1. Introduction
Texas bluegrass (
Poa arachnifera Torr.) is a dioecious, cool-season grass with exceptional heat and drought tolerance; however, it lacks higher levels of tiller density and a finer leaf texture that are desirable for residential and athletic field applications [
1]. Kentucky bluegrass (
Poa pratensis L.) is one of the primary cool-season turf species used on home lawns and sports fields because of its excellent turf quality and its ability to reproduce through apomixis [
2]. The hybridization of Texas bluegrass and Kentucky bluegrass can bring together the traits of heat and drought tolerance with increased turf quality and apomixis [
3,
4,
5]. More recently, Canada bluegrass (
Poa compressa L.)
, Poa ligularis (Nees ex Steud.), and Sandberg bluegrass (
Poa secunda J. Presl) have been hybridized with Texas bluegrass as an alternative means to increase turf quality, abiotic stress tolerance, and sexual reproduction and to allow for breeding of desirable traits in advanced generations [
1,
6,
7]. Confirmation of hybrids can be done through analysis of morphological characteristics; however, molecular markers would greatly increase the efficiency of the breeding process.
Previously, isozyme markers have been used to support the hypothesis that annual bluegrass (
Poa annua L.) formed through the hybridization and duplication of early meadowgrass (
Poa infirma H.B.K.) and supine bluegrass (
Poa supina Schrad.) [
8]. Randomly amplified polymorphic DNA (RAPD) is a cheap, less laborious marker system that has been used to characterize different cultivars of Kentucky bluegrass [
9]. RAPD markers however, can be arduous to reproduce as they are highly dependent on the uniformity of the PCR conditions between experiments [
10]. Amplified fragment length polymorphism (AFLPs) markers were used to map and identified male-specific markers in dioecious Texas bluegrass [
11]. Other scientists have sequenced RAPD markers to develop sequence characterized amplified region (SCAR) markers for Texas x Kentucky bluegrass hybrid confirmation [
3]. More recently, Texas bluegrass ×
Poa spp. hybrids have been confirmed using simple sequence repeats (SSR) and inter simple sequence repeats (ISSR) [
6,
12]. SSR and ISSR markers can become laborious when screening large populations for potential hybrids especially if the locus in the male parent is heterozygous with a null (absent) allele that effectively renders the utility of these markers to be a dominant rather than a co-dominant marker system. An alternative co-dominant marker system, known as conserved-intron scanning primers (CISPs), anneal to the borders of exons and utilize the greater evolutionary constraints for sequence conservation on exons than on introns to amplify sequence variations within the more variable spanning intron. Moreover, gene duplication and subsequent intron sequence divergence between species also enhances the potential to use CISPs in developing species-specific markers. Cross species transferability is a limiting factor in many PCR-based marker systems; however, CISP markers have been successfully transferred across taxa [
13].
Ent-kaurene oxidase (
KO) functions in the gibberellic acid (GA) biosynthetic pathway by converting ent-kaurene to ent-kaurenoic acid [
14]. In rice, the
KO gene has undergone several duplication events resulting in three copies of
KO genes and two
KO-like paralogs that have evolutionarily diverged and may have taken on a new function in phytoalexin synthesis. All five copies are clustered together on chromosome six and are flanked by retroelements. In other grasses, southern blot analysis suggests that the
KO gene may have multiple copies on different chromosomes, for example, on chromosomes two and seven in wheat and chromosomes two, three, and seven in barley [
15]. Overall, these five rice
KO sequences have significant differences in their fragment lengths, primarily due to insertion/deletion events in the introns [
16], which are advantageous for developing CISP markers. In the present study, we cloned and sequenced PCR products amplified by
KO derived primers from seven
Poa species. These sequences were then used to build species-specific CISP markers to identify different
Poa species. These species-specific markers could assist in developing co-dominant markers for inter-specific hybrid identification.
