll fluorescence imaging technique in horticultural research: A review. Sci. Hortic. 2012, 138, 24–35. [Google Scholar] [CrossRef]
- Osorio, M.L.; Osorio, J.; Vieira, A.C.; Goncalves, S.; Romano, A. Influence of enhanced temperature on photosynthesis, photooxidative damage, and antioxidant strategies in Ceratonia siliqua L. seedlings subjected to water deficit and rewatering. Photosynthetica 2011, 49, 3–12. [Google Scholar] [CrossRef]
- Subrahmanyam, D.; Subash, N.; Haris, A.; Sikka, A.K. Influence of water stress on leaf photosynthetic characteristics in wheat cultivars differing in their susceptibility to drought. Photosynthetica 2006, 44, 125–129. [Google Scholar] [CrossRef]
- Razavi, F.; Pollet, B.; Steppe, K.; van Labeke, M.C. Chlorophyll fluorescence as a tool for evaluation of drought stress in strawberry. Photosynthetica 2008, 46, 631–633. [Google Scholar] [CrossRef]
- Efeoglu, B.; Ekmekci, Y.; Cicek, N. Physiological responses of three maize cultivars to drought stress and recovery. S. Afr. J. Bot. 2009, 75, 34–42. [Google Scholar] [CrossRef]
- Terzi, R.; Saglam, A.; Kutlu, N.; Nar, H.; Kadioglu, A. Impact of soil drought stress on photochemical efficiency of Photosystem II and antioxidant enzyme activities of Phaseolus vulgaris cultivars. Turk. J. Bot. 2010, 34, 1–10. [Google Scholar]
- Sun, W.; Van Montagu, M.; Verbruggen, N. Small heat shock proteins and stress tolerance in plants. Biochim. Biophys. Acta 2002, 1577, 1–9. [Google Scholar] [CrossRef]
- Waters, E.R. The evolution, function, structure, and expression of the plant sHSPs. J. Exp. Bot. 2013, 64, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Hayano-Kanashiro, C.; Calderon-Vazquez, C.; Ibarra-Laclette, E.; Herrera-Estrella, L.; Simpson, J. Analysis of gene expression and physiological responses in three Mexican maize landraces under drought stress and recovery irrigation. PLoS ONE 2009, 4, e7531. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Liu, C.; Liu, A.; Zou, D.; Chen, X. Overexpression of OsHsp17.0 and OsHsp23.7 enhances drought and salt tolerance in rice. J. Plant Physiol. 2012, 169, 628–635. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Sun, C.; Li, Z.; Hu, Q.; Han, L.; Luo, H. AsHSP17, a creeping bentgrass small heat shock protein modulates plant photosynthesis and ABA-dependent and independent signalling to attenuate plant response to abiotic stress. Plant Cell Environ. 2016, 39, 1320–1337. [Google Scholar] [CrossRef] [PubMed]
- Grigorova, B.; Vaseva, I.; Demirevska, K.; Feller, U. Combined drought and heat stress in wheat: Changes in some heat shock proteins. Biol. Plant. 2011, 55, 105–111. [Google Scholar] [CrossRef]
- Svoboda, P.; Janska, A.; Spiwok, V.; Prasil, I.T.; Kosova, K.; Vitamvas, P.; Ovesna, J. Global scale transcriptional profiling of two contrasting barley genotypes exposed to moderate drought donditions: Contribution of leaves and crowns to water shortage coping strategies. Front. Plant Sci. 2016, 7, 1958. [Google Scholar] [CrossRef] [PubMed]
- Henikoff, S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat. Rev. Genet. 2008, 9, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Radman-Livaja, M.; Rando, O.J. Nucleosome positioning: How is it established, and why does it matter? Dev. Biol. 2010, 339, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.M.; To, T.K.; Ishida, J.; Matsui, A.; Kimura, H.; Seki, M. Transition of chromatin status during the process of recovery from drought stress in Arabidopsis thaliana. Plant Cell Physiol. 2012, 53, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Roy, D.; Paul, A.; Roy, A.; Ghosh, R.; Ganguly, P.; Chaudhuri, S. Differential Acetylation of Histone H3 at the Regulatory Region of OsDREB1b Promoter Facilitates Chromatin Remodelling and Transcription Activation during Cold Stress. PLoS ONE 2014, 9, e100343. [Google Scholar] [CrossRef] [PubMed]
- Presting, G.G.; Malysheva, L.; Fuchs, J.; Schubert, I. A Ty3/gypsy retrotransposon-like sequence localizes to the centromeric regions of cereal chromosomes. Plant J. 1998, 16, 721–728. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, N.; Boyle, S.; Fiegler, H.; Woodfine, K.; Carter, N.P.; Bickmore, W.A. Chromatin architecture of the human genome: Gene-rich domains are enriched in open chromatin fibers. Cell 2004, 118, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Van Zanten, M.; Tessadori, F.; Peeters, A.J.M.; Fransz, P. Environment-induced chromatin reorganization and plant acclimation. In Signaling and Communication in Plants; Grafi, G., Ohad, N., Eds.; Springer: Berlin, Germany, 2013; pp. 21–40. ISBN 978-3-642-35226-3. [Google Scholar]
