Metabolic Profiling of Phloem Exudates as a Tool to Improve Bread-Wheat Cultivars
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Extraction Method
2.3. Sample Processing
2.4. Data Analysis
3. Results
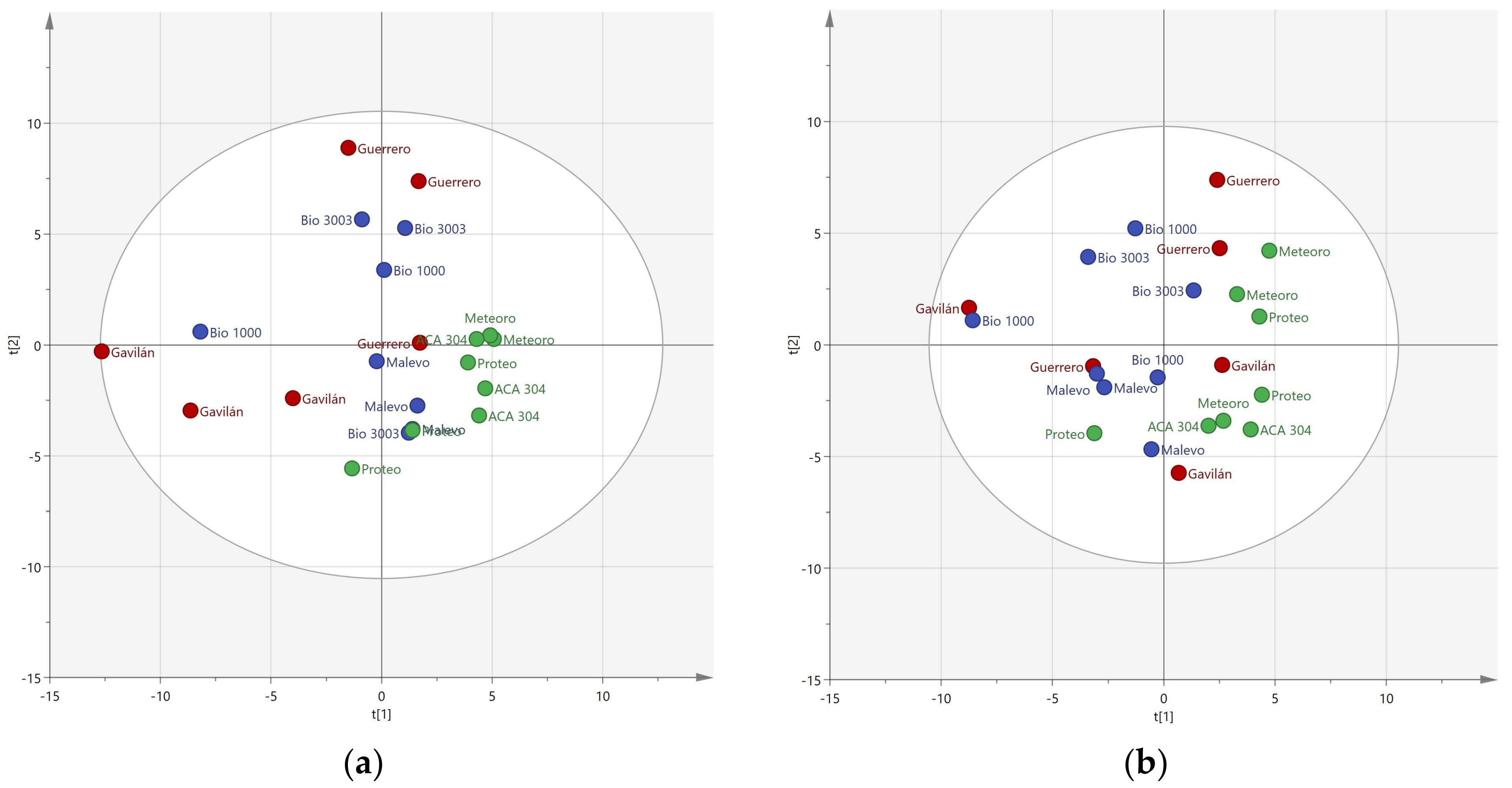
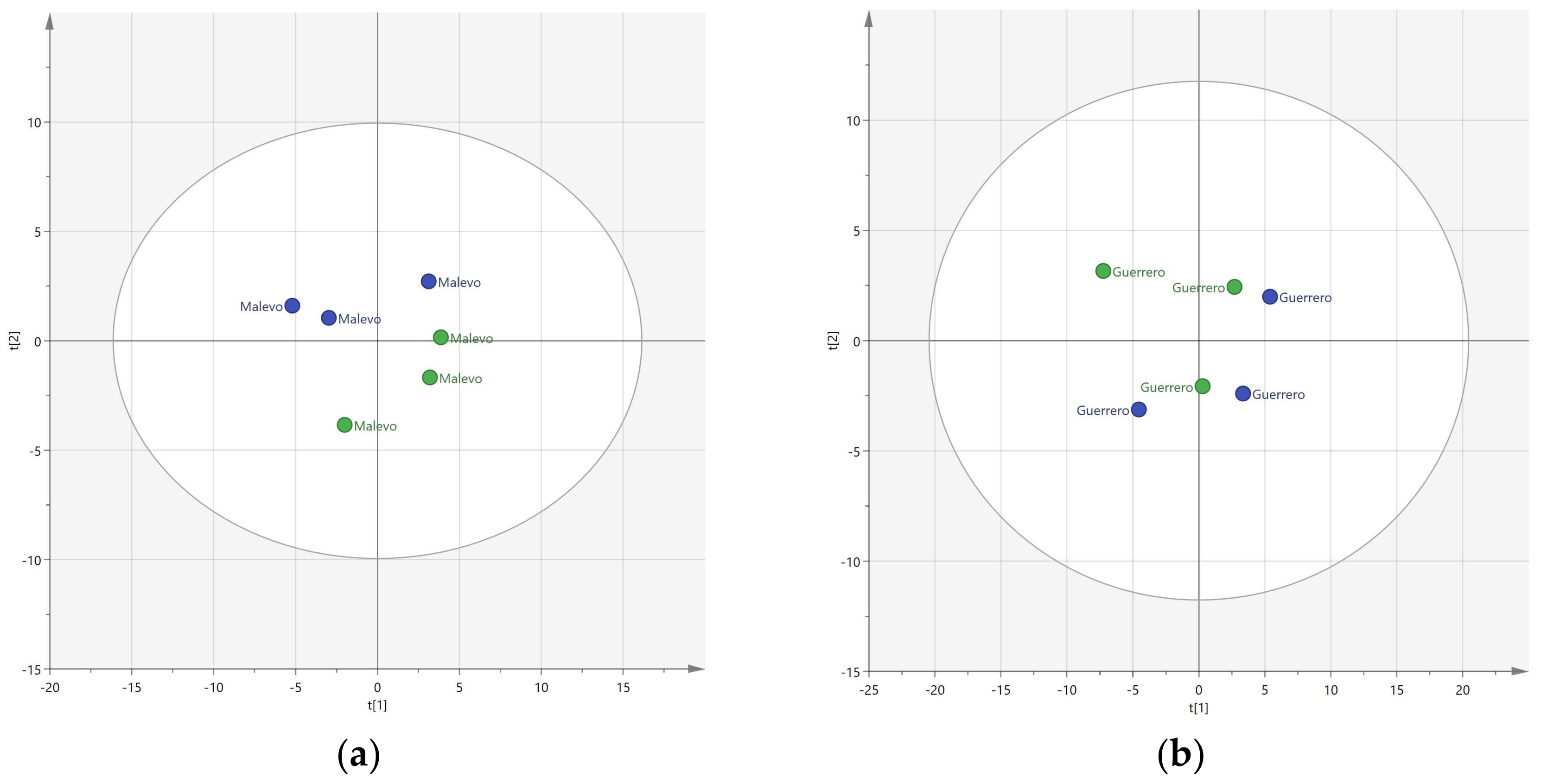
3.1. Resolution of Cultivars
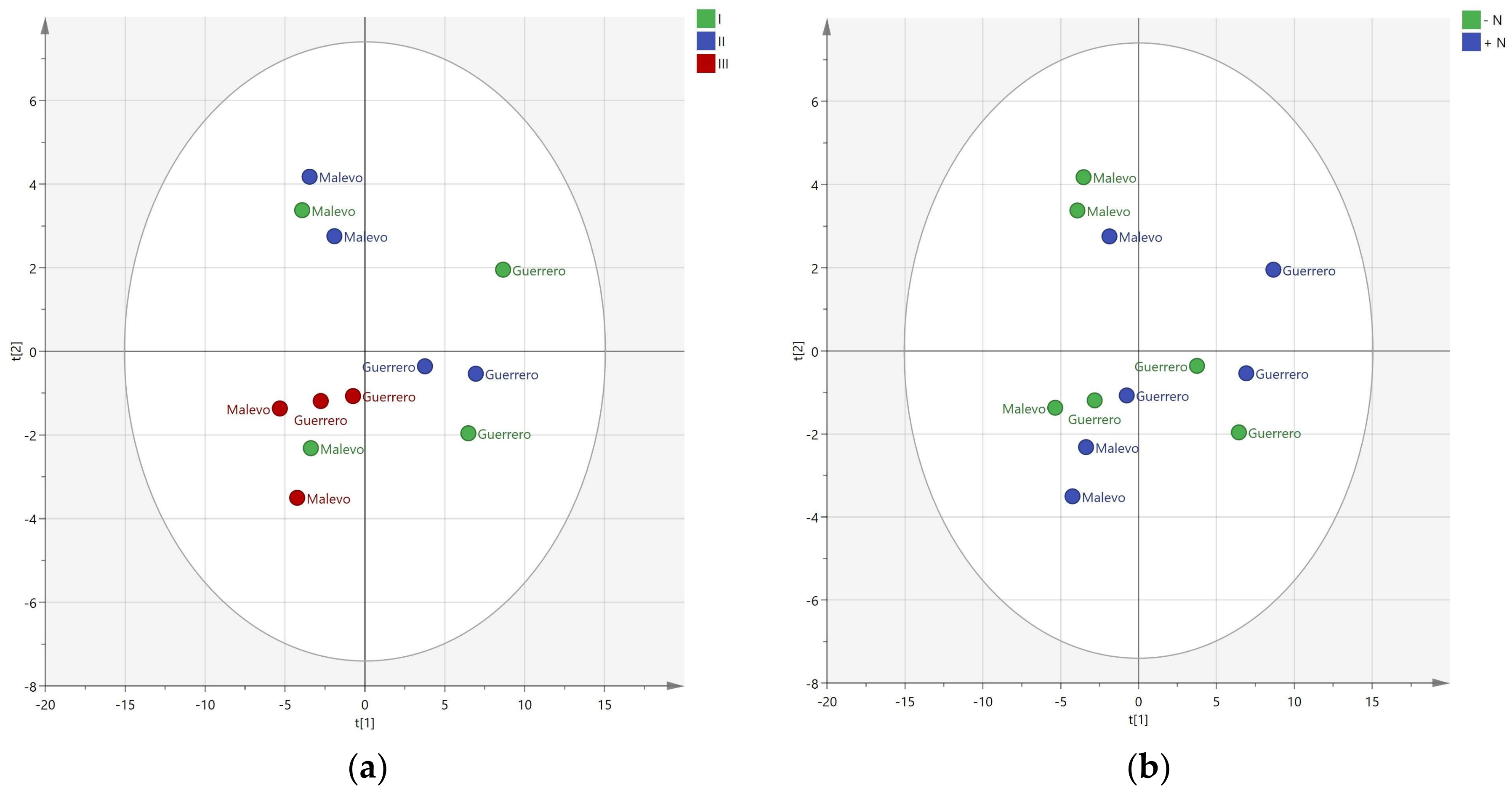
3.2. Effect of Nitrogen
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Barneix, A.J. Physiology and biochemistry of source-regulated protein accumulation in the wheat grain. J. Plant Physiol. 2007, 164, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Hall, R.; Brouwer, I.; Fitzgerald, M. Plant metabolomics and its potential application for human nutrition. Physiol. Plant. 2008, 132, 162–175. [Google Scholar] [CrossRef] [PubMed]
- Overy, S.A.; Walker, H.J.; Malone, S.; Howard, T.P.; Baxter, C.J.; Sweetlove, L.J.; Hill, S.A.; Quick, W.P. Application of metabolite profiling to the identification of traits in a population of tomato introgression lines. J. Exp. Bot. 2005, 56, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Schauer, N.; Zamir, D.; Fernie, A.R. Metabolic profiling of leaves and fruit of wild species tomato: A survey of the Solanumlycopersicum complex. J. Exp. Bot. 2005, 56, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Urbanczyk-Wochniak, E.; Fernie, A.R. Metabolic profiling reveals altered nitrogen nutrient regimes have diverse effects on the metabolism of hydroponically-grown tomato (Solanum lycopersicum) plants. J. Exp. Bot. 2005, 56, 309–321. [Google Scholar] [CrossRef] [PubMed]
- Cuniberti, M. Propuesta de clasificación del trigo argentino. IDIA XXI 2004, 6, 21–25. [Google Scholar]