2. Materials and Methods
Genomic DNA was extracted, according to Chen and Ronald [
17], separately from the following individuals in at least two replicates: Texas bluegrass TAES 6012 (
P. arachnifera), Kentucky bluegrass cv. ‘Huntsville’ and TAES 6013 (
P. pratensis), ‘Reveille’ (
P. arachnifera ×
P. pratensis), Canadian bluegrass cv. ‘Reubens’ (
P. compressa), rough bluegrass cv. ‘Havana’ (
Poa trivialis L.), annual bluegrass TAES 6014 (
P. annua), supine bluegrass TAES 6016 (
P. supina), early meadowgrass TAES 6015 (
P. infirma).
KO gene sequence from
Hordeum vulgare (AY551434.1),
Triticum aestivum (AK334773.1) and
Oryza sativa (AY660664.1) were aligned using the BLASTN function within the NCBI database (
http://www.ncbi.nlm.nih.gov/) [
18]. The forward and reverse primers were designed based on the multiple sequence alignment of
KO gene from
H. vulgar,
T. aestivum and
O. sativa with the forward primer (KO-F) 5′-CCAGCTGTTCCTGGTTTACCG 3′ being homologous to conserved sequences starting at nucleotides 53, 302, and 142, respectively. The reverse primer (KO-R) 5′-ACTCTTGTTTCGAAGCTCTTGTTT 3′ was homologous to conserved sequences starting at nucleotides 696, 945, and 782, respectively. Primers were synthesized by Integrated DNA Technologies (San Diego, CA, USA). PCR amplification was completed using the following conditions; 2.0 ng genomic DNA, 1× GoTaq Buffer (Promega, Madison, WI, USA), 1.5 mM MgCl
2 (Promega), 0.2 mM dNTP, 5 μM of each primer, 2.5 units of Go
Taq Flexi polymerase (Promega) in a 20 μL PCR reaction. Thermocycling conditions were one cycle of 94 °C for 3 min, 32 cycles of 94 °C for 30 s, 61 °C for 60 s, 72 °C for 60 s, and one cycle of 72 °C for 7 min. PCR products were run on 1% agarose gel with ethidium bromide staining. PCR bands were cut out of the gel with QIAquick Gel Extraction Kit (QIAGEN Inc., Valencia, CA, USA). Gel extracted PCR products were inserted into the TOPO TA cloning vector according to the TOPO TA Cloning Kit for Sequencing (Invitrogen, Carlsbad, CA, USA) and electroporated into competent cells using a BioRad MicroPulser (Biorad, Hercules, CA, USA). Transformed cells were plated onto Luria-Bertani-Amp50 with X-Gal and white colonies were transferred into 50-mL tubes containing liquid Luria–Bertani medium with 50 μg/mL of ampicillin. Plasmids from positive colonies were extracted using the StrataPrep Plasmid Miniprep Kit (Stratagene, La Jolla, CA, USA).
Cycle sequencing reactions were performed at the Huck Institute’s Nucleic Acid Facility at Penn State University using 3′ BigDye-labeled dideoxynucleotide triphosphates (Version 3.1 dye terminators; Applied Biosystems, Foster City, CA, USA) and run on an ABI 3730XL DNA Analyzer, using ABI Data Collection Program (Version 2.0, Applied Biosystems, Foster City, CA, USA). Data was analyzed with ABI Sequencing Analysis software (Version 5.1.1, Applied Biosystems, Foster City, CA, USA). Sequencing reactions were performed using protocol # 4303237 from Applied Biosystems. All plasmids were sequenced twice from each orientation with M13 and M13R primers.