- Mieczkowski, J.; Cook, A.; Bowman, S.K.; Mueller, B.; Alver, B.H.; Kundu, S.; Deaton, A.M.; Urban, J.A.; Larschan, E.; Park, P.J.; et al. MNase titration reveals differences between nucleosome occupancy and chromatin accessibility. Nat. Commun. 2016, 7, 11485. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, J.; Schubert, I. Chromosomal distribution and functional interpretation of epigenetic histone marks in plants. In Plant Cytogenetics: Genome Structure and Chromosome Function; Bass, H.W., Birchler, J.A., Eds.; Springer: New York, NY, USA, 2012; Volume 4, pp. 231–253. [Google Scholar]
- Zhou, J.; Wang, X.; He, K.; Charron, J.B.; Elling, A.A.; Deng, X.W. Genome-wide profiling of histone H3 lysine 9 acetylation and dimethylation in Arabidopsis reveals correlation between multiple histone marks and gene expression. Plant Mol. Biol. 2010, 72, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Brusslan, J.A.; Bonora, G.; Rus-Canterbury, A.M.; Tariq, F.; Jaroszewicz, A.; Pellegrini, M. A genome-wide chronological study of gene expression and two histone modifications, H3K4me3 and H3K9ac, during developmental leaf senescence. Plant Physiol. 2015, 168, 1246–1261. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhang, L.; He, S.; Huang, M.; Tan, J.; Zhao, L.; Yan, S.; Li, H.; Zhou, K.; Liang, Y.; et al. Cold stress selectively unsilences tandem repeats in heterochromatin associated with accumulation of H3K9ac. Plant Cell Environ. 2012, 35, 2130–2142. [Google Scholar] [CrossRef] [PubMed]
- Berger, S.L. The complex language of chromatin regulation during transcription. Nature 2007, 447, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Santos-Rosa, H.; Schneider, R.; Bannister, A.J.; Sherriff, J.; Bernstein, B.E.; Emre, N.C.; Schreiber, S.L.; Mellor, J.; Kouzarides, T. Active genes are tri-methylated at K4 of histone H3. Nature 2002, 419, 407–411. [Google Scholar] [CrossRef] [PubMed]
- Van Dijk, K.; Ding, Y.; Malkaram, S.; Riethoven, J.J.; Liu, R.; Yang, J.; Laczko, P.; Chen, H.; Xia, Y.; Ladunga, I.; et al. Dynamic changes in genome-wide histone H3 lysine 4 methylation patterns in response to dehydration stress in Arabidopsis thaliana. BMC Plant Biol. 2010, 10, 238. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Lu, F.; Cui, X.; Cao, X. Histone methylation in higher plants. Annu. Rev. Plant Biol. 2010, 61, 395–420. [Google Scholar] [CrossRef] [PubMed]
- Roudier, F.; Ahmed, I.; Berard, C.; Sarazin, A.; Mary-Huard, T.; Cortijo, S.; Bouyer, D.; Caillieux, E.; Duvernois-Berthet, E.; Al-Shikhley, L.; et al. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 2011, 30, 1928–1938. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, J.; Franzen, R.; Ngyuen, T.H.; Garcia-Maroto, F.; Pozzi, C.; Salamini, F.; Rohde, W. Cloning, mapping and expression analysis of barley MADS-box genes. Plant Mol. Biol. 2000, 42, 899–913. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Fromm, M.; Avramova, Z. H3K27me3 and H3K4me3 chromatin environment at super-induced dehydration stress memory genes of Arabidopsis thaliana. Mol. Plant. 2014, 7, 502–513. [Google Scholar] [CrossRef] [PubMed]
- Misra, A.N.; Misra, M.; Singh, R. Chlorophyll fluorescence in plant biology. In Biophysics; Misra, A.N., Ed.; InTech: Shanghai, China, 2012; pp. 171–192. ISBN 978-953-51-0376-9. [Google Scholar]
- Brestic, M.; Zivcak, M. PSII fluorescence techniques for measurement of drought and high temperature stress signal in crop plants: Protocols and applications. In Molecular Stress Physiology of Plants; Rout, G.R., Das, A.B., Eds.; Springer: New Delhi, India, 2013; pp. 87–131. ISBN 978-81-322-0806-8. [Google Scholar]
- Ay, N.; Janack, B.; Fischer, A.; Reuter, G.; Humbeck, K. Alterations of histone modifications at the senescence-associated gene HvS40 in barley during senescence. Plant Mol. Biol. 2015, 89, 127–141. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Jiang, Q.; Wang, F.; Wang, G.L.; Xu, Z.S.; Xiong, A.S. Selection of suitable reference genes for qPCR normalization under abiotic stresses and hormone stimuli in carrot leaves. PLoS ONE 2015, 10, e0117569. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Temel, A.; Janack, B.; Humbeck, K. Drought Stress-Related Physiological Changes and Histone Modifications in Barley Primary Leaves at HSP17 Gene. Agronomy 2017, 7, 43. https://doi.org/10.3390/agronomy7020043
Temel A, Janack B, Humbeck K. Drought Stress-Related Physiological Changes and Histone Modifications in Barley Primary Leaves at HSP17 Gene. Agronomy. 2017; 7(2):43. https://doi.org/10.3390/agronomy7020043
Chicago/Turabian StyleTemel, Aslihan, Bianka Janack, and Klaus Humbeck. 2017. "Drought Stress-Related Physiological Changes and Histone Modifications in Barley Primary Leaves at HSP17 Gene" Agronomy 7, no. 2: 43. https://doi.org/10.3390/agronomy7020043