- Caputo, C.; Criado, M.V; Roberts, I.N.; Gelso, M.A.; Barneix, A.J. Regulation of glutamine synthetase 1 and amino acids transport in the phloem of young wheat plants. Plant Physiol. Biochem. 2009, 47, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Gai, Y.P.; Han, X.J.; Li, Y.Q.; Yuan, C.Z.; Mo, Y.Y.; Guo, F.Y.; Liu, Q.X.; Ji, X.L. Metabolomic analysis reveals the potential metabolites and pathogenesis involved in mulberry yellow dwarf disease. Plant Cell Environ. 2014, 37, 1474–1490. [Google Scholar] [CrossRef] [PubMed]
- Millet, E.; Zaccai, M.; Feldman, M. Paternal and maternal effects on grain wheat and protein percentages in crosses between hexaploid and tetraploid high protein and low protein wheat genotypes. Genome 1992, 35, 257–260. [Google Scholar] [CrossRef]
- Barneix, A.J.; Guitman, M.R. Leaf regulation of the nitrogen concentration in the grain of wheat plants. J. Exp. Bot. 1993, 44, 1607–1612. [Google Scholar] [CrossRef]
- Poulton, J.E. Cyanogenesis in plants. Plant Physiol. 1990, 94, 401. [Google Scholar] [CrossRef] [PubMed]
- Walker, T.S.; Pal Bais, H.; Halligan, K.M.; Stermitz, F.R.; Vivanco, J.M. Metabolic profiling of root exudates of Arabidopsis thaliana. J. Agric. Food Chem. 2003, 51, 2548–2554. [Google Scholar] [CrossRef] [PubMed]
- Villar, J.; Cencig, G. Evaluación De Cultivares De Trigo 2004/2005 Y Recomendaciones Para La Próxima Campaña; INTA—Estación Experimental Agropecuaria Rafaela, Información técnica de trigo campaña 2005, Nº 103; Publicación Miscelánea, INTA EEA: Rafaela, Argentina, 2005. [Google Scholar]
- Villar, J.; Cencig, G. Evaluación De Cultivares De Trigo 2008 Y Recomendaciones Para La Próxima Campaña; Información técnica de trigo y otros cultivos de invierno campaña 2009, Nº 113; Publicación Miscelánea, INTA EEA: Rafaela, Argentina, 2009. [Google Scholar]
- Villar, J.; Cencig, G. Evaluación De Cultivares De Trigo 2010 Y Recomendaciones Para La Próxima Campaña; Información técnica de trigo y otros cultivos de invierno campaña 2011, Nº 119; Publicación Miscelánea, INTA EEA: Rafaela, Argentina, 2011. [Google Scholar]