All cloned fragments were analyzed for sequence homology to
H. vulgar,
T. aestivum and
O. sativa KO gene sequences. Putative introns were identified based on non-alignment gaps between
Poa genomic sequences and
H. vulgare mRNA using ClustalW in MEGA Version 5.0 (Pennsylvania State University, Philadelphia, PA, USA) [
19]. CISP sequences were designed through the alignment of the
P. arachnifera genomic sequences and each of the other
Poa genomic sequences separately. The CISP primer pair TxKy-F/TxKy-R sequences was highly homologous to all of the
Poa species. These CISPs initiated PCR amplification within each of the first two putative exons and flank the insertion/deletion polymorphisms within intron one (
Figure S1). TxKy-F/TxKy-R was tested on
P. arachnifera with
P. compressa,
P. pratensis, and
P. trivialis respectively for
KO amplification and separation through agarose gel electrophoresis. The reverse primer TxComp-R was designed within the first intron to shorten the amplification product and create better visual separation of the polymorphisms between
P. arachnifera with
P. compressa and
P. trivialis, respectively. Similarly, the reverse primer SUP-R was also designed to shorten the product for band separation on an agarose gel but also to exploit all of the insertion/deletion polymorphism existing between
P. arachnifera and
P. annua.
All additional primers were used under the same PCR conditions with an annealing temperature of 61 °C. (TxKy-F) 5′-CTGTGCTCAACTCAACAGAAGTA 3′, (TxKy-R) 5′-AACCATTGCCTGCAAATACAGGA 3′, (TxComp-R) 5′-TCTGTTCTGCATCGCAGCCAA 3′, (SUP-R) 5′-TCCAATCAACCTCAATTGCGCA 3′.
3. Results
The primer pair KO-F/KO-R allowed for the amplification of the
KO homolog from all seven
Poa species used in the present study.
P. compressa, P. supina, and
P. trivialis each produced a single amplicon ranging in lengths from 1405 bp to 1383 bp while