- Molfese, E.R.; Astiz, V. Calidad Del Trigo Pan En El Sur Bonaerense; Cosecha 2014–2015, Colección divulgación; Ediciones, INTA EEA: Barrow, Argentina, 2015. [Google Scholar]
- Donaire, G.; Masiero, B.; Gutierrez, C.; Conde, B.; Salines, J.; Chiacchera, S.; Bertram, N.; Amigone, M.; Fraschina, J.; Gómez, D.; et al. Caracterización De Cultivares De Trigo Doble Propósito; Planteos ganaderos; Aapresid: Rosario, Argentina, 2010. [Google Scholar]
- Albrecht, R.; Quaino, O.; Perez, D.; Martins, L. Evaluación De Cultivares De Trigo; Campaña 2008/2009 San Fabián—Departamento San Jerónimo—Santa Fe, INTA—Estación Experimental Agropecuaria Rafaela, Información técnica de trigo y otros cultivos de invierno, Campaña, Nº 113; Publicación Miscelánea, INTA EEA: Rafaela, Argentina, 2009. [Google Scholar]
- Bainotti, C.T.; Fraschina, J.; Salines, J.; Alberione, E.; Gómez, D.; Donaire, G.; Nisi, J.; Masiero, B.; Conde, B.; Cuniberti, M.; et al. Evaluación De Cultivares De Trigo En La Eea Marcos Juárez-Campaña 2007/2008; Trigo Actualización 2008, Informe de Actualización Técnica Nº 8; INTA EEA: Marcos Juárez, Argentina, 2008. [Google Scholar]
- Urquhart, A.A.; Joy, K.W. Use of phloem exudate technique in the study of amino acid transport in pea plants. Plant Physiol. 1981, 68, 750–754. [Google Scholar] [CrossRef] [PubMed]
- Caputo, C.; Barneix, A.J. The Relationship between Sugar and Amino Acid Export to the Phloem in Young Wheat Plants. Ann. Bot. 1999, 84, 33–38. [Google Scholar] [CrossRef]
- Cao, T.; Lahiri, I.; Singh, V.; Louis, J.; Shah, J.; Ayre, B.G. Metabolic engineering of raffinose-family oligosaccharides in the phloem reveals alterations in carbon partitioning and enhances resistance to green peach aphid. Front. Plant Sci. 2013, 4, 263. [Google Scholar] [CrossRef] [PubMed]
- Balzarini, M.; Di Renzo, J. Infogen: Software Para Análisis Estadísticos De Marcadores Genéticos; Facultad de Ciencias Agropecuarias, Universidad Nacional de Córdoba: Córdoba, Argentina, 2003. [Google Scholar]
- Perretant, M.R.; Cadalen, T.; Charmet, G.; Sourdille, P; Nicolas, P.; Boeuf, C.; Tixier, M.H.; Branlard, G.; Bernard, S.; Bernard, M. QTL analysis of bread-making quality in wheat using a doubled haploid population. Theor. Appl. Genet. 2000, 100, 1167–1175. [Google Scholar] [CrossRef]
- Blanco, A.; Bellomo, M.P.; Lotti, C.; Maniglio, T.; Pasqualone, A.; Simeone, R.; Troccoli, A.; Di Fonzo, N. Genetic mapping of sedimentation volume across environments using recombinant inbred lines of durum wheat. Plant Breed. 1998, 117, 413–417. [Google Scholar] [CrossRef]
- Rousset, M.; Brabant, P.; Kota, R.S.; Dubcovsky, J.; Dvorak, J. Use of recombinant substitution lines for gene mapping and QTL analysis of bread making quality in wheat. Euphytica 2001, 119, 81–87. [Google Scholar] [CrossRef]
- Groos, C.; Robert, N.; Bervas, E.; Charmet, G. Genetic analysis of grain protein-content, grain yield and thousand-kernel weight in bread wheat. Theor. Appl. Genet. 2003, 106, 1032–1040. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, J.; Cui, F.; Jun, L.; Ding, A.; Zhao, C.; Li, X.; Feng, D.; Gao, J.; Wang, H. Conditional QTL mapping of protein content in wheat with respect to grain yield and its components. J. Genet. 2012, 91, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Cormier, F.; Le Gouis, J.; Dubreuil, P.; Lafarge, S.; Praud, S. A genome-wide identification of chromosomal regions determining nitrogen use efficiency components in wheat (Triticum aestivum L.). Theor. Appl. Genet. 2014, 127, 2679–2693. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Bai, G.; Chao, S.; Carver, B.; Wang, Z. Single nucleotide polymorphisms linked to quantitative trait loci for grain quality traits in wheat. Crop J. 2016, 4. [Google Scholar] [CrossRef]