P. annua, P. arachnifera, P. infirma, and
P. pratensis each produced multiple amplicons ranging in lengths from 1538 bp to 1327 bp (
Table 1).
P. arachnifera and
P. pratensis each produced two amplicons using the KO-F/KO-R primer pair and both pairs were sequenced.
P. infirma and
P. annua produce two and four amplicons, respectively. In total, 10 amplicons were sequenced from seven
Poa species. All of the sequenced amplicons showed significant sequence homology (<e
−50) to the
H. vulgare,
T. aestivum, and
O. sativa KO gene sequence (data not shown). The genomic sequences of
KO homologs from
Poa species were aligned with
H. vulgare KO mRNA sequence using ClustalW in MEGA V5.0. The multiple sequence alignment revealed sequence length variations within three regions of
Poa species that did not align with the
H. vulgare mRNA (
Table 1). For the purpose of this manuscript, we are referring to these variants as putative introns one, two, and three.
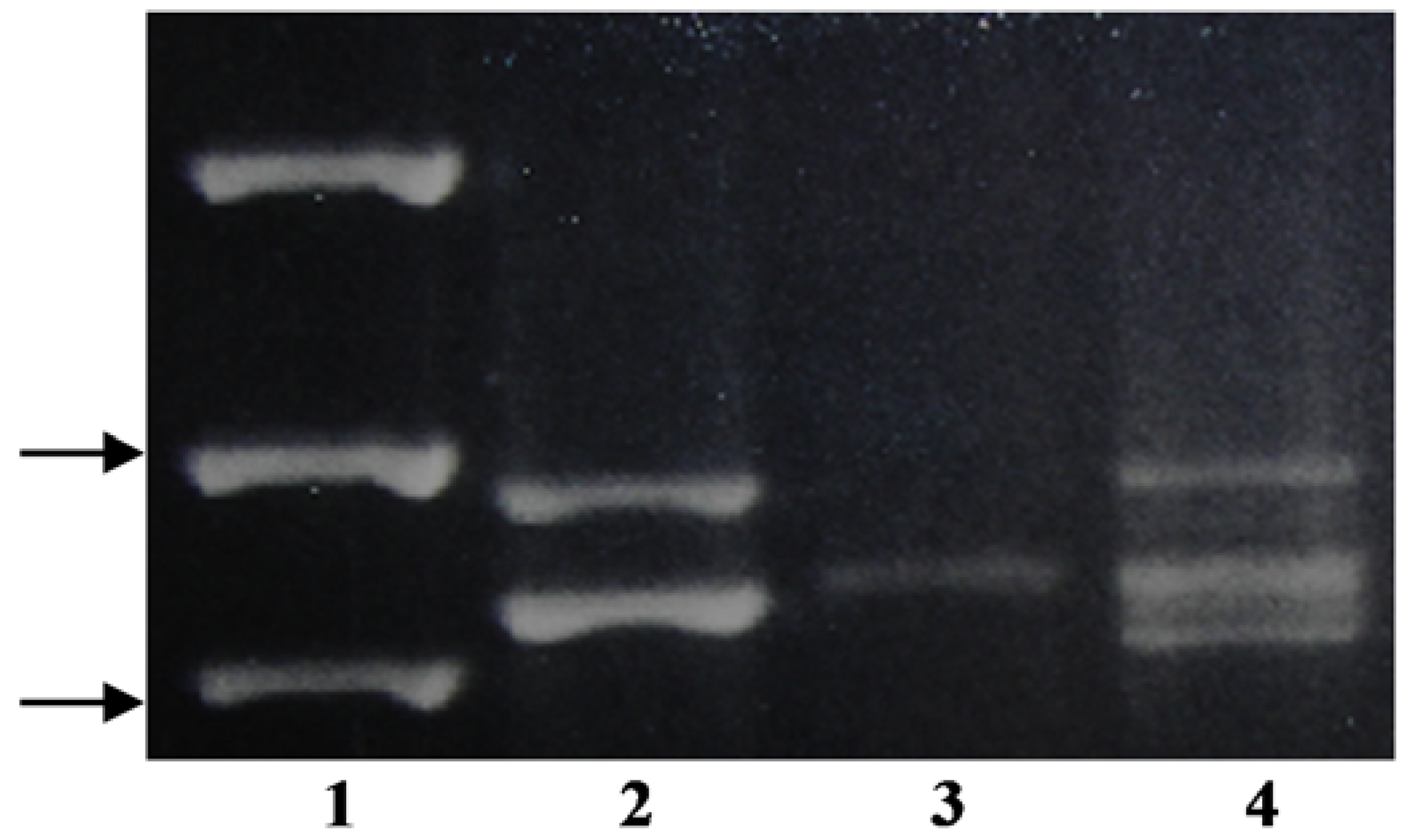
Using the KO-F/KO-R primer pair,
P. annua produced four distinguishable bands, where one was significantly larger than the rest of the amplicons (
Figure 1). The smaller bands from
P. infirma,
P. supina, and
P. annua were similar in size to each other and to both of the bands from
P. arachnifera (
Table 1). Sequencing only one of the two
P. infirma amplicons and two of the four
P. annua bands were necessary to distinguish between
P. infirma, P. supina,
P. annua, and
P. arachnifera (
Table 1) (
Figure 1).
Using the KO-F/KO-R primer pair, only
P. annua and
P. infirma produced bands that could be easily scored as polymorphic to
P. arachnifera (
Table 1)
. New primer pairs were designed through the paired alignment of
P. archnifera and the other respective
Poa species using ClustalW to develop co-dominant CISP markers (
Table 2,
Figure S1).
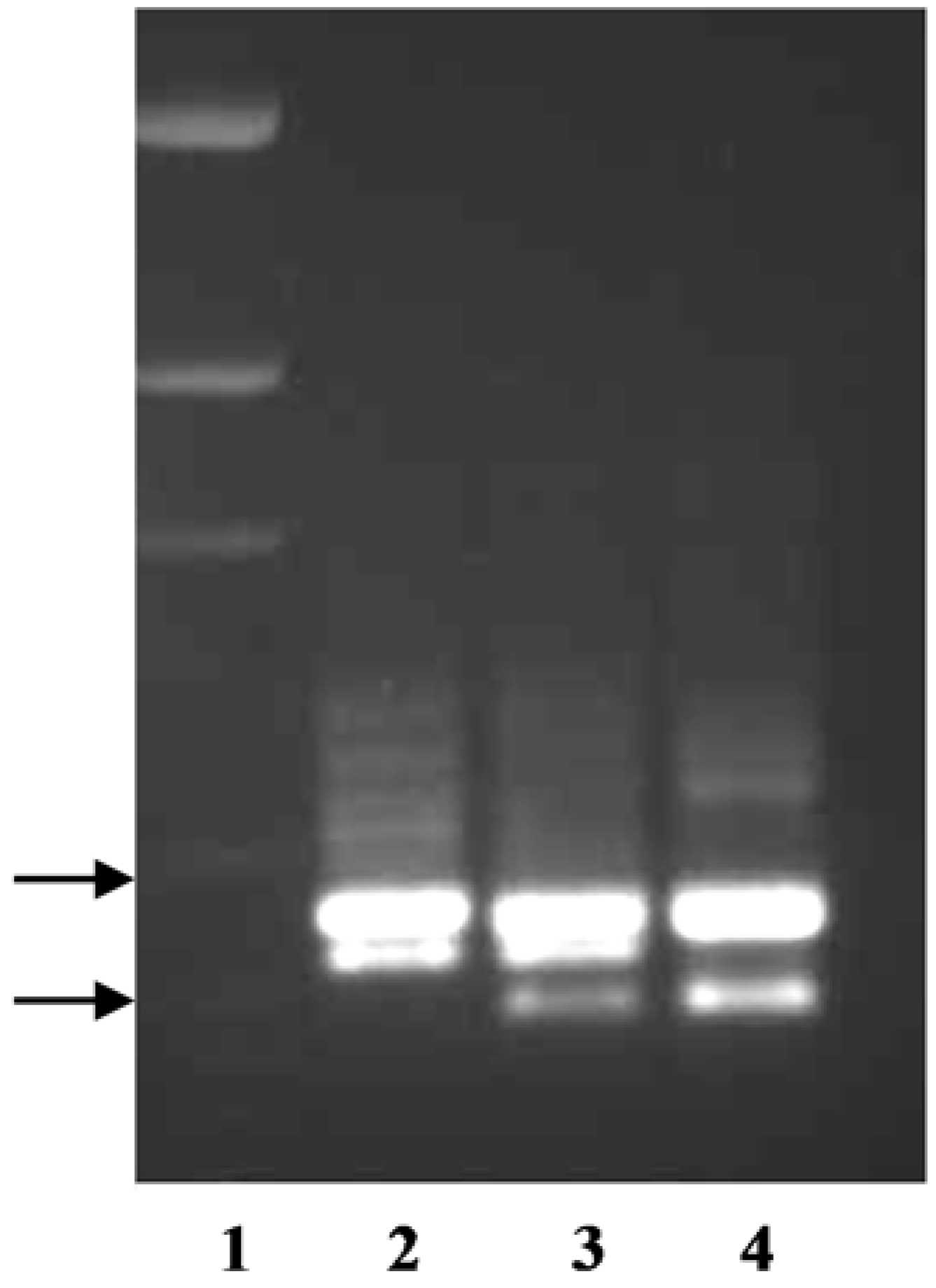
The primer pair TxKy-F/TxKy-R utilized the variations within intron one of
P. arachnifera and
P. pratensis (
Table 2,
Figure S1). This primer pair amplified two bands from each species, where one of the bands was polymorphic and the other was monomorphic (
Figure 2). Texas bluegrass hybrid ‘Reveille’ is a released cultivar from a cross between
P. arachnifera TAES 6012 and
P. pratensis ‘Huntsville’ [
4]. When the primer pair TxKy-F /TxKy-R was run on the parents and the F
1 Reveille, the upper monomorphic band and two lower polymorphic bands from each parent were amplified in Reveille (
Figure 2).