- Krishnappa, G.; Singh, A.M.; Chaudhary, S.; Ahlawat, A.K.; Singh, S.K.; Shukla, R.B.; Jaiswal, J.P.; Singh, G.P.; Singh Solanki, I. Molecular mapping of the grain iron and zinc concentration, protein content and thousand kernel weight in wheat (Triticum aestivum L.). PLoS ONE 2017, 12, e0174972. [Google Scholar] [CrossRef] [PubMed]
- Nedelkou, I.P.; Maurer, A.; Schubert, A.; Léon, J.; Pillen, K. Exotic QTL improve grain quality in the tri-parental wheat population SW84. PLoS ONE 2017, 12, e0179851. [Google Scholar] [CrossRef] [PubMed]
- Monostori, I.; Szira, F.; Tondelli, A.; Árendás, T.; Gierczik, K.; Cattivelli, L.; Galiba, G.; Vágújfalvi, A. Genome-wide association study and genetic diversity analysis on nitrogen use efficiency in a Central European winter wheat (Triticum aestivum L.) collection. PLoS ONE 2017, 12, e0189265. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Feng, B.; Xu, Z.; Fan, X.; Jiang, F.; Jin, X.; Cao, J.; Wang, F.; Liu, Q.; Yang, L.; Wang, T. A genome-wide association study of wheat yield and quality-related traits in southwest China. Mol. Breed. 2018, 38. [Google Scholar] [CrossRef]
- Carrillo, J.M.; Rousset, M.; Qualset, C.O.; Kasarda, D.D. Use of recombinant inbred lines of wheat for study of associations of high-molecular-weight glutenin subunit alleles to quantitative traits. 1. Grain yield and quality prediction tests. Theor. Appl. Genet. 1990, 79, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Silvela, L.; Ayuso, M.G.; Gil-Delgado, L.G.; Solaices, L. Genetic and environmental contributions to bread-wheat flour quality using the SDS sedimentation test as an index. Theor. Appl. Genet. 1993, 86, 889–894. [Google Scholar] [CrossRef] [PubMed]
- Blanco, A.; Mangini, G.; Giancaspro, A.; Giove, S.; Colasuonno, P.; Simeone, R.; Signorile, A.; De Vita, P.; Mastrangelo, A.M.; Cattivelli, L.; et al. Relationships between grain protein content and grain yield components through quantitative trait locus analyses in a recombinant inbred line population derived from two elite durum wheat cultivars. Mol. Breed. 2012, 30, 79–92. [Google Scholar] [CrossRef]
- Rogers, W.J.; Cogliatti, M.; Burrell, M.M.; Steels, C.; Kallenberg, F.; Bongiorno, F.; DallaValle, H. Mejoramiento y metabolómica del cultivo de alpiste (Phalaris canariensis). In Cereales de Invierno. Investigación Científico-Técnica; Steinglein, S.A., Moreno, M.V., Cogliatti, M., Rogers, W.J., Carmona, M.A., Lavado, R.S., Eds.; Universidad Nacional del Centro de la Provincia de Buenos Aires: Tandil/Buenos Aires, Argentina, 2012; pp. 147–154. ISBN 978-950-658-301-9. [Google Scholar]
- Montgomery, D.C.; Runger, G.C. Design and analysis of single-factors experiments: The analysis of variance. In Applied Statistics and Probability for Engineers, 3rd ed.; John Wiley & Sons, Inc.: New York, NY, USA, 2003; Chapter 13; pp. 468–504. ISBN 0-471-20454-4. [Google Scholar]
- Tetyuk, O.; Benning, U.F.; Hoffmann-Benning, S. Collection and Analysis of Arabidopsis Phloem Exudates Using the EDTA-facilitated Method. J. Vis. Exp. 2013, 80, e51111. [Google Scholar] [CrossRef] [PubMed]
| Group 3 from Group 2 | Group 3 from Group 1 | Group 2 from Group 1 |
|---|---|---|
| 203 | 203 | 301.2 |
| 140 | 140 | 471 |
| 239 | 301.2 | 361 |
| 118 | 383 | 157 |
| 383 | 239 | 227 |
| 701.2 | 261 | 173 |
| 204 | 539 | 449 |
| 539 | 441.2 | 329 |
| 156 | 245 | 195 |
| 261 | 156 | 383 |
| Ranking Pattern | Mass Bin | Means | ||
|---|---|---|---|---|
| Group 1 | Group 2 | Group 3 | ||
| 3c > 2b > 1a | 203 | 3.205 | 6.589 | 10.256 |