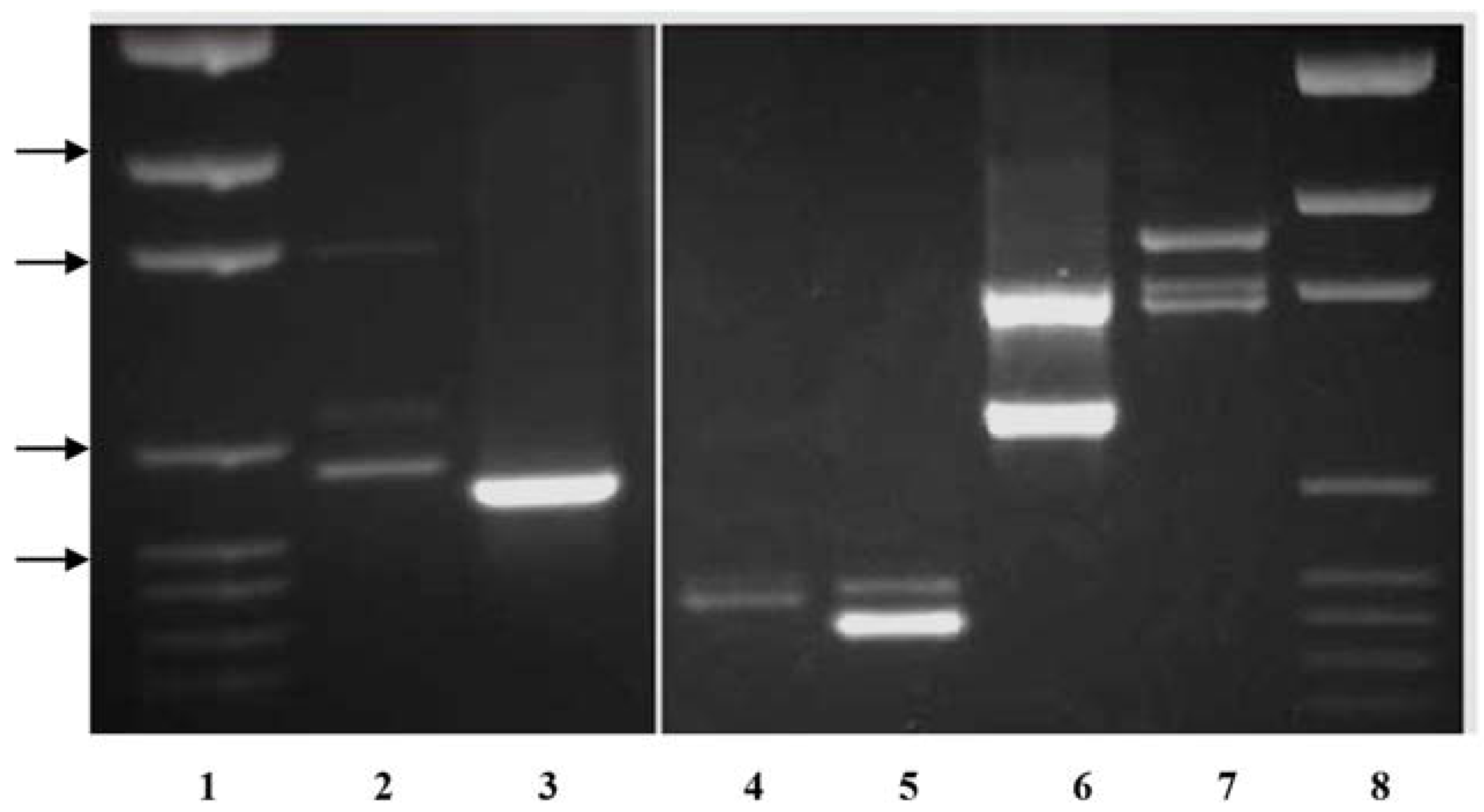
Primer pairs were also developed to amplify the single
KO band in
P. compressa and
P. trivialis that are intermediate in length between the two
P. arachnifera bands. The primer pair TxKy-F/TxComp-R amplified the expected profile of two bands in
P. arachnifera with a polymorphic band in
P. compressa, which is intermediate in length (
Table 2,
Figure 3). This primer combination also produced several lighter polymorphic bands in
P. compressa. The primer pair KO-F/TxComp-R was used to distinguish between
P. trivialis and
P. arachnifera (
Table 2). This primer combination exploits the differences in the first intron sequence length and produced clear polymorphic bands for confirmation of future
P. arachnifera breeding with
P. trivialis (
Figure 3).