| 1c > 2b > 3a | 370.6 | 0.141 | 0.096 | 0.046 |
| 2c > 1b > 3a | 143 | 0.326 | 0.425 | 0.27 |
| 165 | 0.12 | 0.148 | 0.093 | |
| 1b > 2a = 3a | 131 | 0.124 | 0.094 | 0.094 |
| 315 | 10.939 | 5.972 | 5.517 | |
| 651 | 0.202 | 0.099 | 0.061 | |
| 673 | 1.276 | 0.717 | 0.598 | |
| 695 | 3.607 | 2.311 | 1.804 | |
| 696 | 0.581 | 0.27 | 0.242 | |
| 717 | 1.481 | 1.077 | 0.835 | |
| 316 | 1.05 | 0.444 | 0.508 | |
| 375 | 1.144 | 0.661 | 0.668 | |
| 376 | 0.039 | 0.015 | 0.023 | |
| 2b = 1b > 3a | 172 | 0.112 | 0.131 | 0.064 |
| 371 | 0.018 | 0.02 | 0.008 | |
| 382 | 0.404 | 0.415 | 0.278 | |
| 1b = 2b > 3a | 144 | 0.011 | 0.009 | 0.004 |
| 255 | 0.223 | 0.208 | 0.089 | |
| 269 | 0.128 | 0.108 | 0.062 | |
| 287.2 | 0.189 | 0.142 | 0.075 | |
| 370 | 1.097 | 0.894 | 0.402 | |
| 761 | 0.335 | 0.297 | 0.148 | |
| 3b > 1a = 2a | 135 | 0.025 | 0.013 | 0.11 |
| 156 | 0.193 | 0.178 | 0.303 | |
| 204 | 0.228 | 0.111 | 0.405 | |
| 205 | 0.074 | 0.053 | 0.107 | |
| 219 | 0.405 | 0.232 | 0.881 | |
| 261 | 0.3 | 0.188 | 0.517 | |
| 383 | 0.314 | 0.16 | 0.766 | |
| 2b = 3b > 1a | 167 | 0.056 | 0.092 | 0.09 |
| 181 | 0.06 | 0.094 | 0.088 | |
| 307.2 | 0.083 | 0.114 | 0.11 | |
| 3b = 2b > 1a | 140 | 5.687 | 6.743 | 7.397 |
| 183 | 0.041 | 0.07 | 0.075 | |
| 301.2 | 3.458 | 5.521 | 5.763 | |
| 302.2 | 0.428 | 0.572 | 0.583 | |
| 561 | 0.068 | 0.165 | 0.17 | |
| 2b > 1a = 3a | 222 | 0.013 | 0.02 | 0.014 |
| 249.2 | 0.11 | 0.202 | 0.119 | |
| 295.2 | 0.056 | 0.07 | 0.059 | |
| 299.2 | 0.142 | 0.42 | 0.252 | |
| 309.2 | 0.149 | 0.272 | 0.193 | |
| 403 | 0.657 | 1.218 | 0.663 | |
| 180 | 0.01 | 0.014 | 0.008 | |
| 195 | 0.125 | 0.181 | 0.123 | |
| 199 | 0.377 | 0.568 | 0.367 | |
| 300.2 | 0.008 | 0.02 | 0.008 | |
| 329 | 0.111 | 0.202 | 0.085 | |
| 392 | 0.199 | 0.345 | 0.1 | |
| 449 | 0.069 | 0.148 | 0.058 | |
| 560 | 0.162 | 0.272 | 0.073 | |
| 1b = 3b > 2a | 353 | 0.479 | 0.34 | 0.443 |