4. Discussion
KO is fundamental in the gibberellic acid biosynthetic pathway, catalyzing the step(s) from ent-kaurene to ent-kaurenoic acid [
14]. Insertion/deletion events in the introns of
KO genes [
16] are useful for developing CISP markers. Species-specific CISP markers based on
KO sequences allowed for identification of seven different
Poa species.
P. arachnifera,
P. pratensis, and
P. annua each produced multiple amplicons that were highly homologous to the
KO orthologs in rice, barley, and wheat. The fact that these
Poa species are polyploid may suggest that the multiple bands are gene copies from each genome rather than from a locus duplication event [
20].
Poa annua however, produced four amplicons, two of which were most likely inherited from its ancestral diploid maternal parent
P. infirma [
21]. The inheritance of both bands from
P.
infirma to
P. annua suggests that the two copies arose from a duplication event in
P.
infirma prior to the formation of
P.
annua. A third
P. annua band was inherited from its other progenitor,
P. supina. The forth
P. annua band was directly sequenced after extraction from an agarose gel. Sequence analysis revealed a large deletion starting in the second intron and ending in the third intron with complete removal of the third exon (data not shown). The results of using the primer pair TxKy-F/SUP-R on
P. annua and
P. arachnifera indicate that this forth, shorter amplicon can be used as a marker to differentiate between the two species for future hybrid analysis (
Figure 2). It is yet to be determined whether the forth band in
P. annua is an allelic variation of one of three putative
KO copies or it is also resulting from a duplication event after speciation (
Figure 1).
P. annua and
P. supina both display an excellent tolerance to low mowing heights which would improve the utility of
P. arachnifera [
22,
23]. Some variants of
P. annua can be extremely similar to
P. supina in their morphological features, life history traits, and flowering requirements [
8]. The
KO-based CISP primers (KO-F/KO-R), however, were able to produce an identifying banding pattern for the three species (
Figure 1). Interspecific hybrids between
P. supina and
P. annua have been made to improve the turf quality of
P. annua [
23] and could also improve the instability of green-type
P. annua [
24]. The sequence information and marker system could also be adapted to confirm these hybrids as well.
The primer pair TxKy-F/TxComp-R amplified the expected profile of two bands in
P. arachnifera with a single band in
P. compressa, which is intermediate in length. The gel electrophoresis also revealed an additional band in
P. compressa that may be an allelic variant or an additional gene copy that is slightly larger than the upper
P. arachnifera band (
Figure 2).
P.
compressa is also a polyploid, which could explain the additional gene copies [
20]. The primer pair KO-F/TxComp-R was used to distinguish between
P. trivialis and
P. arachnifera. This primer combination exploits the differences in the first intron sequence length and produced clear polymorphic bands for confirmation of future
P. arachnifera breeding with
P. trivialis (
Figure 2).
Texas bluegrass hybrid ‘Reveille’ is a released cultivar from a cross between
P. arachnifera TAES 6012 and
P. pratensis ‘Huntsville’ [
4]. Based on the
KO sequences, the primer pair TxKy-F/TxKy-R should have produced a single band in
P. pratensis that was intermediate to the two
P. arachnifera bands (
Table 1). A smaller, unexpected, band was amplified in
P. pratensis while the expected
P. arachnifera band of 556 bp and
P. pratensis bands at 543 bp and 540 bp were indistinguishable. In the F
1 Reveille, the lower bands from each parent were inherited which indicates at a molecular level that Reveille is a true hybrid. The
P. pratensis parent, cv. Huntsville, produced a lower band that was not expected based on the sequenced data. These results indicate that this
P. pratensis accession may have additional putative
KO sequences. Never the less, the sequencing of the putative
KO gene and development of the primers TxKy-F/TxKy-R confirms the hybrid nature of Reveille and demonstrates the utility of these CISP markers.