| 2b > 1ab > 3a | 99 | 0.011 | 0.012 | 0.006 |
| 102 | 0.021 | 0.028 | 0.013 | |
| 189 | 0.112 | 0.127 | 0.093 | |
| 263.2 | 0.064 | 0.074 | 0.053 | |
| 269.2 | 0.064 | 0.073 | 0.054 | |
| 279 | 0.445 | 0.491 | 0.362 | |
| 3b > 2ab > 1a | 118 | 1.887 | 2.135 | 2.553 |
| 539 | 0.233 | 0.383 | 0.529 | |
| 2b > 3ab > 1a | 193.2 | 0.159 | 0.222 | 0.19 |
| 251.2 | 0.187 | 0.254 | 0.23 | |
| 267.2 | 0.207 | 0.28 | 0.23 | |
| 303.2 | 0.031 | 0.043 | 0.036 | |
| 321.2 | 0.071 | 0.106 | 0.088 | |
| 1b > 2ab > 3a | 257 | 0.933 | 0.76 | 0.556 |
| 359 | 14.285 | 12.586 | 10.842 | |
| 739 | 1.107 | 0.9 | 0.6 | |
| Mass Bin | Means | |
|---|---|---|
| −N | +N | |
| 91 | 0.053a | 0.111b |
| 149 | 0.087a | 0.114b |
| 183 | 0.059a | 0.065b |
| 301.2 | 4.482a | 5.346b |
| 339 | 0.146a | 0.211b |
| 442.2 | 0.404a | 0.435b |
| 211 | 0.200a | 0.252b |
| 329 | 0.117a | 0.148b |
| 255 | 0.148a | 0.198b |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Basile, S.M.L.; Burrell, M.M.; Walker, H.J.; Cardozo, J.A.; Steels, C.; Kallenberg, F.; Tognetti, J.A.; DallaValle, H.R.; Rogers, W.J. Metabolic Profiling of Phloem Exudates as a Tool to Improve Bread-Wheat Cultivars. Agronomy 2018, 8, 45. https://doi.org/10.3390/agronomy8040045
Basile SML, Burrell MM, Walker HJ, Cardozo JA, Steels C, Kallenberg F, Tognetti JA, DallaValle HR, Rogers WJ. Metabolic Profiling of Phloem Exudates as a Tool to Improve Bread-Wheat Cultivars. Agronomy. 2018; 8(4):45. https://doi.org/10.3390/agronomy8040045
Chicago/Turabian StyleBasile, S. Marisol L., Mike M. Burrell, Heather J. Walker, Jorge A. Cardozo, Chloe Steels, Felix Kallenberg, Jorge A. Tognetti, Horacio R. DallaValle, and W. John Rogers. 2018. "Metabolic Profiling of Phloem Exudates as a Tool to Improve Bread-Wheat Cultivars" Agronomy 8, no. 4: 45. https://doi.org/10.3390/agronomy8040045





